Oligosaccharide Presentation Modulates the Molecular Recognition of Glycolipids by Galectins on Membrane Surfaces
Abstract
:1. Introduction
2. Results
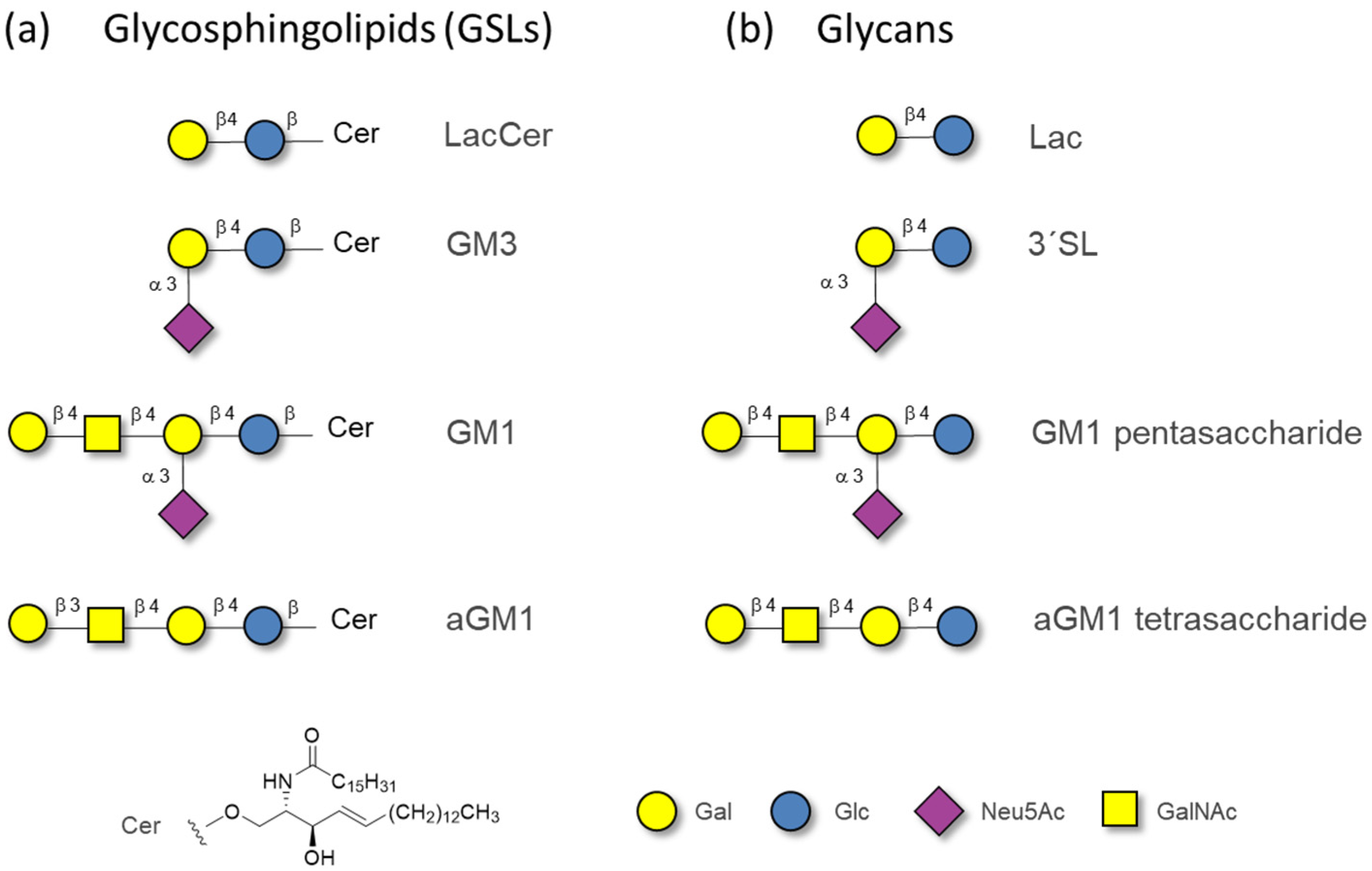
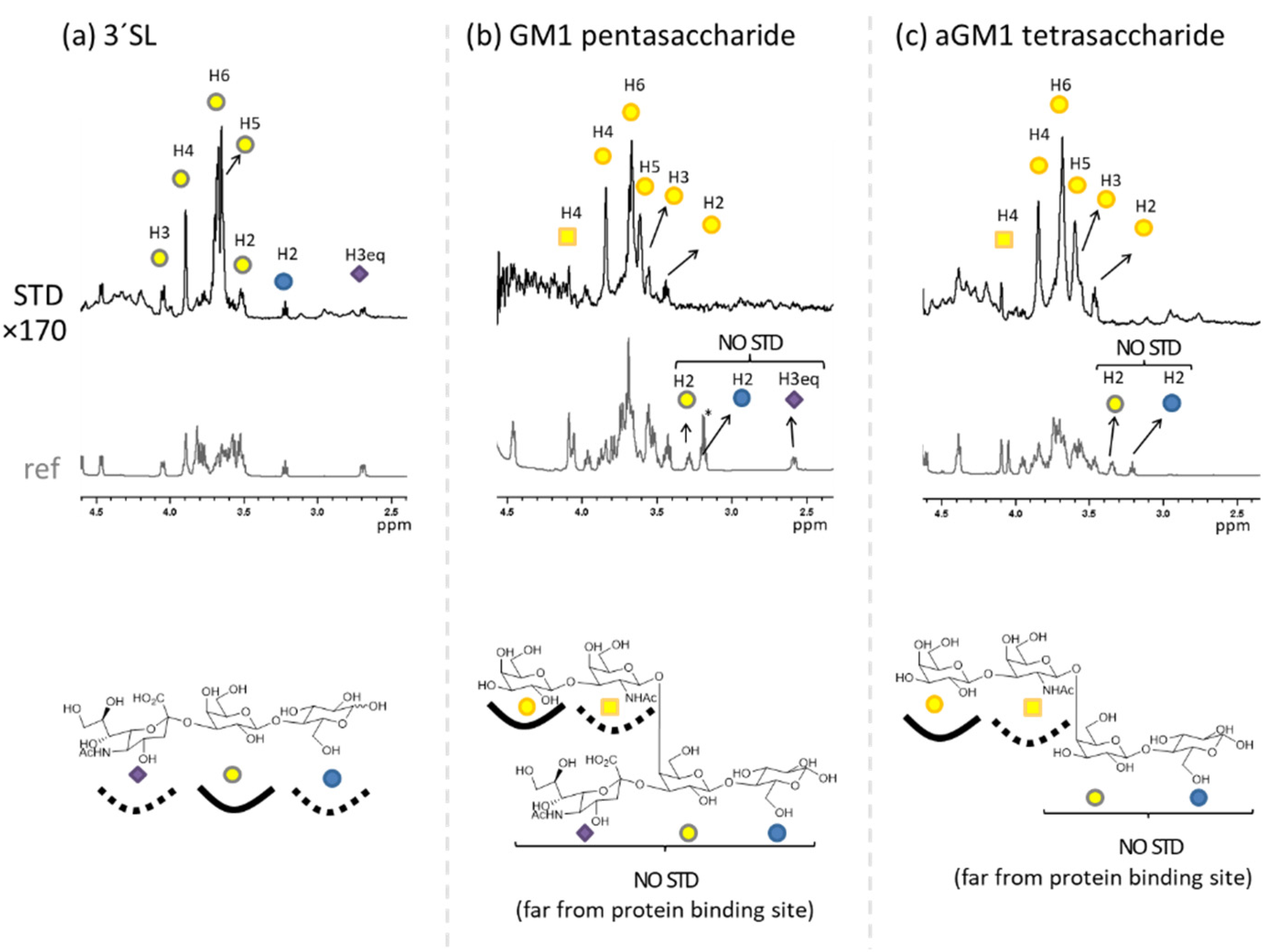
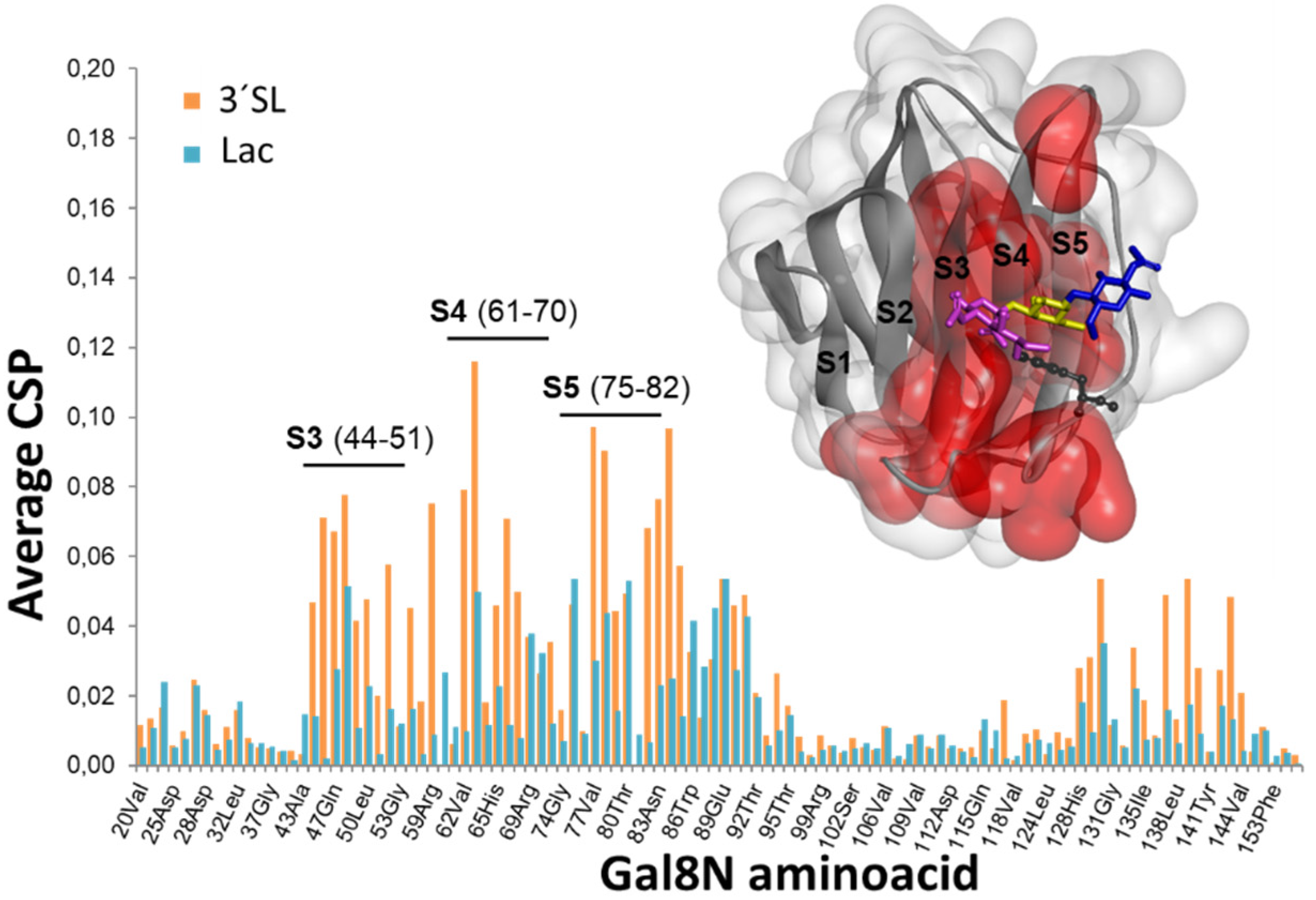
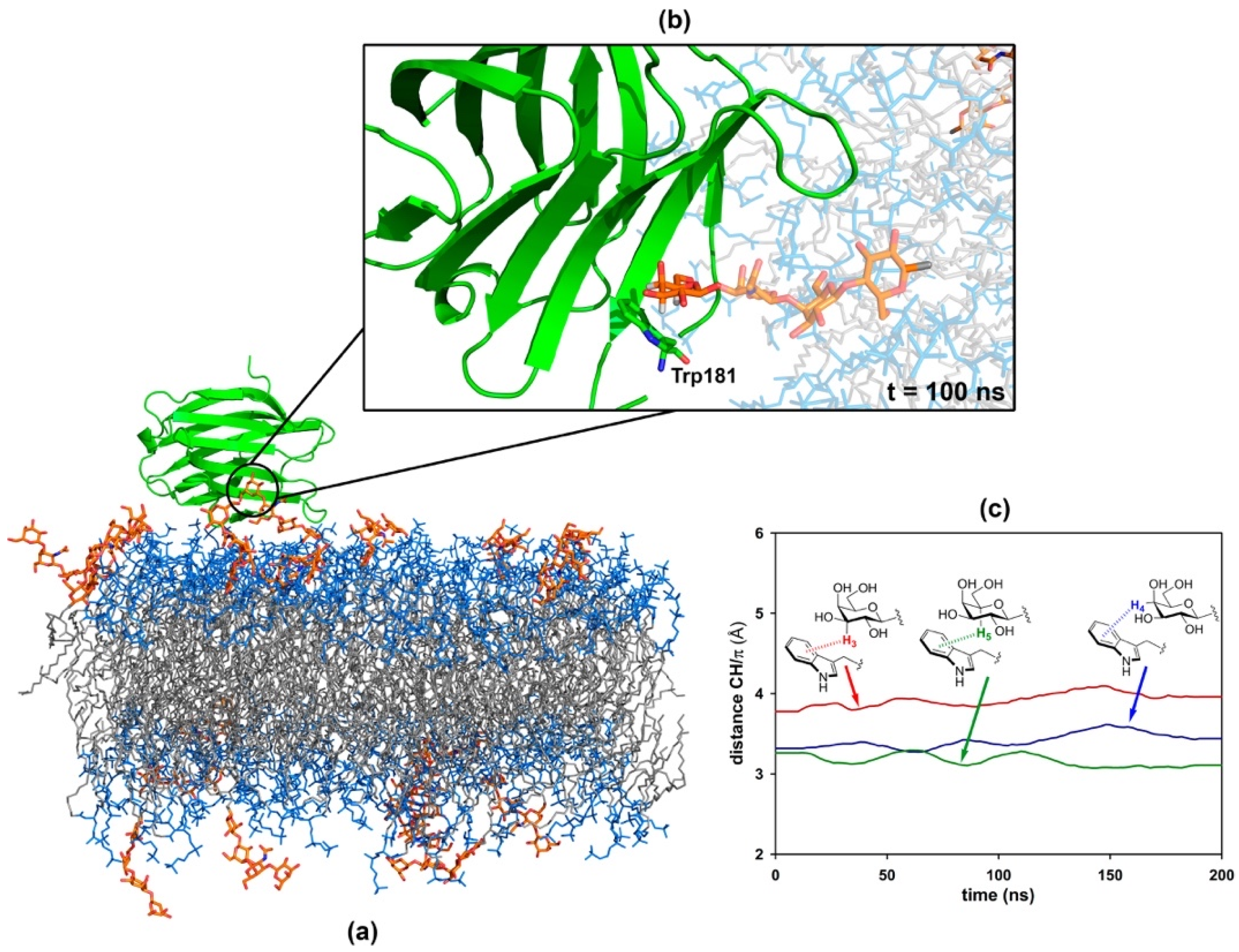
2.1. The Binding of the Sugar Moieties to Galectin 3-and N-Terminal Domain of Galectin-8
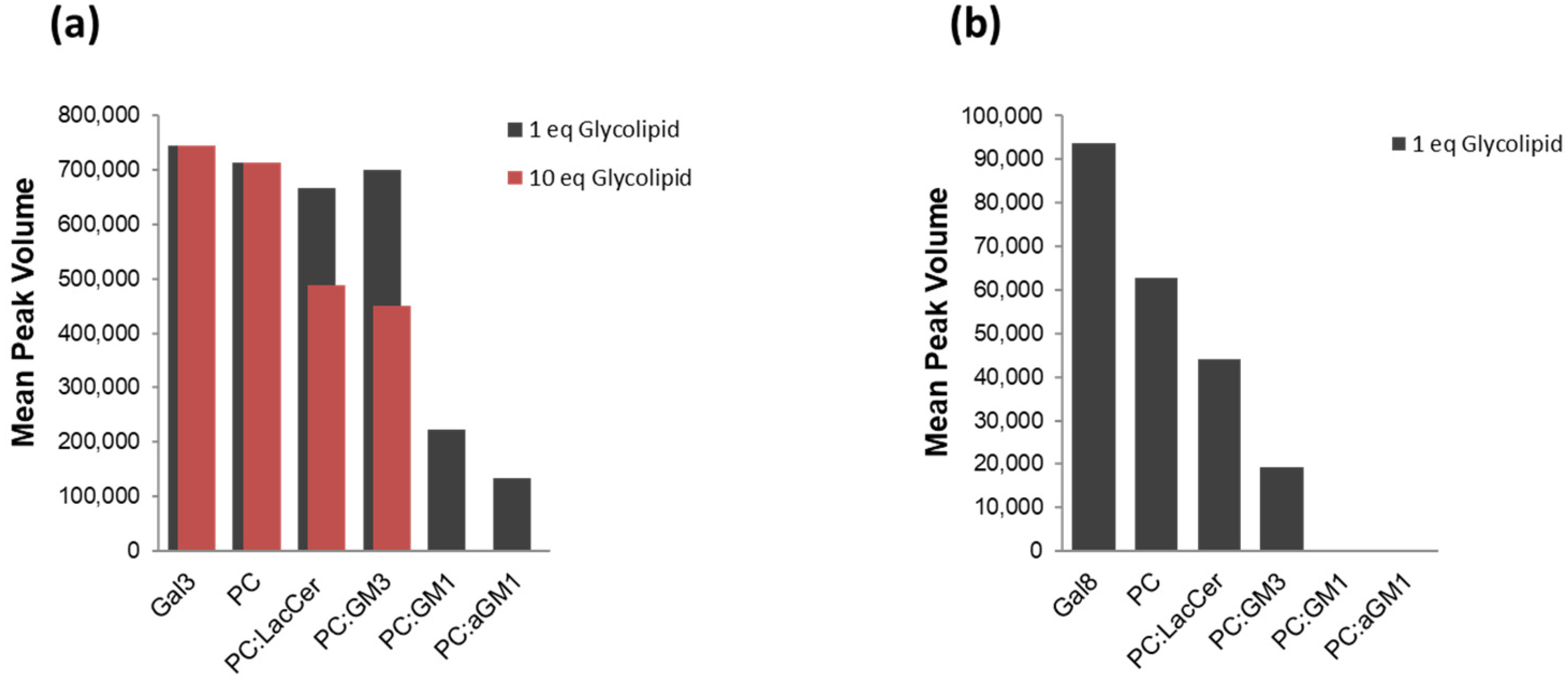
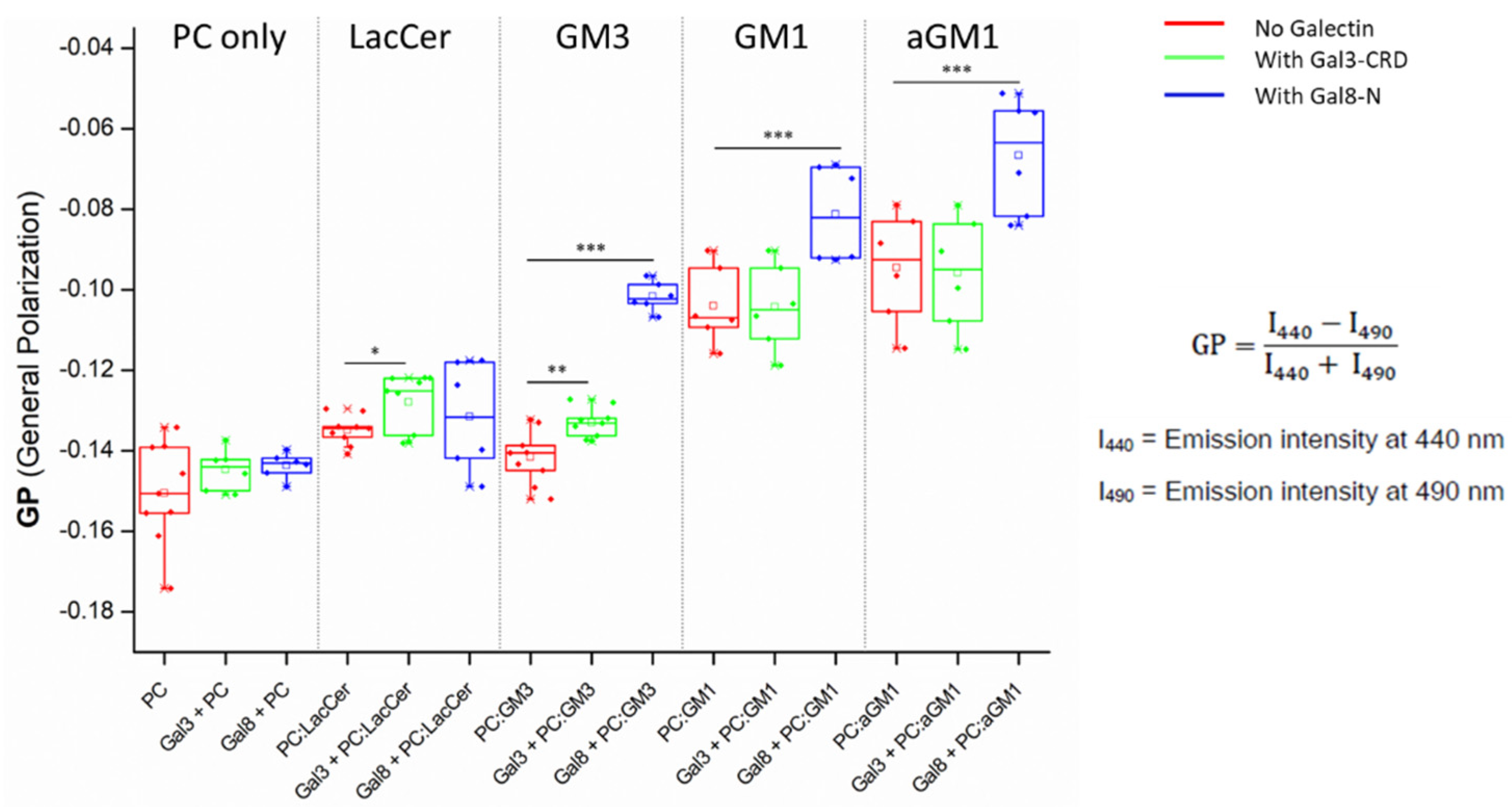
2.2. The Binding of Galectins to Gangliosides in Model Membranes
2.3. Membrane Perturbation
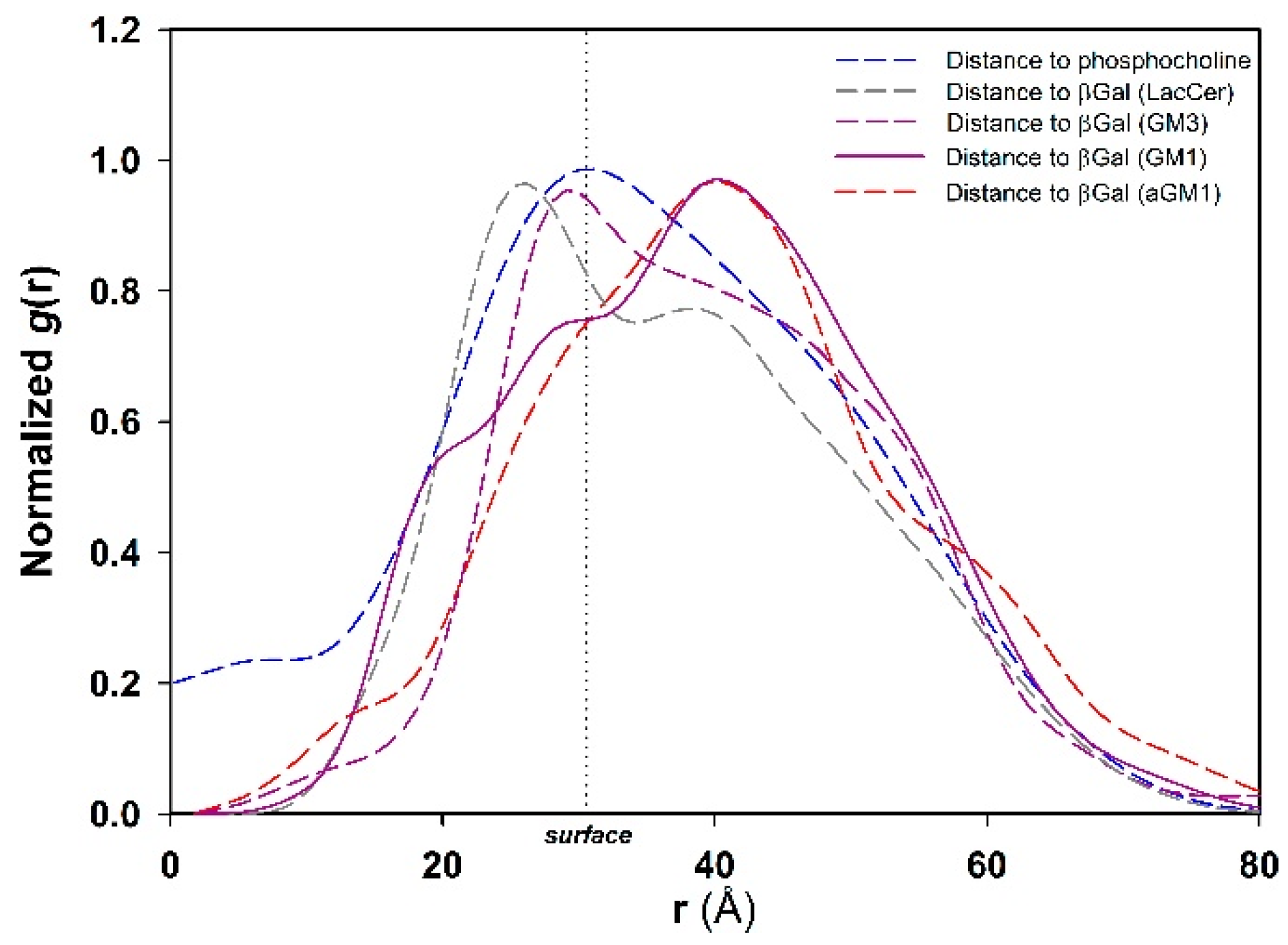
2.4. Molecular Simulation of Membranes Containing Glycolipids
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Protein Expression and Purification
4.3. Liposome Preparation
4.4. NMR Experiments
4.5. Membrane Polarization
4.6. Model Building and MD Details
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taylor, M.E.; Drickamer, K. Mammalian sugar-binding receptors: Known functions and unexplored roles. FEBS J. 2019, 286, 1800–1814. [Google Scholar] [CrossRef] [PubMed]
- Crook, S.J.; Boggs, J.M.; Vistnes, A.I.; Koshy, K.M. Factors affecting surface expression of glycolipids: Influence of lipid environment and ceramide composition on antibody recognition of cerebroside sulfate in liposomes. Biochemistry 1986, 25, 7488–7494. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, M.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H.; et al. (Eds.) Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Krengel, U.; Bousquet, P.A. Molecular recognition of gangliosides and their potential for cancer immunotherapies. Front. Immunol. 2014, 5, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakomori, S.I. Structure and function of glycosphingolipids and sphingolipids: Recollections and future trends. Biochim. Biophys. Acta 2008, 1780, 325–346. [Google Scholar] [CrossRef] [Green Version]
