Major Achievements in the Design of Quadruplex-Interactive Small Molecules
Abstract
:1. Introduction
1.1. Structures of G-Quadruplexes and i-Motifs


1.2. Quadruplexes as Therapeutic Targets
1.2.1. Quadruplexes in Cancer


| G4 Ligand (Chemotype) | Target in Cancer Cells | Effects | Refs. |
|---|---|---|---|
| BRACO-19 (acridine) | Telomere G4 in glioblastoma cells | Induces telomere uncapping; inhibits telomerase activity; induces DNA damage, apoptosis and senescence. | [79] |
| RHSP4 (acridinium) | Telomere G4 in glioblastoma cells | Radiosensitizing agent in a glioblastoma multiforme xenograft model, inducing telomere dysfunction. | [80] |
| SchCoD (quinazolinone) | Telomere G4 in cervical squamous cancer | It induces telomere uncapping, leading to a DNA damage response and inhibits tumor growth in a xenograft model. | [81] |
| QN-1 (quinoxaline) | c-MYC promoter G4 in triple-negative breast cancer | Downregulation of c-MYC transcription in vitro and in vivo. Anticancer activity in a xenograft model. | [88] |
| APTO-253 (phenanthroline) | c-MYC promoter G4 in acute myeloid leukemia | Inhibits c-Myc expression, induces cell cycle arrest and apoptosis in acute myeloid leukemia cells; induces the Krüppel-like factor 4 (KLF4) tumor suppressor. | [89,90] |
| In Phase I clinical trials. | [97] | ||
| IQb2 (indoloquinoline) | k-RAS promoter G4 in colon cancer cells | Decrease wild-type k-RAS promoter activity; preferentially downregulation of k-RAS expression compared to other G4-containing genes; induces apoptosis and expression of tumor suppressor p53 in HCT116 cell line. | [94] |
| GTC365 (acridine) | hTERT promoter G4 in breast cancer cells | Binds to G-quartet and duplex stem loop of G4, restoring wild-type folding topology; Induces apoptosis and senescence in cancer cells. | [95] |
| TA20 (triarylpyridine) | Several DNA G4 in cervical cancer cells | Stabilizes several G4s in vitro. Transcriptome analysis of treated cancer cells revealed that most affected genes are those related with DNA damage, cell growth and autophagy; genes enriched in G4-forming sequences are preferentially downregulated. | [93] |
| CM03 (naphthalene diimide) | Several DNA G4 in pancreatic cancer | Stabilizes several G4s in vitro and increases G4 foci in treated cells; Transcriptome analysis indicates a preferential downregulation of genes with G4-forming sequences in promoters and implicated in cancer cells survival, development of metastasis and drug resistance; Anticancer activity in a xenograft model. | [92] |
| RGB-1 (thienopyrimidinone) | 5′-UTR G4 of NRAS mRNA in breast cancer | Downregulation of NRAS expression in cancer cell lines. | [77] |
| Quarfloxin/CX-3543 (quinolone) | Ribosomal DNA G4 in Carcinoid/neuroendocrine tumors | Inhibits rRNA biogenesis; induces DNA damage and apoptosis. | [98] |
| Reached Phase II clinical trials. | [99] | ||
| CX-5461 (quinolone) | Ribosomal DNA G4 in BRCA1/2-deficient tumors | Inhibits RNA polymerase I and Topoisomerase II. | [100,101] |
| Induces G4-mediated DNA damage. | [102] | ||
| In Phase I Clinical trials. | [103] |
1.2.2. G4s in Human-Infecting Parasites and Bacteria
| IC50/µM | |||||
|---|---|---|---|---|---|
| G4 Ligand | P. falciparum | T. brucei | L. major | M. tuberculosis | Human Cells |
| TMPyP4 | 35 [114]; 0.62 [51] | >10 [115] | 21 [115] | 6.25 [116] | >25 [115] |
| TMPyP2 | 11 [51] | ||||
| Telomestatin | 5 [114] | ||||
| 3AQN | 1.8–2.5 [117] | 170 [117] | |||
| 6AQN | 1.5–2.7 [117] | 170 [117] | |||
| PDC-360A | 0.9–1.2 [117] | 120 [117] | |||
| Carb-NDI6 | 0.275 [115] | 0.017 [115] | 0.537 [115] | 0.71 [115] | |
| Pyridostatin | 2.65 [115]; 5.2 [53] | 7.82 [115] | 5.00 [115] | 5.38 [115] | |
| BRACO-19 (a) | 9.70 [115] | 5.51 [115] | 12.7 [115] | <12.5 [118] | 8.33 [115] |
| Quarfloxin (a) | 0.11 [51] | ||||
| c-exNDI 2 | <2.5 [118] | ||||
| PIPER-d3 | 0.027 [119] | >100 [119] | 2.5–53 [119] | ||
1.2.3. G4s in Human Viruses
| G4 Ligand (Chemotype) | Structure in Figure | Virus | Effects | Refs. |
|---|---|---|---|---|
| TMPyP4 (porhyrine) | 6 | HIV-1 | Blocked viral replication in lymphocyte T cells with established HIV-1 latency. | [154] |
| Inhibition of viral infectivity. | [155] | |||
| Enhanced killing of latently infected cells when in combination with latency reversing agents. | [156] | |||
| KSHV | Inhibited viral DNA replication; reduced in 60% the viral episome copy numbers; inhibited LANA1 translation in KSHV infected cells. | [157,158] | ||
| HCV | Promoted viral RNA polymerase stalling. | [159] | ||
| EBOV | Reduced transcription of L gene (encodes for viral RNA-dependent RNA polymerase) and impaired replication of viral genome. | [160] | ||
| HSV-1 | Showed good antiviral activity at microM concentrations; did not inhibited virus DNA replication or entry but inhibited virus release by the cells. | [161] | ||
| ZIKV | Inhibited viral growth, genome replication and protein expression. | [162] | ||
| SARS-CoV-2 | Inhibited replication and gene expression of virus RNA G4-forming sequences in in-vitro assays | [145] | ||
| Acridine C8 | 7 | HPV | The exposure of cervical cells to C8 at 0.25 microM induced a >100-fold decrease in HPV18 viral titre; C8 probably affects viral genome encapsidation rather than genome amplification. | [163] |
| BRACO-19 (acridine) | 4 | HIV-1 | Reduced viral titre to undetectable levels in latently infected cells. | [154] |
| Blocked RT progression in-vitro, which was counteract by viral Ncp7, a protein known to unfold RNA G4s. | [164,165] | |||
| Antiviral activity at microM concentrations. | [164] | |||
| Reduced proviral LTR promoter activity. | [165] | |||
| Enhanced killing of latently infected cells when in combination with latency reversing agents. | [156] | |||
| HSV-1 | Antiviral activity (IC50~8 µM) with inhibition of viral DNA synthesis. | [166] | ||
| HHV-6 | Reduction of viral genome integration in human chromosomes at telomeres. | [167] | ||
| EBV | Reduced viral genome copy numbers in infected lymphocytes; reduced transcription of viral proteins EBNA2 and EBNA3A; reduced EBNA1-dependent DNA replication. | [168] | ||
| HBV | Enhanced preS2/S gene promoter activity, which product regulates production of the HBV surface antigen and virion secretion. | [169] | ||
| ZIKV | Inhibited viral growth, genome replication and protein expression. | [162] | ||
| SARS-CoV-2 | Inhibited replication and gene expression of virus RNA G4-forming sequences in in-vitro assays | [145] | ||
| c-exNDI (naphetalene Diimide) | 6 | HIV-1 | Strong antiviral activity (IC50 < 25 nM). | [170] |
| HSV-1 | Antiviral activity (IC50~18 nM) with inhibition of viral DNA synthesis. | [171] | ||
| Pyridostatin | 6 | EBV | Reduced EBNA1 synthesis and recognition of EBV-infected cells by virus-specific T cells. | [137] |
| HBV | Enhanced preS2/S gene promoter activity. | [169] | ||
| ZIKV | Inhibited mRNA synthesis, virus cytopathic effect and viral NS2B-NS3 protease activity in infected Vero cells, particularly during postinfection treatment. | [172] | ||
| PDP (pyridostatin) | 7 | HCV | Promoted viral RNA polymerase stalling. In-vivo G4-mediated antiviral activity in the low microM range. | [159] |
| SARS- CoV-2 | Inhibited translation of nucleocapsid protein N, in-vitro and in-vivo. | [147] | ||
| CX-5461 (quinolone) | 4 | CMV | Reduced viral titre by 2 log, acting at the viral DNA replication stage. | [173] |
| PhenDC3 (phenanthroline) | 6 | EBV | Inhibited nucleolin binding to EBNA1 mRNA G4s and increased the endogenous EBNA1 levels in EBV-infected cells. | [139] |
| KSHV | Inhibited viral DNA replication by stalling the replication fork at the TR level; consequent reduction of viral episome copy numbers. | [157] | ||
| HCV | Inhibited viral replication in cells. | [174] | ||
| SARS-CoV-2 | Inhibited in-vitro the SUD-NM/TRF2 G4 interaction with an IC50 = 51 nM. | [146] | ||
| BzSeX (Benzoseleno xanthene) | 7 | IAV | Reduced viral titers in-vitro, with downregulation of TMPRSS2 expression, a transmembrane serine protease essential for virus entry into the host cells | [156] |
| ribavirin | 7 | SARS-CoV-2 | Antiviral activity; reduced expression of TMPRSS2 and AC2; inhibited TMPRSS2 enzymatic activity. | [152] |
1.3. Approaches in the Design of Quadruplex-Interactive Small Molecules
2. G4-Ligand Complexes and Intermolecular Interaction Modes
2.1. Fused Polycyclic Ligands
2.1.1. Acridines
2.1.2. Indoloquinolines
2.1.3. Berberines

2.1.4. Anthracyclines

2.1.5. Naphthalene Diimides
2.1.6. Phenanthrolines and Quinacridines
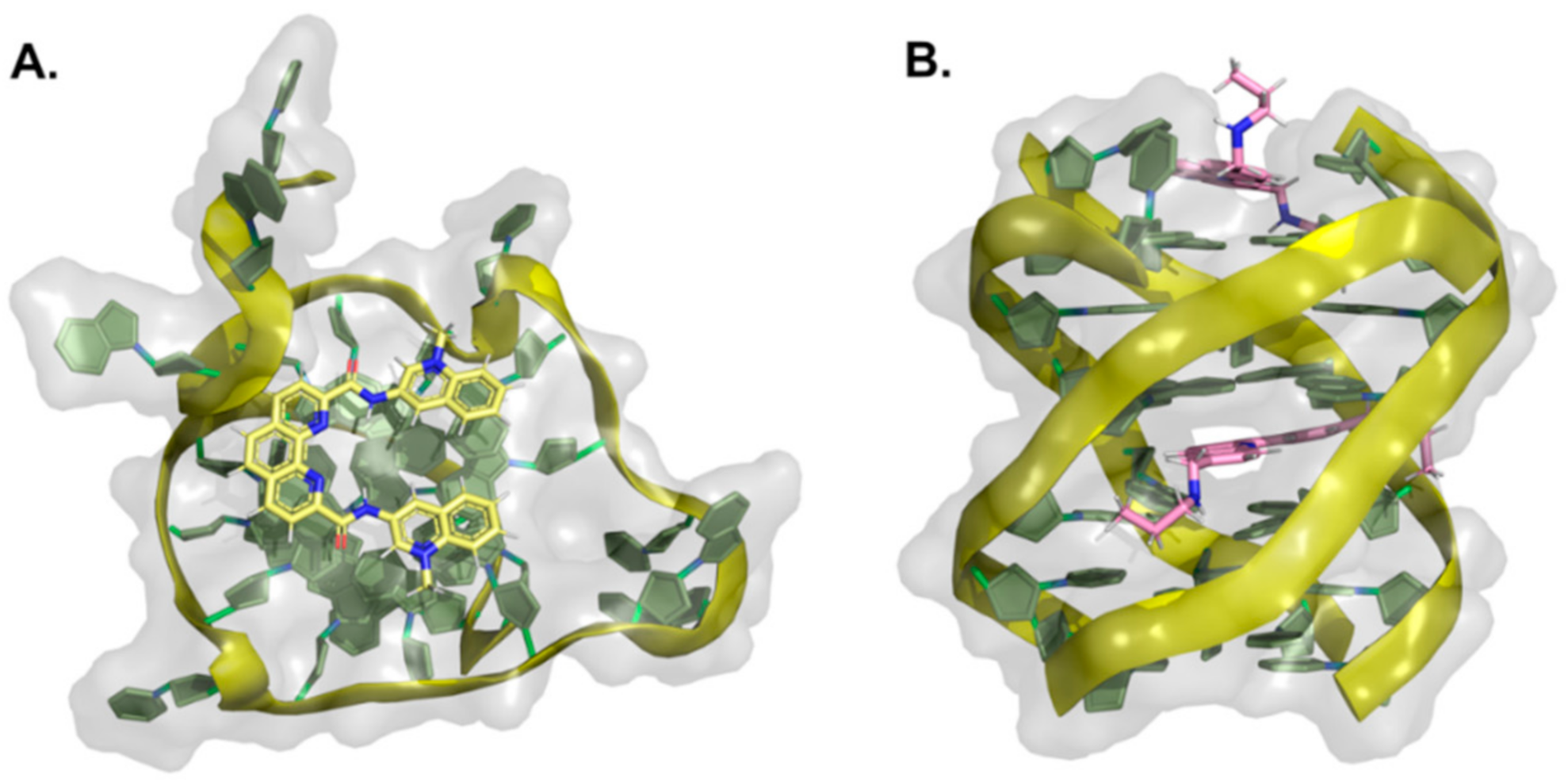
| PDB ID | Method | Ligand Chemotype | Quadruplex | Refs. |
|---|---|---|---|---|