- Connell, T.D. Cholera toxin, LT-I, LT-IIa and LT-IIb: The critical role of ganglioside binding in immunomodulation by type I and type II heat-labile enterotoxins. Expert Rev. Vaccines 2007, 6, 821–834. [Google Scholar] [CrossRef] [Green Version]
- Johannes, L. Shiga Toxin-A Model for Glycolipid-Dependent and Lectin-Driven Endocytosis. Toxins 2017, 9, 340. [Google Scholar] [CrossRef]
- Nilsson, E.C.; Storm, R.J.; Bauer, J.; Johansson, S.M.; Lookene, A.; Angstrom, J.; Hedenstrom, M.; Eriksson, T.L.; Frangsmyr, L.; Rinaldi, S.; et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nat. Med. 2011, 17, 105–109. [Google Scholar] [CrossRef]
- Neu, U.; Woellner, K.; Gauglitz, G.; Stehle, T. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. USA 2008, 105, 5219–5224. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Son, K.Y.; Koo, K.M.; Kim, J.Y.; Alfajaro, M.M.; Park, J.G.; Hosmillo, M.; Soliman, M.; Baek, Y.B.; Cho, E.H.; et al. Porcine Sapelovirus Uses alpha2,3-Linked Sialic Acid on GD1a Ganglioside as a Receptor. J. Virol. 2016, 90, 4067–4077. [Google Scholar] [CrossRef] [Green Version]
- Miljan, E.A.; Bremer, E.G. Regulation of growth factor receptors by gangliosides. Sci. STKE 2002, 2002, re15. [Google Scholar] [CrossRef]
- Wang, J.; Yu, R.K. Association of Glycolipids and Growth Factor Receptors in Lipid Rafts. Methods Mol. Biol. 2021, 2187, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Triet, H.M.; Ryu, S.H. Regulation of EGFR activation and signaling by lipids on the plasma membrane. Prog. Lipid Res. 2021, 83, 101115. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Feng, W.; Wang, Y.J.; Sun, Y.; Shi, G.; Yu, Q. Galectins as potential emerging key targets in different types of leukemia. Eur. J. Pharmacol. 2019, 844, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Groux-Degroote, S.; Delannoy, P. Cancer-Associated Glycosphingolipids as Tumor Markers and Targets for Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 6145. [Google Scholar] [CrossRef]