| 1K2L | X-ray | Acridine | Four-way DNA junctions | [227] |
| 1NZM | NMR | Acridine | Parallel hTelo DNA G4 | [228] |
| 1L1H | X-ray | Acridine | Antiparallel DNA G4 from Oxytricha nova | [210] |
| 3CE5 | X-ray | Acridine (BRACO-19) | Parallel hTelo DNA G4 | [229] |
| 3EUM | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| 3EUI | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| 3EM2 | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| 3EQW | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| ERU | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| 3ES0 | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| 3ET8 | X-ray | Acridine | Oxytricha nova bimolecular G4 | [230] |
| 3NZ7 | X-ray | Acridine | Antiparallel DNA G4 from Oxytricha nova | [231] |
| 3NYP | X-ray | Acridine | Antiparallel DNA G4 from Oxytricha nova | [231] |
| 3MIJ | X-ray | Acridine | Parallel hTelo RNA (TERRA) | [196] |
| 3QCR | X-ray | Acridine | Parallel hTelo RNA (TERRA) | [196] |
| 5LIG | NMR | Acridine | c-Myc promoter parallel DNA G4 | [232] |
| 2L7V | NMR | indoloquinoline | c-Myc promoter parallel DNA G4 | [197] |
| 3R6R | X-ray | Berberine | Parallel hTelo DNA G4 | [206] |
| 4P1D | X-ray | Berberine | Bimolecular hTelo DNA G4 | [233] |
| 5CDB | X-ray | Berberine | Antiparallel hTelo DNA G4 | [234] |
| 6CCW | NMR | Berberine | Hybrid hTelo DNA G4 | [235] |
| 6S15 | X-ray | Berberine | Bimolecular hTelo DNA G4 | [207] |
| 6JWD | NMR | Berberine | RET promoter parallel DNA G4 | [236] |
| 1O0K | X-ray | Anthracyclin | Intermolecular parallel Telo DNA G4 | [211] |
| 3TVB | X-ray | Anthracyclin | Parallel Telo DNA G4 | [215] |
| 6FC9 | NMR | Anthracyclin | Quadruplex-duplex junction | [237] |
| 6KXZ | NMR | Anthracyclin | Parallel hTelo DNA G4 | [214] |
| 6KN4 | NMR | Anthracyclin | Parallel hTelo DNA G4 | [213] |
| 3SC8 | X-ray | Naphthalene diimide | Parallel hTelo DNA G4 | [221] |
| 3T5E | X-ray | Naphthalene diimide | Parallel hTelo DNA G4 | [221] |
| 3UYH | X-ray | Naphthalene diimide | Parallel hTelo DNA G4 | [179] |
| 4DA3 | X-ray | Naphthalene diimide | Parallel hTelo DNA G4 | [179] |
| 4DAQ | X-ray | Naphthalene diimide | Parallel hTelo DNA G4 | [179] |
| 3CDM | X-ray | Phenantroline | Parallel hTelo DNA G4 | [222] |
| 3CCO | X-ray | Phenantroline | Parallel hTelo DNA G4 | [222] |
| 2MGN | NMR | Phenantroline (PhenDC-3) | c-Myc promoter parallel DNA G4 | [225] |
| 2JWQ | NMR | Quinacridine | Tetramolecular G4 derived from hTelo | [226] |
| 6JJ0 | NMR | Carbazole | c-Myc promoter parallel DNA G4 | [238] |
| 6O2L | NMR | Carbazole | c-Myc promoter parallel DNA G4 | [238] |
2.1.7. Carbazoles
2.2. Modular Ligands
2.2.1. Ligands with Aryl–Aryl Bonds
2.2.2. Ligands with Carboxamide Based Linkers
2.2.3. Quinoline Derivatives with Olefin Linkers
2.3. Macrocycles
2.3.1. Telomestatin
2.3.2. Porphyrins
| PDB ID | Method | Ligand chemotype | Quadruplex | Refs. |
|---|---|---|---|---|
| 6KFJ | NMR | tri-arylamine | Hybrid hTelo DNA g4 | [246] |
| 6KFI | NMR | tri-arylamine | Hybrid hTelo DNA G4 | [246] |
| 2JT7 | NMR | pyrrole carboxamide | Parallel DNA G4 | [248] |
| 2KVY | NMR | pyrrole carboxamide | Antiparallel dimeric DNA G4 | [249] |
| 5W77 | NMR | benzofuran (DC34) | c-MYC parallel DNA G4 | [181] |
| 6SX3 | NMR | pyridine carboxamide (360A) | AGCGA- DNA quadruplex | [250] |
| 7KBX | NMR | quinoline with olefin linker | c-MYC 2345 T23 mutant DNA G4 | [182] |
| 7KBW | NMR | quinoline with olefin linker | c-MYC 2345 parallel DNA G4 | [182] |
| 2MB3 | NMR | Telomestatin (3,3-L2H2-6OTD) | Parallel hTelo DNA G4 | [259] |
| 2A5R | NMR | Porphyrin (TMPyP4) | c-MYC parallel DNA G4 | [261] |
| 2HRI | X-ray | Porphyrin | Parallel hTelo DNA G4 | [264] |
| 4G0F | X-ray | Mesoporphyrin | Parallel hTelo DNA G4 | [262] |
| 4FXM | X-ray | Mesoporphyrin | Parallel hTelo DNA G4 | [262] |
| 6JJI | X-ray | Mesoporphyrin | Two-quartet parallel RNA G4 | [265] |
| 6JJH | X-ray | Mesoporphyrin | Two-quartet parallel RNA G4 | [265] |
| 6PNK | X-ray | Mesoporphyrin | Parallel DNA G4 dimer | [263] |
| 6P45 | X-ray | Mesoporphyrin | Parallel DNA G4 dimer | [263] |
3. Ligand-Induced Effects
3.1. Ligand-Induced Quadruplex Stabilization or Destabilization


| Ligand (Chemotype) | Effect on G4 | Effect on iM | Potential Application | Refs. |
|---|---|---|---|---|
| Berberine | Stabilization (DNA G4) | Low and unspecific interaction | Anticancer drug | [270,278,281,282,283] |
| Berb8 (berberine) | Stabilization (DNA G4) | - | Anticancer drug | [274] |
| SYUIQ-5 (indolo[3,2-b]quinoline) | Stabilization (DNA G4) | - | Anticancer drug | [290] |
| IQb2 (indolo[3,2-b]quinoline) | Stabilization (DNA G4) | - | Anticancer drug | [94] |
| IQc3d (indolo[3,2-c]quinoline) | Stabilization (DNA and RNA G4) | - | Anticancer drug | [297] |
| BRACO-19 (acridine) | Stabilization (DNA G4) | Destabilization | Anticancer drug | [282,299,301] |
| PhenDC3 (Phenanthroline) | Stabilization (DNA G4) | Destabilization | Anticancer drug G4 probe | [282] |
| Nitidine (Benzophenanthridine) | Stabilization (DNA G4) | Destabilization | Anticancer drug | [108] |
| 3,3-L2H2-6OTD (oxazole-based) | Stabilization (DNA G4) | - | Anticancer drug | [305] |
| L1BOD-7OTD (oxazole-based) | Stabilization (DNA G4) | - | Fluorescent probe | [307] |
| L2H2-4OTD (oxazole-based) | Low stabilization (DNA G4) | Stabilization | - | [107] |
| TMPyP4 (porphyrine) | Stabilization/destabilization (DNA G4) Destabilization (RNA G4) | Promotion and stabilization | Low potential application due to its low selectivity to quadruplexes | [308,313,318,319,320,321,322] |
| pyridostatin | Stabilization (DNA G4) | No effect | G4 probe | [282] |
| CarboxyPDS (pyridostatin) | Stabilization (RNA G4) | - | Anticancer drug | [59] |
| TA17 (Terpyridine) | Stabilization at 1 µM (DNA G4) Destabilization at >10 µM (c-KIT2 G4) | - | Anticancer drug | [324,325] |
| PB1 (pyridine dicarboxamide) | Low stabilization | Induction and stabilization | Therapeutic agent | [110] |
| PB2 (pyridine dicarboxamide) | Induction and stabilization (DNA G4) | Low stabilization | Anticancer drug | [110] |
| ICM-48 (steroid) | - | Stabilization | Therapeutic agent | [280] |
| ICM-76 (steroid) | - | Destabilization | Anticancer drug | [280] |
| B19 (acridone) | No interaction | Stabilization | Anticancer drug | [109] |
| STI (stilbene) | Regulates the folding/unfolding of the telomeric G4 in a photoresponsive manner | - | In nanodevices | [327] |
| PhpC (pyrrolopyrimidine) | Destabilization | - | Therapeutic agent | [329] |

3.2. Ligand-Induced G4 Topology Switches


3.3. Alkylating G4 Ligands

4. Challenges in the Design of Therapeutically Useful G4 Targeting Small Molecules
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, D.; Gilbert, W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Smith, F.W.; Feigon, J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature 1992, 356, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Aboul-Ela, F.; Murchie, A.I.H.; Lilley, D.M.J. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T). Nature 1992, 360, 280–282. [Google Scholar] [CrossRef]
- Laughlan, G.; Murchie, A.I.H.; Norman, D.G.; Moore, M.H.; Moody, P.C.E.; Lilley, D.M.J.; Luisi, B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science 1994, 265, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Plückthun, A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehring, K.; Leroy, J.-L.; Guéron, M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 1993, 363, 561–565. [Google Scholar] [CrossRef]
- Chen, L.; Cai, L.; Zhang, X.; Rich, A. Crystal Structure of a Four-Stranded Intercalated DNA: D(C4). Biochemistry 1994, 33, 13540–13546. [Google Scholar] [CrossRef]
- Mergny, J.L.; Sen, D. DNA quadruple helices in nanotechnology. Chem. Rev. 2019, 119, 6290–6325. [Google Scholar] [CrossRef] [PubMed]
- Dzatko, S.; Krafcikova, M.; Hänsel-Hertsch, R.; Fessl, T.; Fiala, R.; Loja, T.; Krafcik, D.; Mergny, J.L.; Foldynova-Trantirkova, S.; Trantirek, L. Evaluation of the Stability of DNA i-Motifs in the Nuclei of Living Mammalian Cells. Angew. Chem. Int. Ed. 2018, 57, 2165–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.L.; Kendrick, S. The I-motif as a molecular target: More than a complementary DNA secondary structure. Pharmaceuticals 2021, 14, 96. [Google Scholar] [CrossRef]
- Palma, E.; Carvalho, J.; Cruz, C.; Paulo, A. Metal-Based G-Quadruplex Binders for Cancer Theranostics. Pharmaceuticals 2021, 14, 605. [Google Scholar] [CrossRef] [PubMed]
- Kench, T.; Vilar, R. Metal complexes as G-quadruplex binders. Annu. Rep. Med. Chem. 2020, 54, 485–515. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, M.W. Experimental demonstration of T:(G:G:G:G):T hexad and T:A:A:T tetrad alignments within a DNA quadruplex stem. Biochemistry 2005, 44, 3754–3764. [Google Scholar] [CrossRef]
- Lim, K.W.; Alberti, P.; Guédin, A.; Lacroix, L.; Riou, J.-F.; Royle, N.J.; Mergny, J.-L.; Phan, A.T. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G·C·G·C tetrad. Nucleic Acids Res. 2009, 37, 6239–6248. [Google Scholar] [CrossRef]
- Largy, E.; Mergny, J.-L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. In Metal Ions in Life Sciences; Walter de Gruyter GmbH: Berlin, Germany, 2016; Volume 16, pp. 203–258. [Google Scholar]
- Lyu, K.; Chow, E.Y.C.; Mou, X.; Chan, T.F.; Kwok, C.K. RNA G-quadruplexes (rG4s): Genomics and biological functions. Nucleic Acids Res. 2021, 49, 5426–5450. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Yang, D. Polymorphism of human telomeric quadruplex structures. Biochimie 2008, 90, 1172–1183. [Google Scholar] [CrossRef] [Green Version]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef]
- You, H.; Zeng, X.; Xu, Y.; Lim, C.J.; Efremov, A.K.; Phan, A.T.; Yan, J. Dynamics and stability of polymorphic human telomeric G-quadruplex under tension. Nucleic Acids Res. 2014, 42, 8789–8795. [Google Scholar] [CrossRef] [Green Version]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef] [Green Version]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef] [PubMed]
- Karsisiotis, A.I.; O’Kane, C.; Webba da Silva, M. DNA quadruplex folding formalism—A tutorial on quadruplex topologies. Methods 2013, 64, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)(n) among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bochman, M.; Paeschke, K.; Zakian, V. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, H.; Wu, J.; Shao, F.; Yan, J. Stability and Kinetics of c-MYC Promoter G-Quadruplexes Studied by Single-Molecule Manipulation. J. Am. Chem. Soc. 2015, 137, 2424–2427. [Google Scholar] [CrossRef]
- Phan, A.T.; Modi, Y.S.; Patel, D.J. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004, 126, 8710–8716. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [Green Version]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Burrows, C.J. Oxidative stress-mediated epigenetic regulation by G-quadruplexes. NAR Cancer 2021, 3, zcab038. [Google Scholar] [CrossRef] [PubMed]
- Ducani, C.; Bernardinelli, G.; Högberg, B.; Keppler, B.K.; Terenzi, A. Interplay of three G-quadruplex units in the KIT promoter. J. Am. Chem. Soc. 2019, 141, 10205–10213. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Paulo, A. Oncogene Expression Modulation in Cancer Cell Lines by DNA G-Quadruplex-Interactive Small Molecules. Curr. Med. Chem. 2016, 24, 4873–4904. [Google Scholar] [CrossRef]
- Lago, S.; Nadai, M.; Ruggiero, E.; Tassinari, M.; Marušič, M.; Tosoni, B.; Frasson, I.; Cernilogar, F.M.; Pirota, V.; Doria, F.; et al. The MDM2 inducible promoter folds into four-tetrad antiparallel G-quadruplexes targetable to fight malignant liposarcoma. Nucleic Acids Res. 2021, 49, 847–863. [Google Scholar] [CrossRef]
- Wei, D.; Husby, J.; Neidle, S. Flexibility and structural conservation in a c-KIT G-quadruplex. Nucleic Acids Res. 2015, 43, 629–644. [Google Scholar] [CrossRef] [Green Version]
- Mergny, J.L.; Lacroix, L.; Han, X.; Leroy, J.L.; Hélène, C. Intramolecular Folding of Pyrimidine Oligodeoxynucleotides into an i-DNA Motif. J. Am. Chem. Soc. 1995, 117, 8887–8898. [Google Scholar] [CrossRef]
- Rajendran, A.; Nakano, S.; Sugimoto, N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 2010, 46, 1299–1301. [Google Scholar] [CrossRef]
- Miyoshi, D.; Nakao, A.; Sugimoto, N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry 2002, 41, 15017–15024. [Google Scholar] [CrossRef]
- Kan, Z.; Yao, Y.; Wang, P.; Li, X.; Hao, Y.; Tan, Z. Molecular Crowding Induces Telomere G-Quadruplex Formation under Salt-Deficient Conditions and Enhances its Competition with Duplex Formation. Angew. Chemie Int. Ed. 2006, 45, 1629–1632. [Google Scholar] [CrossRef]
- Todd, A.K.; Johnston, M.; Neidle, S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [Green Version]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohálová, N.; Mergny, J.-L.; Brázda, V. Novel G-quadruplex prone sequences emerge in the complete assembly of the human X chromosome. Biochimie 2021, 191, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Ahn, J.H.; Kim, K.K. Unraveling the Regulatory G-Quadruplex Puzzle: Lessons from Genome and Transcriptome-Wide Studies. Front. Genet. 2019, 10, 1002. [Google Scholar] [CrossRef]
- Mishra, S.K.; Jain, N.; Shankar, U.; Tawani, A.; Sharma, T.K.; Kumar, A. Characterization of highly conserved G-quadruplex motifs as potential drug targets in Streptococcus pneumoniae. Sci. Rep. 2019, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaya, E.; Smithgall, T.E.; Palumbo, M.; Palù, G.; Richter, S.N. A Dynamic G-Quadruplex Region Regulates the HIV-1 Long Terminal Repeat Promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef] [Green Version]
- Amrane, S.; Kerkour, A.; Bedrat, A.; Vialet, B.; Andreola, M.-L.; Mergny, J.-L. Topology of a DNA G-Quadruplex Structure Formed in the HIV-1 Promoter: A Potential Target for Anti-HIV Drug Development. J. Am. Chem. Soc. 2014, 136, 5249–5252. [Google Scholar] [CrossRef]
- Yadav, V.; Hemansi; Kim, N.; Tuteja, N.; Yadav, P. G quadruplex in plants: A ubiquitous regulatory element and its biological relevance. Front. Plant Sci. 2017, 8, 1163. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.M.; Monsell, K.R.; Noulin, F.; Famodimu, M.T.; Smargiasso, N.; Damblon, C.; Horrocks, P.; Merrick, C.J. G-quadruplex DNA motifs in the malaria parasite plasmodium falciparum and their potential as novel antimalarial drug targets. Antimicrob. Agents Chemother. 2018, 62, e01828-17. [Google Scholar] [CrossRef] [Green Version]
- Dumetz, F.; Merrick, C.J. Parasitic protozoa: Unusual roles for g-quadruplexes in early-diverging eukaryotes. Molecules 2019, 24, 1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazanion, E.; Lacroix, L.; Alberti, P.; Gurung, P.; Wein, S.; Cheng, M.; Mergny, J.-L.; Gomes, A.R.; Lopez-Rubio, J.-J. Genome wide distribution of G-quadruplexes and their impact on gene expression in malaria parasites. PLoS Genet. 2020, 16, e1008917. [Google Scholar] [CrossRef] [PubMed]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Luo, Y.; Bartas, M.; Kaura, P.; Porubiaková, O.; Šťastný, J.; Pečinka, P.; Verga, D.; Da Cunha, V.; Takahashi, T.S.; et al. G-Quadruplexes in the Archaea Domain. Biomolecules 2020, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Di Antonio, M.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [Green Version]
- Salgado, G.F.; Cazenave, C.; Kerkour, A.; Mergny, J.L. G-quadruplex DNA and ligand interaction in living cells using NMR spectroscopy. Chem. Sci. 2015, 6, 3314–3320. [Google Scholar] [CrossRef]
- Lam, E.Y.N.; Beraldi, D.; Tannahill, D.; Balasubramanian, S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 2013, 4, 1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [Green Version]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Asamitsu, S.; Takeuchi, M.; Ikenoshita, S.; Imai, Y. Perspectives for Applying G-Quadruplex Structures in Neurobiology and Neuropharmacology. Int. J. Mol. Sci. 2019, 20, 2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabor, N.; Ngwa, C.; Mitteaux, J.; Meyer, M.D.; Moruno-Manchon, J.F.; Zhu, L.; Liu, F.; Monchaud, D.; Mccullough, L.D.; Andrey, S. Differential responses of neurons, astrocytes, and microglia to G-quadruplex stabilization. Aging 2021, 13, 15917–15941. [Google Scholar] [CrossRef]
- Cave, J.W.; Willis, D.E. G-quadruplex regulation of neural gene expression. FEBS J. 2021, 1–20. [Google Scholar] [CrossRef]
- Johnson, F.B. Fundamentals of G-quadruplex biology. Annu. Rep. Med. Chem. 2020, 54, 3–44. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Roy, S.S.; Chowdhury, S. Non-duplex G-Quadruplex DNA Structure: A Developing Story from Predicted Sequences to DNA Structure-Dependent Epigenetics and Beyond. Acc. Chem. Res. 2021, 54, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Balaratnam, S.; Schneekloth, J.S. Transcriptional regulation of MYC through G-quadruplex structures. Annu. Rep. Med. Chem. 2020, 54, 361–407. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. Biochimica et Biophysica Acta G4 DNA in ras genes and its potential in cancer therapy. Biochim. Biophys. Acta 2016, 1859, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. G-quadruplex, Friend or Foe: The Role of the G-quartet in Anticancer Strategies. Trends Mol. Med. 2020, 26, 848–861. [Google Scholar] [CrossRef]
- Paulo, A.; Castillo, C.C.; Neidle, S. Targeting Promoter Quadruplex Nucleic Acids for Cancer Therapy. In Comprehensive Medicinal Chemistry III; Elsevier: Oxford, UK, 2017; Volume 5, pp. 308–340. [Google Scholar]