- Sasaki, N.; Toyoda, M.; Ishiwata, T. Gangliosides as Signaling Regulators in Cancer. Int. J. Mol. Sci. 2021, 22, 5076. [Google Scholar] [CrossRef]
- Bartish, M.; Del Rincon, S.V.; Rudd, C.E.; Saragovi, H.U. Aiming for the Sweet Spot: Glyco-Immune Checkpoints and gammadelta T Cells in Targeted Immunotherapy. Front. Immunol. 2020, 11, 564499. [Google Scholar] [CrossRef]
- Ledeen, R.W.; Kopitz, J.; Abad-Rodriguez, J.; Gabius, H.J. Glycan Chains of Gangliosides: Functional Ligands for Tissue Lectins (Siglecs/Galectins). Prog. Mol. Biol. Transl. Sci. 2018, 156, 289–324. [Google Scholar] [CrossRef]
- Novak, J.; Kriston-Pal, E.; Czibula, A.; Deak, M.; Kovacs, L.; Monostori, E.; Fajka-Boja, R. GM1 controlled lateral segregation of tyrosine kinase Lck predispose T-cells to cell-derived galectin-1-induced apoptosis. Mol. Immunol. 2014, 57, 302–309. [Google Scholar] [CrossRef] [Green Version]
- Fajka-Boja, R.; Blasko, A.; Kovacs-Solyom, F.; Szebeni, G.J.; Toth, G.K.; Monostori, E. Co-localization of galectin-1 with GM1 ganglioside in the course of its clathrin- and raft-dependent endocytosis. Cell Mol. Life Sci. 2008, 65, 2586–2593. [Google Scholar] [CrossRef]
- Boscher, C.; Zheng, Y.Z.; Lakshminarayan, R.; Johannes, L.; Dennis, J.W.; Foster, L.J.; Nabi, I.R. Galectin-3 protein regulates mobility of N-cadherin and GM1 ganglioside at cell-cell junctions of mammary carcinoma cells. J. Biol. Chem. 2012, 287, 32940–32952. [Google Scholar] [CrossRef] [Green Version]