- Ros, S.D.; Nicoletto, G.; Rigo, R.; Ceschi, S.; Zorzan, E.; Dacasto, M.; Giantin, M.; Sissi, C. G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter. Int. J. Mol. Sci. 2020, 22, 329. [Google Scholar] [CrossRef] [PubMed]
- Rigo, R.; Sissi, C. Characterization of G4-G4 Crosstalk in the c-KIT Promoter Region. Biochemistry 2017, 56, 4309–4312. [Google Scholar] [CrossRef] [PubMed]
- Zell, J.; Sperti, F.R.; Britton, S.; Monchaud, D. DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem. Biol. 2021, 2, 47–76. [Google Scholar] [CrossRef]
- Kim, N. The Interplay between G-quadruplex and Transcription. Curr. Med. Chem. 2019, 26, 2898–2917. [Google Scholar] [CrossRef]
- Katsuda, Y.; Sato, S.; Asano, L.; Morimura, Y.; Furuta, T.; Sugiyama, H.; Hagihara, M.; Uesugi, M. A Small Molecule That Represses Translation of G-Quadruplex-Containing mRNA. J. Am. Chem. Soc. 2016, 138, 9037–9040. [Google Scholar] [CrossRef] [PubMed]
- Teng, F.Y.; Jiang, Z.Z.; Guo, M.; Tan, X.Z.; Chen, F.; Guang, X. G-quadruplex DNA: A novel target for drug design. Cell. Mol. Life Sci. 2021, 78, 6557–6583. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, X.; Li, Y.; Xu, S.; Ma, C.; Wu, X.; Cheng, Y.; Yu, Z.; Zhao, G.; Chen, Y.; et al. Telomere targeting with a novel G-quadruplex-interactive ligand BRACO-19 induces T-loop disassembly and telomerase displacement in human glioblastoma cells. Oncotarget 2016, 7, 14925–14939. [Google Scholar] [CrossRef] [Green Version]
- Berardinelli, F.; Tanori, M.; Muoio, D.; Buccarelli, M.; di Masi, A.; Leone, S.; Ricci-Vitiani, L.; Pallini, R.; Mancuso, M.; Antoccia, A. G-quadruplex ligand RHPS4 radiosensitizes glioblastoma xenograft in vivo through a differential targeting of bulky differentiated- and stem-cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 311. [Google Scholar] [CrossRef]
- Che, T.; Chen, S.B.; Tu, J.L.; Wang, B.; Wang, Y.Q.; Zhang, Y.; Wang, J.; Wang, Z.Q.; Zhang, Z.P.; Ou, T.M.; et al. Discovery of Novel Schizocommunin Derivatives as Telomeric G-Quadruplex Ligands That Trigger Telomere Dysfunction and the Deoxyribonucleic Acid (DNA) Damage Response. J. Med. Chem. 2018, 61, 3436–3453. [Google Scholar] [CrossRef]
- Todd, A.K.; Neidle, S. The relationship of potential G-quadruplex sequences in cis -upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res. 2008, 36, 2700–2704. [Google Scholar] [CrossRef] [Green Version]
- Cogoi, S.; Shchekotikhin, A.E.; Xodo, L.E. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014, 42, 8379–8388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigo, R.; Palumbo, M.; Sissi, C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, O.; Bourdoncle, A.; Boulé, J.B.; Brosh, R.M., Jr.; Mergny, J. G-quadruplexes and helicases. Nucleic Acids Res. 2016, 44, 1989–2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Martin, V.; Soriano, M.; Garcia-Salcedo, J.A. Quadruplex Ligands in Cancer Therapy. Cancers 2021, 13, 3156. [Google Scholar] [CrossRef]
- Hu, M.H.; Wu, T.Y.; Huang, Q.; Jin, G. New substituted quinoxalines inhibit triple-negative breast cancer by specifically downregulating the c-MYC transcription. Nucleic Acids Res. 2019, 47, 10529–10542. [Google Scholar] [CrossRef]
- Local, A.; Zhang, H.; Benbatoul, K.D.; Folger, P.; Sheng, X.; Tsai, C.Y.; Howell, S.B.; Rice, W.G. APTO-253 stabilizes G-quadruplex DNA, inhibits MYC expression, and induces DNA damage in acute myeloid leukemia cells. Mol. Cancer Ther. 2018, 17, 1177–1186. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Local, A.; Benbatoul, K.; Folger, P.; Sheng, S.; Esquivies, L.; Lightfoot, J.; Vellanki, A.; Rice, W.G. Inhibition of c-Myc by Apto-253 As an Innovative Therapeutic Approach to Induce Cell Cycle Arrest and Apoptosis in Acute Myeloid Leukemia. Blood 2016, 128, 1716. [Google Scholar] [CrossRef]
- Robinson, J.; Raguseo, F.; Nuccio, S.P.; Liano, D.; Antonio, M. Di DNA G-quadruplex structures: More than simple roadblocks to transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [Google Scholar] [CrossRef]
- Marchetti, C.; Zyner, K.G.; Ohnmacht, S.A.; Robson, M.; Haider, S.M.; Morton, J.P.; Marsico, G.; Vo, T.; Laughlin-Toth, S.; Ahmed, A.A.; et al. Targeting Multiple Effector Pathways in Pancreatic Ductal Adenocarcinoma with a G-Quadruplex-Binding Small Molecule. J. Med. Chem. 2018, 61, 2500–2517. [Google Scholar] [CrossRef]
- Beauvarlet, J.; Bensadoun, P.; Darbo, E.; Labrunie, G.; Rousseau, B.; Richard, E.; Draskovic, I.; Londono-Vallejo, A.; Dupuy, J.-W.; Nath Das, R.; et al. Modulation of the ATM/autophagy pathway by a G-quadruplex ligand tips the balance between senescence and apoptosis in cancer cells. Nucleic Acids Res. 2019, 47, 2739–2756. [Google Scholar] [CrossRef] [PubMed]
- Brito, H.; Martins, A.C.; Lavrado, J.; Mendes, E.; Francisco, A.P.; Santos, S.A.; Ohnmacht, S.A.; Kim, N.S.; Rodrigues, C.M.P.; Moreira, R.; et al. Targeting KRAS oncogene in colon cancer cells with 7-carboxylate Indolo[3,2-b] quinoline tri-alkylamine derivatives. PLoS ONE 2015, 10, e0126891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.-J.; Cui, Y.; Yin, H.; Scheid, A.; Hendricks, W.P.D.; Schmidt, J.; Sekulic, A.; Kong, D.; Trent, J.M.; Gokhale, V.; et al. A Pharmacological Chaperone Molecule Induces Cancer Cell Death by Restoring Tertiary DNA Structures in Mutant hTERT Promoters. J. Am. Chem. Soc. 2016, 138, 13673–13692. [Google Scholar] [CrossRef] [Green Version]
- Kharel, P.; Balaratnam, S.; Beals, N.; Basu, S. The role of RNA G-quadruplexes in human diseases and therapeutic strategies. Wiley Interdiscip. Rev. RNA 2020, 11, e1568. [Google Scholar] [CrossRef]
- A Study of APTO-253 in Patients with Relapsed or Refractory AML or MDS. Available online: https://clinicaltrials.gov/ct2/show/NCT02267863 (accessed on 10 August 2021).
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [Green Version]
- Quarfloxin in Patients with Low to Intermediate Grade Neuroendocrine Carcinoma. Available online: https://clinicaltrials.gov/ct2/show/NCT00780663 (accessed on 29 July 2021).
- Haddach, M.; Schwaebe, M.K.; Michaux, J.; Nagasawa, J.; O’Brien, S.E.; Whitten, J.P.; Pierre, F.; Kerdoncuff, P.; Darjania, L.; Stansfield, R.; et al. Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. ACS Med. Chem. Lett. 2012, 3, 602. [Google Scholar] [CrossRef] [Green Version]
- Bruno, P.M.; Lu, M.; Dennis, K.A.; Inam, H.; Moore, C.J.; Sheehe, J.; Elledge, S.J.; Hemann, M.T.; Pritchard, J.R. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc. Natl. Acad. Sci. USA 2020, 117, 4053–4060. [Google Scholar] [CrossRef]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Dos Santos, N.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- The Canadian Cancer Trials Group. A Phase I Study of CX5461. 2020. Available online: https://clinicaltrials.gov/ct2/show/NCT02719977 (accessed on 20 January 2022).
- Brooks, T.A.; Hurley, L.H. Targeting MYC Expression through G-Quadruplexes. Genes Cancer 2010, 1, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Miglietta, G.; Russo, M.; Duardo, C.; Capranico, G. G-quadruplex binders as cytostatic modulators of innate immune genes in cancer cells. Nucleic Acids Res. 2021, 49, 6673–6686. [Google Scholar] [CrossRef] [PubMed]
- Zeraati, M.; Langley, D.B.; Schofield, P.; Moye, A.L.; Rouet, R.; Hughes, W.E.; Bryan, T.M.; Dinger, M.E.; Christ, D. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018, 10, 631–637. [Google Scholar] [CrossRef]
- Masoud, S.S.; Nagasawa, K. I-motif-binding ligands and their effects on the structure and biological functions of I-motif. Chem. Pharm. Bull. 2018, 66, 1091–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, C.E.; Van Ert, N.A.; Agrawal, P.; Chawla, R.; Yang, D.; Hurley, L.H. Insight into the Complexity of the i-Motif and G-Quadruplex DNA Structures Formed in the KRAS Promoter and Subsequent Drug-Induced Gene Repression. J. Am. Chem. Soc. 2017, 139, 8522–8536. [Google Scholar] [CrossRef]
- Shu, B.; Cao, J.; Kuang, G.; Qiu, J.; Zhang, M.; Zhang, Y.; Wang, M.; Li, X.; Kang, S.; Ou, T.M.; et al. Syntheses and evaluation of new acridone derivatives for selective binding of oncogene c-: Myc promoter i-motifs in gene transcriptional regulation. Chem. Commun. 2018, 54, 2036–2039. [Google Scholar] [CrossRef] [PubMed]
- Debnath, M.; Ghosh, S.; Chauhan, A.; Paul, R.; Bhattacharyya, K.; Dash, J. Preferential targeting of i-motifs and G-quadruplexes by small molecules. Chem. Sci. 2017, 8, 7448–7456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. World Malaria Report 2021. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021 (accessed on 20 January 2022).