- Lakshminarayan, R.; Wunder, C.; Becken, U.; Howes, M.T.; Benzing, C.; Arumugam, S.; Sales, S.; Ariotti, N.; Chambon, V.; Lamaze, C.; et al. Galectin-3 drives glycosphingolipid-dependent biogenesis of clathrin-independent carriers. Nat. Cell Biol. 2014, 16, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Ideo, H.; Seko, A.; Ishizuka, I.; Yamashita, K. The N-terminal carbohydrate recognition domain of galectin-8 recognizes specific glycosphingolipids with high affinity. Glycobiology 2003, 13, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Johannes, L.; Wunder, C.; Shafaq-Zadah, M. Glycolipids and Lectins in Endocytic Uptake Processes. J. Mol. Biol. 2016, 428, 4792–4818. [Google Scholar] [CrossRef] [PubMed]
- Ivashenka, A.; Wunder, C.; Chambon, V.; Sandhoff, R.; Jennemann, R.; Dransart, E.; Podsypanina, K.; Lombard, B.; Loew, D.; Lamaze, C.; et al. Glycolipid-dependent and lectin-driven transcytosis in mouse enterocytes. Commun. Biol. 2021, 4, 173. [Google Scholar] [CrossRef]
- Johannes, L. The Cellular and Chemical Biology of Endocytic Trafficking and Intracellular Delivery-The GL-Lect Hypothesis. Molecules 2021, 26, 3299. [Google Scholar] [CrossRef]
- Collins, P.M.; Bum-Erdene, K.; Yu, X.; Blanchard, H. Galectin-3 interactions with glycosphingolipids. J. Mol. Biol. 2014, 426, 1439–1451. [Google Scholar] [CrossRef]
- Bum-Erdene, K.; Leffler, H.; Nilsson, U.J.; Blanchard, H. Structural characterization of human galectin-4 C-terminal domain: Elucidating the molecular basis for recognition of glycosphingolipids, sulfated saccharides and blood group antigens. FEBS J. 2015, 282, 3348–3367. [Google Scholar] [CrossRef]
- Ideo, H.; Seko, A.; Yamashita, K. Galectin-4 binds to sulfated glycosphingolipids and carcinoembryonic antigen in patches on the cell surface of human colon adenocarcinoma cells. J. Biol. Chem. 2005, 280, 4730–4737. [Google Scholar] [CrossRef] [Green Version]
- Hirabayashi, J.; Hashidate, T.; Arata, Y.; Nishi, N.; Nakamura, T.; Hirashima, M.; Urashima, T.; Oka, T.; Futai, M.; Muller, W.E.; et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim. Biophys. Acta 2002, 1572, 232–254. [Google Scholar] [CrossRef]