- Balaji, S.N.; Deshmukh, R.; Trivedi, V. Severe malaria: Biology, clinical manifestation, pathogenesis and consequences. J. Vector Borne Dis. 2020, 57, 1–13. [Google Scholar] [PubMed]
- Smargiasso, N.; Gabelica, V.; Damblon, C.; Rosu, F.; De Pauw, E.; Teulade-Fichou, M.-P.; Rowe, J.A.; Claessens, A. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genom. 2009, 10, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo, E.P.; Wasserman, M. G-Quadruplex ligands: Potent inhibitors of telomerase activity and cell proliferation in Plasmodium falciparum. Mol. Biochem. Parasitol. 2016, 207, 33–38. [Google Scholar] [CrossRef]
- Belmonte-Reche, E.; Martínez-García, M.; Guédin, A.; Zuffo, M.; Arévalo-Ruiz, M.; Doria, F.; Campos-Salinas, J.; Maynadier, M.; López-Rubio, J.J.; Freccero, M.; et al. G-Quadruplex Identification in the Genome of Protozoan Parasites Points to Naphthalene Diimide Ligands as New Antiparasitic Agents. J. Med. Chem. 2018, 61, 1231–1240. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.K.; Shankar, U.; Jain, N.; Sikri, K.; Tyagi, J.S.; Sharma, T.K.; Mergny, J.L.; Kumar, A. Characterization of G-Quadruplex Motifs in espB, espK, and cyp51 Genes of Mycobacterium tuberculosis as Potential Drug Targets. Mol. Ther.-Nucleic Acids 2019, 16, 698–706. [Google Scholar] [CrossRef] [Green Version]
- Anas, M.; Sharma, R.; Dhamodharan, V.; Pradeepkumar, P.I.; Manhas, A.; Srivastava, K.; Ahmed, S.; Kumar, N. Investigating Pharmacological Targeting of G-Quadruplexes in the Human Malaria Parasite. Biochemistry 2017, 56, 6691–6699. [Google Scholar] [CrossRef] [PubMed]
- Perrone, R.; Lavezzo, E.; Riello, E.; Manganelli, R.; Palù, G.; Toppo, S.; Provvedi, R.; Richter, S.N. Mapping and characterization of G-quadruplexes in Mycobacterium tuberculosis gene promoter regions. Sci. Rep. 2017, 7, 5743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Street, S.T.G.; Peñalver, P.; O’Hagan, M.P.; Hollingworth, G.J.; Morales, J.C.; Galan, M.C. Imide Condensation as a Strategy for the Synthesis of Core-Diversified G-Quadruplex Ligands with Anticancer and Antiparasitic Activity. Chem. Eur. J. 2021, 27, 7712–7721. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Grellier, P.; Mouray, E.; Depoix, D.; Bertrand, H.; Monchaud, D.; Teulade-Fichou, M.-P.; Mergny, J.-L.; Alberti, P. Plasmodium Telomeric Sequences: Structure, Stability and Quadruplex Targeting by Small Compounds. ChemBioChem 2008, 9, 2730–2739. [Google Scholar] [CrossRef]
- Kerry, L.E.; Pegg, E.E.; Cameron, D.P.; Budzak, J.; Poortinga, G.; Hannan, K.M.; Hannan, R.D.; Rudenko, G. Selective inhibition of RNA polymerase I transcription as a potential approach to treat African trypanosomiasis. PLoS Negl. Trop. Dis. 2017, 11, e0005432. [Google Scholar] [CrossRef]
- Rawal, P.; Kummarasetti, V.B.R.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S.K.; Chowdhury, S. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006, 16, 644–655. [Google Scholar] [CrossRef] [Green Version]
- Holder, I.T.; Hartig, J.S. A matter of location: Influence of G-quadruplexes on escherichia coli gene expression. Chem. Biol. 2014, 21, 1511–1521. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, O.I.; Berber, B.; Hekim, N.; Doluca, O. G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch. Nucleic Acids Res. 2016, 44, 9083–9095. [Google Scholar] [CrossRef] [Green Version]
- Kota, S.; Dhamodharan, V.; Pradeepkumar, P.I.; Misra, H.S. G-quadruplex forming structural motifs in the genome of Deinococcus radiodurans and their regulatory roles in promoter functions. Appl. Microbiol. Biotechnol. 2015, 99, 9761–9769. [Google Scholar] [CrossRef]
- Rehm, C.; Wurmthaler, L.A.; Li, Y.; Frickey, T.; Hartig, J.S. Investigation of a Quadruplex-Forming Repeat Sequence Highly Enriched in Xanthomonas and Nostoc sp. PLoS ONE 2015, 10, e0144275. [Google Scholar] [CrossRef] [Green Version]
- Seifert, H.S. Above and Beyond Watson and Crick: Guanine Quadruplex Structures and Microbes. Annu. Rev. Microbiol. 2018, 8, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Belmonte-Reche, E.; Pirota, V.; De Jong, A.; Morales, J.C.; Freccero, M.; Doria, F.; Kuipers, O.P. G-Quadruplex DNA as a Target in Pathogenic Bacteria: Efficacy of an Extended Naphthalene Diimide Ligand and Its Mode of Action. J. Med. Chem. 2021. [Google Scholar] [CrossRef]
- Cahoon, L.A.; Seifert, H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 2009, 325, 764–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahoon, L.A.; Seifert, H.S. Transcription of a cis-acting, Noncoding, Small RNA Is Required for Pilin Antigenic Variation in Neisseria gonorrhoeae. PLoS Pathog. 2013, 9, e1003074. [Google Scholar] [CrossRef] [Green Version]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLOS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef] [Green Version]
- Brázda, V.; Porubiaková, O.; Cantara, A.; Bohálová, N.; Coufal, J.; Bartas, M.; Fojta, M.; Mergny, J.-L. G-quadruplexes in H1N1 influenza genomes. BMC Genom. 2021, 22, 77. [Google Scholar] [CrossRef]
- Bartas, M.; Brázda, V.; Bohálová, N.; Cantara, A.; Volná, A.; Stachurová, T.; Malachová, K.; Jagelská, E.B.; Porubiaková, O.; Červeň, J.; et al. In-Depth Bioinformatic Analyses of Nidovirales Including Human SARS-CoV-2, SARS-CoV, MERS-CoV Viruses Suggest Important Roles of Non-canonical Nucleic Acid Structures in Their Lifecycles. Front. Microbiol. 2020, 11, 1583. [Google Scholar] [CrossRef]
- Belmonte-Reche, E.; Serrano-Chacón, I.; Gonzalez, C.; Gallo, J.; Bañobre-López, M. Potential G-quadruplexes and i-Motifs in the SARS-CoV-2. PLoS ONE 2021, 16, e0250654. [Google Scholar] [CrossRef] [PubMed]
- Peck, K.M.; Lauring, A.S. Complexities of Viral Mutation Rates. J. Virol. 2018, 92, e01031-17. [Google Scholar] [CrossRef] [Green Version]
- Ruggiero, E.; Richter, S.N. Viral G-quadruplexes: New frontiers in virus pathogenesis and antiviral therapy. Annu. Rep. Med. Chem. 2020, 54, 101–131. [Google Scholar] [CrossRef]
- Murat, P.; Zhong, J.; Lekieffre, L.; Cowieson, N.P.; Clancy, J.L.; Preiss, T.; Balasubramanian, S.; Khanna, R.; Tellam, J. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014, 10, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Kim, Y.-E.; Bansal, V.; Ghosh, A.; Hur, J.; Subramani, V.K.; Pradhan, S.; Lee, M.K.; Kim, K.K.; Ahn, J.-H. Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLOS Pathog. 2018, 14, e1007334. [Google Scholar] [CrossRef] [PubMed]
- Lista, M.J.; Martins, R.P.; Billant, O.; Contesse, M.A.; Findakly, S.; Pochard, P.; Daskalogianni, C.; Beauvineau, C.; Guetta, C.; Jamin, C.; et al. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat. Commun. 2017, 8, 16043. [Google Scholar] [CrossRef] [PubMed]
- Reznichenko, O.; Quillévéré, A.; Martins, R.P.; Loaëc, N.; Kang, H.; Lista, M.J.; Beauvineau, C.; González-García, J.; Guillot, R.; Voisset, C.; et al. Novel cationic bis(acylhydrazones) as modulators of Epstein–Barr virus immune evasion acting through disruption of interaction between nucleolin and G-quadruplexes of EBNA1 mRNA. Eur. J. Med. Chem. 2019, 178, 13–29. [Google Scholar] [CrossRef]
- Ji, D.; Juhas, M.; Tsang, C.M.; Kwok, C.K.; Li, Y.; Zhang, Y. Discovery of G-quadruplex-forming sequences in SARS-CoV-2. Brief. Bioinform. 2021, 22, 1150–1160. [Google Scholar] [CrossRef]
- Bezzi, G.; Piga, E.J.; Binolfi, A.; Armas, P. CNBP Binds and Unfolds In Vitro G-Quadruplexes Formed in the SARS-CoV-2 Positive and Negative Genome Strands. Int. J. Mol. Sci. 2021, 22, 2614. [Google Scholar] [CrossRef] [PubMed]
- Panera, N.; Tozzi, A.E.; Alisi, A. The G-Quadruplex/Helicase World as a Potential Antiviral Approach Against COVID-19. Drugs 2020, 80, 941–946. [Google Scholar] [CrossRef]
- Zhang, R.; Xiao, K.; Gu, Y.; Liu, H.; Sun, X. Whole Genome Identification of Potential G-Quadruplexes and Analysis of the G-Quadruplex Binding Domain for SARS-CoV-2. Front. Genet. 2020, 11, 587829. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, L. G-Quadruplexes Are Present in Human Coronaviruses Including SARS-CoV-2. Front. Microbiol. 2020, 11, 2570. [Google Scholar] [CrossRef]
- Lavigne, M.; Helynck, O.; Rigolet, P.; Boudria-Souilah, R.; Nowakowski, M.; Baron, B.; Brülé, S.; Hoos, S.; Raynal, B.; Guittat, L.; et al. SARS-CoV-2 Nsp3 unique domain SUD interacts with guanine quadruplexes and G4-ligands inhibit this interaction. Nucleic Acids Res. 2021, 49, 7695–7712. [Google Scholar] [CrossRef]
- Zhao, C.; Qin, G.; Niu, J.; Wang, Z.; Wang, C.; Ren, J.; Qu, X. Targeting RNA G-Quadruplex in SARS-CoV-2: A Promising Therapeutic Target for COVID-19? Angew. Chemie Int. Ed. 2021, 60, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhou, J.; To, K.K.W.; Chu, H.; Li, C.; Wang, D.; Yang, D.; Zheng, S.; Hao, K.; Bossé, Y.; et al. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J. Infect. Dis. 2015, 212, 1214–1221. [Google Scholar] [CrossRef] [Green Version]
- Sakai, K.; Ami, Y.; Tahara, M.; Kubota, T.; Anraku, M.; Abe, M.; Nakajima, N.; Sekizuka, T.; Shirato, K.; Suzaki, Y.; et al. The Host Protease TMPRSS2 Plays a Major Role in In Vivo Replication of Emerging H7N9 and Seasonal Influenza Viruses. J. Virol. 2014, 88, 5608–5616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, L.W.; Qian, M.Q.; Yu, K.; Narva, S.; Yu, F.; Wu, Y.L.; Zhang, W. Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci. Rep. 2020, 10, 7635. [Google Scholar] [CrossRef] [PubMed]