- Grant, O.C.; Smith, H.M.; Firsova, D.; Fadda, E.; Woods, R.J. Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data. Glycobiology 2014, 24, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Dam, T.K.; Brewer, C.F. Maintenance of cell surface glycan density by lectin-glycan interactions: A homeostatic and innate immune regulatory mechanism. Glycobiology 2010, 20, 1061–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimeno, A.; Reichardt, N.C.; Canada, F.J.; Perkams, L.; Unverzagt, C.; Jimenez-Barbero, J.; Arda, A. NMR and Molecular Recognition of N-Glycans: Remote Modifications of the Saccharide Chain Modulate Binding Features. ACS Chem. Biol. 2017, 12, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, M.L.; Woods, R.J. Atomic-resolution conformational analysis of the GM3 ganglioside in a lipid bilayer and its implications for ganglioside-protein recognition at membrane surfaces. Glycobiology 2009, 19, 344–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demarco, M.L.; Woods, R.J.; Prestegard, J.H.; Tian, F. Presentation of membrane-anchored glycosphingolipids determined from molecular dynamics simulations and NMR paramagnetic relaxation rate enhancement. J. Am. Chem. Soc. 2010, 132, 1334–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi-Utsumi, M.; Kato, K. Structural and dynamic views of GM1 ganglioside. Glycoconj. J. 2015, 32, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Emmenegger, C.; Xiao, Q.; Kostina, N.Y.; Sherman, S.E.; Rahimi, K.; Partridge, B.E.; Li, S.; Sahoo, D.; Reveron Perez, A.M.; Buzzacchera, I.; et al. Encoding biological recognition in a bicomponent cell-membrane mimic. Proc. Natl. Acad. Sci. USA 2019, 116, 5376–5382. [Google Scholar] [CrossRef] [Green Version]
- Mayer, M.; Meyer, B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Meyer, B.; Peters, T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 2003, 42, 864–890. [Google Scholar] [CrossRef]
- Bian, C.F.; Zhang, Y.; Sun, H.; Li, D.F.; Wang, D.C. Structural basis for distinct binding properties of the human galectins to Thomsen-Friedenreich antigen. PLoS ONE 2011, 6, e25007. [Google Scholar] [CrossRef] [Green Version]
- Furukawa, A.; Konuma, T.; Yanaka, S.; Sugase, K. Quantitative analysis of protein-ligand interactions by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 96, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Salomonsson, E.; Larumbe, A.; Tejler, J.; Tullberg, E.; Rydberg, H.; Sundin, A.; Khabut, A.; Frejd, T.; Lobsanov, Y.D.; Rini, J.M.; et al. Monovalent interactions of galectin-1. Biochemistry 2010, 49, 9518–9532. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, S.; Oberg, C.T.; Carlsson, M.C.; Sundin, A.; Nilsson, U.J.; Smith, D.; Cummings, R.D.; Almkvist, J.; Karlsson, A.; Leffler, H. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology 2007, 17, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Santarsia, S.; Grosso, A.S.; Trovao, F.; Jimenez-Barbero, J.; Carvalho, A.L.; Nativi, C.; Marcelo, F. Molecular Recognition of a Thomsen-Friedenreich Antigen Mimetic Targeting Human Galectin-3. ChemMedChem 2018, 13, 2030–2036. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.; Farris, F.J. Synthesis and spectral properties of a hydrophobic fluorescent probe: 6-propionyl-2-(dimethylamino)naphthalene. Biochemistry 1979, 18, 3075–3078. [Google Scholar] [CrossRef]
- Bertuzzi, S.; Quintana, J.I.; Arda, A.; Gimeno, A.; Jimenez-Barbero, J. Targeting Galectins with Glycomimetics. Front. Chem. 2020, 8, 593. [Google Scholar] [CrossRef]
- Asensio, J.L.; Arda, A.; Canada, F.J.; Jimenez-Barbero, J. Carbohydrate-aromatic interactions. Acc. Chem. Res. 2013, 46, 946–954. [Google Scholar] [CrossRef] [Green Version]