- Unal, M.A.; Bitirim, C.V.; Summak, G.Y.; Bereketoglu, S.; Zeytin, I.C.; Besbinar, O.; Gurcan, C.; Aydos, D.; Goksoy, E.; Kocakaya, E.; et al. Ribavirin shows antiviral activity against SARS-CoV-2 and downregulates the activity of tmprss2 and the expression of ace2 in vitro. Can. J. Physiol. Pharmacol. 2021, 99, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Borowski, P.; Lang, M.; Niebuhr, A.; Haag, A.; Schmitz, H.; zur Wiesch, J.S.; Choe, J.; Siwecka, M.A.; Kulikowski, T. Inhibition of the helicase activity of HCV NTPase/helicase by 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide-5′-triphosphate (ribavirin-TP). Acta Biochim. Pol. 2001, 48, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Piekna-Przybylska, D.; Sharma, G.; Maggirwar, S.B.; Bambara, R.A. Deficiency in DNA damage response, a new characteristic of cells infected with latent HIV-1. Cell Cycle 2017, 16, 968–978. [Google Scholar] [CrossRef] [Green Version]
- Perrone, R.; Nadai, M.; Poe, J.A.; Frasson, I.; Palumbo, M.; Palù, G.; Smithgall, T.E.; Richter, S.N. Formation of a Unique Cluster of G-Quadruplex Structures in the HIV-1 nef Coding Region: Implications for Antiviral Activity. PLoS ONE 2013, 8, e73121. [Google Scholar] [CrossRef] [Green Version]
- Piekna-Przybylska, D.; Bambara, R.A.; Maggirwar, S.B.; Dewhurst, S. G-quadruplex ligands targeting telomeres do not inhibit HIV promoter activity and cooperate with latency reversing agents in killing latently infected cells. Cell Cycle 2020, 19, 2298–2313. [Google Scholar] [CrossRef]
- Madireddy, A.; Purushothaman, P.; Loosbroock, C.P.; Robertson, E.S.; Schildkraut, C.L.; Verma, S.C. G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 2016, 44, 3675–3694. [Google Scholar] [CrossRef] [Green Version]
- Dabral, P.; Babu, J.; Zareie, A.; Verma, S.C. LANA and hnRNP A1 Regulate the Translation of LANA mRNA through G-Quadruplexes. J. Virol. 2020, 94, e01508–e01519. [Google Scholar] [CrossRef]
- Wang, S.-R.; Min, Y.-Q.; Wang, J.-Q.; Liu, C.-X.; Fu, B.-S.; Wu, F.; Wu, L.-Y.; Qiao, Z.-X.; Song, Y.-Y.; Xu, G.-H.; et al. A highly conserved G-rich consensus sequence in hepatitis C virus core gene represents a new anti–hepatitis C target. Sci. Adv. 2016, 2, e1501535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.R.; Zhang, Q.Y.; Wang, J.Q.; Ge, X.Y.; Song, Y.Y.; Wang, Y.F.; Li, X.D.; Fu, B.S.; Xu, G.H.; Shu, B.; et al. Chemical Targeting of a G-Quadruplex RNA in the Ebola Virus L Gene. Cell Chem. Biol. 2016, 23, 1113–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Artusi, S.; Ruggiero, E.; Nadai, M.; Tosoni, B.; Perrone, R.; Ferino, A.; Zanin, I.; Xodo, L.; Flamand, L.; Richter, S.N. Antiviral Activity of the G-Quadruplex Ligand TMPyP4 against Herpes Simplex Virus-1. Viruses 2021, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Majee, P.; Pattnaik, A.; Sahoo, B.R.; Shankar, U.; Pattnaik, A.K.; Kumar, A.; Nayak, D. Inhibition of Zika virus replication by G-quadruplex-binding ligands. Mol. Ther.-Nucleic Acids 2021, 23, 691–701. [Google Scholar] [CrossRef]
- Carvalho, J.; Lopes-Nunes, J.; Campello, M.P.C.; Paulo, A.; Milici, J.; Meyers, C.; Mergny, J.L.; Salgado, G.F.; Queiroz, J.A.; Cruz, C. Human Papillomavirus G-Rich Regions as Potential Antiviral Drug Targets. Nucleic Acid Ther. 2021, 31, 68–81. [Google Scholar] [CrossRef]
- Perrone, R.; Butovskaya, E.; Daelemans, D.; Palù, G.; Pannecouque, C.; Richter, S.N. Anti-HIV-1 activity of the G-quadruplex ligand BRACO-19. J. Antimicrob. Chemother. 2014, 69, 3248–3258. [Google Scholar] [CrossRef] [Green Version]
- Butovskaya, E.; Soldà, P.; Scalabrin, M.; Nadai, M.; Richter, S.N. HIV-1 Nucleocapsid Protein Unfolds Stable RNA G-Quadruplexes in the Viral Genome and Is Inhibited by G-Quadruplex Ligands. ACS Infect. Dis. 2019, 52127–52135. [Google Scholar] [CrossRef] [Green Version]
- Artusi, S.; Nadai, M.; Perrone, R.; Biasolo, M.A.; Palù, G.; Flamand, L.; Calistri, A.; Richter, S.N. The Herpes Simplex Virus-1 genome contains multiple clusters of repeated G-quadruplex: Implications for the antiviral activity of a G-quadruplex ligand. Antiviral Res. 2015, 118, 123. [Google Scholar] [CrossRef] [Green Version]
- Gilbert-Girard, S.; Gravel, A.; Artusi, S.; Richter, S.N.; Wallaschek, N.; Kaufer, B.B.; Flamand, L. Stabilization of Telomere G-Quadruplexes Interferes with Human Herpesvirus 6A Chromosomal Integration. J. Virol. 2017, 91, e00402–e00417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norseen, J.; Johnson, F.B.; Lieberman, P.M. Role for G-Quadruplex RNA Binding by Epstein-Barr Virus Nuclear Antigen 1 in DNA Replication and Metaphase Chromosome Attachment. J. Virol. 2009, 83, 10336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, B.; Kandpal, M.; Vivekanandan, P. A G-quadruplex motif in an envelope gene promoter regulates transcription and virion secretion in HBV genotype B. Nucleic Acids Res. 2017, 45, 11268–11280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrone, R.; Doria, F.; Butovskaya, E.; Frasson, I.; Botti, S.; Scalabrin, M.; Lago, S.; Grande, V.; Nadai, M.; Freccero, M.; et al. Synthesis, Binding and Antiviral Properties of Potent Core-Extended Naphthalene Diimides Targeting the HIV-1 Long Terminal Repeat Promoter G-Quadruplexes. J. Med. Chem. 2015, 58, 9639–9652. [Google Scholar] [CrossRef]
- Callegaro, S.; Perrone, R.; Scalabrin, M.; Doria, F.; Palù, G.; Richter, S.N. A core extended naphtalene diimide G-quadruplex ligand potently inhibits herpes simplex virus 1 replication. Sci. Rep. 2017, 7, 2341. [Google Scholar] [CrossRef]
- Zou, M.; Li, J.Y.; Zhang, M.J.; Li, J.H.; Huang, J.T.; You, P.D.; Liu, S.W.; Zhou, C.Q. G-quadruplex binder pyridostatin as an effective multi-target ZIKV inhibitor. Int. J. Biol. Macromol. 2021, 190, 178–188. [Google Scholar] [CrossRef]
- Westdorp, K.N.; Terhune, S.S. Impact of RNA polymerase I inhibitor CX-5461 on viral kinase-dependent and -independent cytomegalovirus replication. Antivir. Res. 2018, 153, 33–38. [Google Scholar] [CrossRef]
- Jaubert, C.; Bedrat, A.; Bartolucci, L.; Di Primo, C.; Ventura, M.; Mergny, J.L.; Amrane, S.; Andreola, M.L. RNA synthesis is modulated by G-quadruplex formation in Hepatitis C virus negative RNA strand. Sci. Rep. 2018, 8, 8120. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef]
- Ohnmacht, S.A.; Neidle, S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 2602–2612. [Google Scholar] [CrossRef]
- Savva, L.; Georgiades, S.N. Recent developments in small-molecule ligands of medicinal relevance for harnessing the anticancer potential of g-quadruplexes. Molecules 2021, 26, 841. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.R.; Cadoni, E.; Ressurreição, A.S.; Moreira, R.; Paulo, A. Design of Modular G-quadruplex Ligands. ChemMedChem 2018, 13, 869–893. [Google Scholar] [CrossRef]
- Micco, M.; Collie, G.W.; Dale, A.G.; Ohnmacht, S.A.; Pazitna, I.; Gunaratnam, M.; Reszka, A.P.; Neidle, S. Structure-based design and evaluation of naphthalene diimide G-quadruplex ligands as telomere targeting agents in pancreatic cancer cells. J. Med. Chem. 2013, 56, 2959–2974. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Angell, R.; Oxenford, S.; Worthington, J.; Williams, N.; Barton, N.; Fowler, T.G.; O’Flynn, D.E.; Sunose, M.; McConville, M.; et al. Asymmetrically Substituted Quadruplex-Binding Naphthalene Diimide Showing Potent Activity in Pancreatic Cancer Models. ACS Med. Chem. Lett. 2020, 11, 1634–1644. [Google Scholar] [CrossRef]
- Calabrese, D.R.; Chen, X.; Leon, E.C.; Gaikwad, S.M.; Phyo, Z.; Hewitt, W.M.; Alden, S.; Hilimire, T.A.; He, F.; Michalowski, A.M.; et al. Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat. Commun. 2018, 9, 4229. [Google Scholar] [CrossRef] [Green Version]
- Dickerhoff, J.; Dai, J.; Yang, D. Structural recognition of the MYC promoter G-quadruplex by a quinoline derivative: Insights into molecular targeting of parallel G-quadruplexes. Nucleic Acids Res. 2021, 49, 5905–5915. [Google Scholar] [CrossRef] [PubMed]
- Cadoni, E.; Magalh, P.R.; Em, R.M.; Mendes, E.; Jorge, V.; Carvalho, J.; Cruz, C.; Victor, B.L.; Paulo, A. New (Iso) quinolinyl-pyridine-2,6-dicarboxamide G-Quadruplex Stabilizers. A Structure-Activity Relationship Study. Pharmaceuticals 2021, 14, 669. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S. Challenges in Developing Small-Molecule Quadruplex Therapeutics, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; Volume 54, ISBN 9780128210178. [Google Scholar]
- Neidle, S. Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids. Pharmaceuticals 2022, 15, 7. [Google Scholar] [CrossRef]
- Monsen, R.C.; Trent, J.O. G-quadruplex virtual drug screening: A review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef]
- Ma, D.L.; Lai, T.S.; Chan, F.Y.; Chung, W.H.; Abagyan, R.; Leung, Y.C.; Wong, K.Y. Discovery of a drug-like G-quadruplex binding ligand by high-throughput docking. ChemMedChem 2008, 3, 881–884. [Google Scholar] [CrossRef]
- Ohnmacht, S.A.; Varavipour, E.; Nanjunda, R.; Pazitna, I.; Di Vita, G.; Gunaratnam, M.; Kumar, A.; Ismail, M.A.; Boykin, D.W.; Wilson, W.D.; et al. Discovery of new G-quadruplex binding chemotypes. Chem. Commun. 2014, 50, 960–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, K.M.; Tizkova, K.; Reszka, A.P.; Neidle, S.; Thurston, D.E. Identification of novel telomeric G-quadruplex-targeting chemical scaffolds through screening of three NCI libraries. Bioorg. Med. Chem. Lett. 2012, 22, 3006–3010. [Google Scholar] [CrossRef]
- Wu, G.; Tillo, D.; Ray, S.; Chang, T.C.; Schneekloth, J.S.; Vinson, C.; Yang, D. Custom G4 microarrays reveal selective G-quadruplex recognition of small molecule BMVC: A large-scale assessment of ligand binding selectivity. Molecules 2020, 25, 3465. [Google Scholar] [CrossRef]