- Gu, R.X.; Ingolfsson, H.I.; de Vries, A.H.; Marrink, S.J.; Tieleman, D.P. Ganglioside-Lipid and Ganglioside-Protein Interactions Revealed by Coarse-Grained and Atomistic Molecular Dynamics Simulations. J. Phys. Chem. B 2017, 121, 3262–3275. [Google Scholar] [CrossRef] [Green Version]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J. Cell Sci. 2018, 131, jcs208884. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Redondo, M.; Delgado, S.; Núñez-Franco, R.; Jiménez-Osés, G.; Ardá, A.; Jiménez-Barbero, J.; Gimeno, A. The two domains of human galectin-8 bind sialyl- and fucose-containing oligosaccharides in an independent manner. A 3D view by using NMR. RSC Chem. Biol. 2021, 2, 932–941. [Google Scholar] [CrossRef]
- Gimeno, A.; Delgado, S.; Valverde, P.; Bertuzzi, S.; Berbis, M.A.; Echavarren, J.; Lacetera, A.; Martin-Santamaria, S.; Surolia, A.; Canada, F.J.; et al. Minimizing the Entropy Penalty for Ligand Binding: Lessons from the Molecular Recognition of the Histo Blood-Group Antigens by Human Galectin-3. Angew. Chem. Int. Ed. Engl. 2019, 58, 7268–7272. [Google Scholar] [CrossRef] [Green Version]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Case, D.A.; Belfon, K.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Giambasu, G.; et al. AMBER2020; University of California: San Francisco, CA, USA, 2020. [Google Scholar]
- Kucerka, N.; Nieh, M.P.; Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta 2011, 1808, 2761–2771. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; Gonzalez-Outeirino, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [Green Version]
- Tessier, M.B.; Demarco, M.L.; Yongye, A.B.; Woods, R.J. Extension of the GLYCAM06 Biomolecular Force Field to Lipids, Lipid Bilayers and Glycolipids. Mol. Simul. 2008, 34, 349–363. [Google Scholar] [CrossRef] [Green Version]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Dickson, C.J.; Madej, B.D.; Skjevik, A.A.; Betz, R.M.; Teigen, K.; Gould, I.R.; Walker, R.C. Lipid14: The Amber Lipid Force Field. J. Chem. Theory Comput. 2014, 10, 865–879. [Google Scholar] [CrossRef]