- Ray, S.; Tillo, D.; Boer, R.E.; Assad, N.; Barshai, M.; Wu, G.; Orenstein, Y.; Yang, D.; Schneekloth, J.S.; Vinson, C. Custom DNA Microarrays Reveal Diverse Binding Preferences of Proteins and Small Molecules to Thousands of G-Quadruplexes. ACS Chem. Biol. 2020, 15, 925–935. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Gensicka-Kowalewska, M.; Cholewiński, G.; Dzierzbicka, K. Recent developments in the synthesis and biological activity of acridine/acridone analogues. RSC Adv. 2017, 7, 15776–15804. [Google Scholar] [CrossRef] [Green Version]
- Goodell, J.R.; Ougolkov, A.V.; Hiasa, H.; Kaur, H.; Remmel, R.; Billadeau, D.D.; Ferguson, D.M. Acridine-based agents with topoisomerase II activity inhibit pancreatic cancer cell proliferation and induce apoptosis. J. Med. Chem. 2008, 51, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Liu, W.J.; Guo, K.; Rusche, J.J.; Ebbinghaus, S.; Gokhale, V.; Hurley, L.H. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex–interactive agents. Mol. Cancer Ther. 2008, 7, 880–889. [Google Scholar] [CrossRef] [Green Version]
- Collie, G.W.; Sparapani, S.; Parkinson, G.N.; Neidle, S. Structural basis of telomeric RNA quadruplex-acridine ligand recognition. J. Am. Chem. Soc. 2011, 133, 2721–2728. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Hurley, L.H.; Yang, D. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: Insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011, 133, 17673–17680. [Google Scholar] [CrossRef] [Green Version]
- Neag, M.A.; Mocan, A.; Echeverría, J.; Pop, R.M.; Bocsan, C.I.; Crisan, G.; Buzoianu, A.D. Berberine: Botanical Occurrence, traditional uses, extraction methods, and relevance in cardiovascular, metabolic, hepatic, and renal disorders. Front. Pharmacol. 2018, 9, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Z.; Li, K.; Maskey, A.R.; Huang, W.; Toutov, A.A.; Yang, N.; Srivastava, K.; Geliebter, J.; Tiwari, R.; Miao, M.; et al. A small molecule compound berberine as an orally active therapeutic candidate against COVID-19 and SARS: A computational and mechanistic study. FASEB J. 2021, 35, e21360. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, L.M.G.; Lombardi, P.; Tillhon, M.; Scovassi, A.I. Berberine, an Epiphany Against Cancer. Molecules 2014, 19, 12349–12367. [Google Scholar] [CrossRef]
- Bhadra, K.; Suresh Kumar, G. Isoquinoline Alkaloids and their Binding with DNA: Calorimetry and Thermal Analysis Applications. Mini-Rev. Med. Chem. 2012, 10, 1235–1247. [Google Scholar] [CrossRef]
- Kuo, C.L.; Chi, C.W.; Liu, T.Y. The anti-inflammatory potential of berberine in vitro and in vivo. Cancer Lett. 2004, 203, 127–137. [Google Scholar] [CrossRef]
- Maiti, M.; Kumar, G.S. Molecular aspects on the interaction of protoberberine, benzophenanthridine, and aristolochia group of alkaloids with nucleic acid structures and biological perspectives. Med. Res. Rev. 2007, 27, 649–695. [Google Scholar] [CrossRef] [PubMed]
- Maiti, M.; Kumar, G.S. Polymorphic nucleic acid binding of bioactive isoquinoline alkaloids and their role in cancer. J. Nucleic Acids 2010, 593408. [Google Scholar] [CrossRef] [Green Version]
- Satou, T.; Akao, N.; Matsuhashi, R.; Koike, K.; Fujita, K.; Nikaido, T. Inhibitory effect of isoquinoline alkaloids on movement of second-stage larvae of Toxocara canis. Biol. Pharm. Bull. 2002, 25, 1651–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazzicalupi, C.; Ferraroni, M.; Bilia, A.R.; Scheggi, F.; Gratteri, P. The crystal structure of human telomeric DNA complexed with berberine: An interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013, 41, 632–638. [Google Scholar] [CrossRef]
- Papi, F.; Bazzicalupi, C.; Ferraroni, M.; Ciolli, G.; Lombardi, P.; Khan, A.Y.; Kumar, G.S.; Gratteri, P. Pyridine Derivative of the Natural Alkaloid Berberine as Human Telomeric G4-DNA Binder: A Solution and Solid-State Study. ACS Med. Chem. Lett. 2020, 11, 645–650. [Google Scholar] [CrossRef]
- Kurzątkowska, K.; Pazos, M.A.; Herschkowitz, J.I.; Hepel, M. Cancer-targeted controlled delivery of chemotherapeutic anthracycline derivatives using apoferritin nanocage carriers. Int. J. Mol. Sci. 2021, 22, 1362. [Google Scholar] [CrossRef] [PubMed]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular advances and pharmacologie developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haider, S.M.; Parkinson, G.N.; Neidle, S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003, 326, 117–125. [Google Scholar] [CrossRef]
- Clark, G.R.; Pytel, P.D.; Squire, C.J.; Neidle, S. Structure of the First Parallel DNA Quadruplex-Drug Complex. J. Am. Chem. Soc. 2003, 125, 4066–4067. [Google Scholar] [CrossRef]
- Manet, I.; Manoli, F.; Zambelli, B.; Andreano, G.; Masi, A.; Cellai, L.; Monti, S. Affinity of the anthracycline antitumor drugs Doxorubicin and Sabarubicin for human telomeric G-quadruplex structures. Phys. Chem. Chem. Phys. 2011, 13, 540–551. [Google Scholar] [CrossRef]
- Barthwal, R.; Raje, S.; Pandav, K. Structural basis for stabilization of human telomeric G-quadruplex [d-(TTAGGGT)]4 by anticancer drug adriamycin. J. Biomol. Struct. Dyn. 2021, 39, 795–815. [Google Scholar] [CrossRef]
- Barthwal, R.; Raje, S.; Pandav, K. Structural basis for stabilization of human telomeric G-quadruplex [d-(TTAGGGT)]4 by anticancer drug epirubicin. Bioorg. Med. Chem. 2020, 28, 115761. [Google Scholar] [CrossRef]
- Clark, G.R.; Pytel, P.D.; Squire, C.J. The high-resolution crystal structure of a parallel intermolecular DNA G-4 quadruplex/drug complex employing syn glycosyl linkages. Nucleic Acids Res. 2012, 40, 5731–5738. [Google Scholar] [CrossRef] [Green Version]
- Bhosale, S.V.; Jani, C.H.; Langford, S.J. Chemistry of naphthalene diimides. Chem. Soc. Rev. 2008, 37, 331–342. [Google Scholar] [CrossRef]
- Würthner, F. Perylene bisimide dyes as versatile building blocks for functional supramolecular architectures. Chem. Commun. 2004, 4, 1564–1579. [Google Scholar] [CrossRef]
- Cuenca, F.; Greciano, O.; Gunaratnam, M.; Haider, S.; Munnur, D.; Nanjunda, R.; Wilson, W.D.; Neidle, S. Tri- and tetra-substituted naphthalene diimides as potent G-quadruplex ligands. Bioorg. Med. Chem. Lett. 2008, 18, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, S.A.; Marchetti, C.; Gunaratnam, M.; Besser, R.J.; Haider, S.M.; Di Vita, G.; Lowe, H.L.; Mellinas-Gomez, M.; Diocou, S.; Robson, M.; et al. A G-quadruplex-binding compound showing anti-tumour activity in an in vivo model for pancreatic cancer. Sci. Rep. 2015, 5, 11385. [Google Scholar] [CrossRef] [Green Version]
- Platella, C.; Napolitano, E.; Riccardi, C.; Musumeci, D.; Montesarchio, D. Disentangling the Structure-Activity Relationships of Naphthalene Diimides as Anticancer G-Quadruplex-Targeting Drugs. J. Med. Chem. 2021, 64, 3578–3603. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Promontorio, R.; Hampel, S.M.; Micco, M.; Neidle, S.; Parkinson, G.N. Structural basis for telomeric G-quadruplex targeting by naphthalene diimide ligands. J. Am. Chem. Soc. 2012, 134, 2723–2731. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Cuenca, F.; Neidle, S. Topology Conservation and Loop Flexibility in Quadruplex-Drug Recognition: Crystal Structures of Inter- and Intramolecular Telomeric DNA Quadruplex-Drug Complexes. J. Mol. Biol. 2008, 381, 1145–1156. [Google Scholar] [CrossRef]
- Gil, A.; Sanchez-Gonzalez, A.; Branchadell, V. Unraveling the Modulation of the Activity in Drugs Based on Methylated Phenanthroline When Intercalating between DNA Base Pairs. J. Chem. Inf. Model. 2019, 59, 3989–3995. [Google Scholar] [CrossRef]
- Teulade-Fichou, M.P.; Carrasco, C.; Guittat, L.; Bailly, C.; Alberti, P.; Mergny, J.L.; David, A.; Lehn, J.M.; Wilson, W.D. Selective recognition of G-quadruplex telomeric DNA by a bis(quinacridine) macrocycle. J. Am. Chem. Soc. 2003, 125, 4732–4740. [Google Scholar] [CrossRef]
- Chung, W.J.; Heddi, B.; Hamon, F.; Teulade-Fichou, M.P.; Phan, A.T. Solution structure of a G-quadruplex bound to the bisquinolinium compound phen-DC(3). Angew. Chem. Int. Ed. 2014, 53, 999–1002. [Google Scholar] [CrossRef]
- Hounsou, C.; Guittat, L.; Monchaud, D.; Jourdan, M.; Saettel, N.; Mergny, J.L.; Teulade-Fichou, M.P. G-quadruplex recognition by quinacridines: A SAR, NMR, and biological study. ChemMedChem 2007, 2, 655–666. [Google Scholar] [CrossRef]
- Teixeira, S.C.M.; Thorpe, J.H.; Todd, A.K.; Powell, H.R.; Adams, A.; Wakelin, L.P.G.; Denny, W.A.; Cardin, C.J. Structural Characterisation of Bisintercalation in Higher-order DNA at a Junction-like Quadruplex. J. Mol. Biol. 2002, 323, 167–171. [Google Scholar] [CrossRef]
- Gavathiotis, E.; Heald, R.A.; Stevens, M.F.G.; Searle, M.S. Drug recognition and stabilisation of the parallel-stranded DNA quadruplex d(TTAGGGT)4 containing the human telomeric repeat. J. Mol. Biol. 2003, 334, 25–36. [Google Scholar] [CrossRef]
- Campbell, N.H.; Parkinson, G.N.; Reszka, A.P.; Neidle, S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008, 130, 6722–6724. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Patel, M.; Tofa, A.B.; Ghosh, R.; Parkinson, G.N.; Neidle, S. Selectivity in ligand recognition of G-quadruplex loops. Biochemistry 2009, 48, 1675–1680. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.H.; Smith, D.L.; Reszka, A.P.; Neidle, S.; O’Hagan, D. Fluorine in medicinal chemistry: β-fluorination of peripheral pyrrolidines attached to acridine ligands affects their interactions with G-quadruplex DNA. Org. Biomol. Chem. 2011, 9, 1328–1331. [Google Scholar] [CrossRef] [PubMed]
- Kotar, A.; Wang, B.; Shivalingam, A.; Gonzalez-Garcia, J.; Vilar, R.; Plavec, J. NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex. Angew. Chem. Int. Ed. 2016, 55, 12508–12511. [Google Scholar] [CrossRef] [PubMed]
- Ferraroni, M.; Bazzicalupi, C.; Gratteri, P. RCSB PDB-4P1D: Structure of the Complex of a Bimolecular Human Telomeric DNA with Coptisine. Available online: https://www.rcsb.org/structure/4p1d (accessed on 6 January 2022).