- Quigley, D.; Probert, M.I. Langevin dynamics in constant pressure extended systems. J. Chem. Phys. 2004, 120, 11432–11441. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]



| Glycan | Gal3-CRD | Gal8-N | ||||
|---|---|---|---|---|---|---|
| NMR | (a) | (b) | NMR | (b) | (c) | |
| Lactose | 125 ± 13 | 2.8 | 260 | 108 ± 15 | 130 | 3.1/1.7 |
| 3′SL | 70 ± 8 | 1.7 | 230 | - i | 0.6 | 0.05 |
| GM1 Pentasaccharide | 180 ± 27 | 130 | 152 ± 26 | 30 | 4.1 | |
| aGM1 Tetrasaccharide | 307 ± 35 | 180 | 82 ± 10 | 46 | 6.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lete, M.G.; Franconetti, A.; Delgado, S.; Jiménez-Barbero, J.; Ardá, A. Oligosaccharide Presentation Modulates the Molecular Recognition of Glycolipids by Galectins on Membrane Surfaces. Pharmaceuticals 2022, 15, 145. https://doi.org/10.3390/ph15020145
Lete MG, Franconetti A, Delgado S, Jiménez-Barbero J, Ardá A. Oligosaccharide Presentation Modulates the Molecular Recognition of Glycolipids by Galectins on Membrane Surfaces. Pharmaceuticals. 2022; 15(2):145. https://doi.org/10.3390/ph15020145
Chicago/Turabian StyleLete, Marta G., Antonio Franconetti, Sandra Delgado, Jesús Jiménez-Barbero, and Ana Ardá. 2022. "Oligosaccharide Presentation Modulates the Molecular Recognition of Glycolipids by Galectins on Membrane Surfaces" Pharmaceuticals 15, no. 2: 145. https://doi.org/10.3390/ph15020145
APA StyleLete, M. G., Franconetti, A., Delgado, S., Jiménez-Barbero, J., & Ardá, A. (2022). Oligosaccharide Presentation Modulates the Molecular Recognition of Glycolipids by Galectins on Membrane Surfaces. Pharmaceuticals, 15(2), 145. https://doi.org/10.3390/ph15020145