- Ferraroni, M.; Bazzicalupi, C.; Papi, F.; Fiorillo, G.; Guamán-Ortiz, L.M.; Nocentini, A.; Scovassi, A.I.; Lombardi, P.; Gratteri, P. Solution and Solid-State Analysis of Binding of 13-Substituted Berberine Analogues to Human Telomeric G-quadruplexes. Chem. Asian J. 2016, 11, 1107–1115. [Google Scholar] [CrossRef]
- Lin, C.; Wu, G.; Wang, K.; Onel, B.; Sakai, S.; Shao, Y.; Yang, D. Molecular Recognition of the Hybrid-2 Human Telomeric G-Quadruplex by Epiberberine: Insights into Conversion of Telomeric G-Quadruplex Structures. Angew. Chem. Int. Ed. 2018, 57, 10888–10893. [Google Scholar] [CrossRef]
- Wang, F.; Wang, C.; Liu, Y.; Lan, W.; Han, H.; Wang, R.; Huang, S.; Cao, C. Colchicine selective interaction with oncogene: RET G-quadruplex revealed by NMR. Chem. Commun. 2020, 56, 2099–2102. [Google Scholar] [CrossRef]
- Díaz-Casado, L.; Serrano-Chacón, I.; Montalvillo-Jiménez, L.; Corzana, F.; Bastida, A.; Santana, A.G.; González, C.; Asensio, J.L. De Novo Design of Selective Quadruplex–Duplex Junction Ligands and Structural Characterisation of Their Binding Mode: Targeting the G4 Hot-Spot. Chem. Eur. J. 2021, 27, 6204–6212. [Google Scholar] [CrossRef]
- Liu, W.; Lin, C.; Wu, G.; Dai, J.; Chang, T.C.; Yang, D. Structures of 1:1 and 2:1 complexes of BMVC and MYC promoter G-quadruplex reveal a mechanism of ligand conformation adjustment for G4-recognition. Nucleic Acids Res. 2019, 47, 11931–11942. [Google Scholar] [CrossRef]
- Nielsen, M.C.; Borch, J.; Ulven, T. Design, synthesis and evaluation of 4,7-diamino-1,10-phenanthroline G-quadruplex ligands. Bioorg. Med. Chem. 2009, 17, 8241–8246. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Wu, J.Y.; Chien, C.W.; Wu, W.S.; Liu, H.; Kang, C.C.; Yu, L.J.; Chang, T.C. A Fluorescent Carbazole Derivative: High Sensitivity for Quadruplex DNA. Anal. Chem. 2003, 75, 6177–6183. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Kuo, I.C.; Ling, I.F.; Chen, C.T.; Chen, H.C.; Lou, P.J.; Lin, J.J.; Chang, T.C. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004, 76, 4490–4494. [Google Scholar] [CrossRef]
- Chang, C.C.; Chu, J.F.; Kao, F.J.; Chiu, Y.C.; Lou, P.J.; Chen, H.C.; Chang, T.C. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006, 78, 2810–2815. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Kuo, I.C.; Lin, J.J.; Lu, Y.C.; Chenc, C.T.; Back, H.T.; Lou, P.J.; Chang, T.C. A novel carbazole derivative, BMVC: A potential antitumor agent and fluorescence marker of cancer cells. Chem. Biodivers. 2004, 1, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.C.; Chang, C.C.; Chang, T.C.; Liao, L.J.; Lou, P.J.; Xie, W.; Yeung, E.S. A handheld device for potential point-of-care screening of cancer. Analyst 2007, 132, 745–749. [Google Scholar] [CrossRef]
- Yang, T.L.; Lin, L.; Lou, P.J.; Chang, T.C.; Young, T.H. Detection of cell carcinogenic transformation by a quadruplex DNA binding fluorescent probe. PLoS ONE 2014, 9, e0086143. [Google Scholar] [CrossRef]
- Liu, L.Y.; Liu, W.; Wang, K.N.; Zhu, B.C.; Xia, X.Y.; Ji, L.N.; Mao, Z.W. Quantitative Detection of G-Quadruplex DNA in Live Cells Based on Photon Counts and Complex Structure Discrimination. Angew. Chem. Int. Ed. 2020, 59, 9719–9726. [Google Scholar] [CrossRef]
- Pagano, B.; Virno, A.; Mattia, C.A.; Mayol, L.; Randazzo, A.; Giancola, C. Targeting DNA quadruplexes with distamycin A and its derivatives: An ITC and NMR study. Biochimie 2008, 90, 1224–1232. [Google Scholar] [CrossRef]
- Martino, L.; Virno, A.; Pagano, B.; Virgilio, A.; di Micco, S.; Galeone, A.; Giancola, C.; Bifulco, G.; Mayol, L.; Randazzo, A. Structural and Thermodynamic Studies of the Interaction of Distamycin A with the Parallel Quadruplex Structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007, 129, 16048–16056. [Google Scholar] [CrossRef]
- Cosconati, S.; Marinelli, L.; Trotta, R.; Virno, A.; De Tito, S.; Romagnoli, R.; Pagano, B.; Limongelli, V.; Giancola, C.; Baraldi, P.G.; et al. Structural and Conformational Requisites in DNA Quadruplex Groove Binding: Another Piece to the Puzzle. J. Am. Chem. Soc. 2010, 132, 6425–6433. [Google Scholar] [CrossRef] [PubMed]
- Kotar, A.; Kocman, V.; Plavec, J. Intercalation of a Heterocyclic Ligand between Quartets in a G-Rich Tetrahelical Structure. Chem. Eur. J. 2020, 26, 814–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granotier, C.; Pennarun, G.; Riou, L.; Hoffschir, F.; Gauthier, L.R.; De Cian, A.; Gomez, D.; Mandine, E.; Riou, J.F.; Mergny, J.L.; et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005, 33, 4182–4190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saintomé, C.; Alberti, P.; Guinot, N.; Lejault, P.; Chatain, J.; Mailliet, P.; Riou, J.F.; Bugaut, A. Binding properties of mono- and dimeric pyridine dicarboxamide ligands to human telomeric higher-order G-quadruplex structures. Chem. Commun. 2018, 54, 1897–1900. [Google Scholar] [CrossRef]
- Marchand, A.; Granzhan, A.; Iida, K.; Tsushima, Y.; Ma, Y.; Nagasawa, K.; Teulade-Fichou, M.P.; Gabelica, V. Ligand-induced conformational changes with cation ejection upon binding to human telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2015, 137, 750–756. [Google Scholar] [CrossRef] [Green Version]
- Bončina, M.; Hamon, F.; Islam, B.; Teulade-Fichou, M.P.; Vesnaver, G.; Haider, S.; Lah, J. Dominant Driving Forces in Human Telomere Quadruplex Binding-Induced Structural Alterations. Biophys. J. 2015, 108, 2903–2911. [Google Scholar] [CrossRef] [Green Version]
- Shin-ya, K.; Wierzba, K.; Matsuo, K.; Ohtani, T.; Yamada, Y.; Furihata, K.; Hayakawa, Y.; Seto, H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001, 123, 1262–1263. [Google Scholar] [CrossRef]
- De Cian, A.; Guittat, L.; Kaiser, M.; Saccà, B.; Amrane, S.; Bourdoncle, A.; Alberti, P.; Teulade-Fichou, M.P.; Lacroix, L.; Mergny, J.L. Fluorescence-based melting assays for studying quadruplex ligands. Methods 2007, 42, 183–195. [Google Scholar] [CrossRef]
- Sakuma, M.; Ma, Y.; Tsushima, Y.; Iida, K.; Hirokawa, T.; Nagasawa, K. Design and synthesis of unsymmetric macrocyclic hexaoxazole compounds with an ability to induce distinct G-quadruplex topologies in telomeric DNA. Org. Biomol. Chem. 2016, 14, 5109–5116. [Google Scholar] [CrossRef]
- Ma, Y.; Iida, K.; Sasaki, S.; Hirokawa, T.; Heddi, B.; Phan, A.T.; Nagasawa, K. Synthesis and telomeric G-quadruplex-stabilizing ability of macrocyclic hexaoxazoles bearing three side chains. Molecules 2019, 24, 263. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.J.; Heddi, B.; Tera, M.; Iida, K.; Nagasawa, K.; Phan, A.T. Solution structure of an intramolecular (3 + 1) human telomeric G-quadruplex bound to a telomestatin derivative. J. Am. Chem. Soc. 2013, 135, 13495–13501. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.H.; Nie, X.; Liu, H.Y.; Fang, Y.M.; Zhao, Y.; Xia, L.X. TMPyP4 promotes cancer cell migration at low doses, but induces cell death at high doses. Sci. Rep. 2016, 6, 26592. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Kuryavyi, V.; Gaw, H.Y.; Patel, D.J. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005, 1, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Nicoludis, J.M.; Miller, S.T.; Jeffrey, P.D.; Barrett, S.P.; Rablen, P.R.; Lawton, T.J.; Yatsunyk, L.A. Optimized end-stacking provides specificity of N-methyl mesoporphyrin IX for human telomeric G-quadruplex DNA. J. Am. Chem. Soc. 2012, 134, 20446–20456. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.Y.; McCarthy, S.; Powell, B.M.; Manurung, Y.; Xiang, I.M.; Dean, W.L.; Chaires, B.; Yatsunyk, L.A. Biophysical and X-ray structural studies of the (GGGTT)3GGG G-quadruplex in complex with N-methyl mesoporphyrin IX. PLoS ONE 2020, 15, e0241513. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Ghosh, R.; Neidle, S. Structural basis for binding of porphyrin to human telomeres. Biochemistry 2007, 46, 2390–2397. [Google Scholar] [CrossRef]
- Zhang, Y.; El Omari, K.; Duman, R.; Liu, S.; Haider, S.; Wagner, A.; Parkinson, G.N.; Wei, D. Native de novo structural determinations of non-canonical nucleic acid motifs by X-ray crystallography at long wavelengths. Nucleic Acids Res. 2020, 48, 9886–9898. [Google Scholar] [CrossRef]
- O’Hagan, M.P.; Morales, J.C.; Galan, M.C. Binding and Beyond: What Else Can G-Quadruplex Ligands Do? Eur. J. Org. Chem. 2019, 2019, 4995–5017. [Google Scholar] [CrossRef]
- Zhao, J.; Zhai, Q. Recent advances in the development of ligands specifically targeting telomeric multimeric G-quadruplexes. Bioorg. Chem. 2020, 103, 104229. [Google Scholar] [CrossRef]
- Santos, T.; Salgado, G.F.; Cabrita, E.J.; Cruz, C. G-Quadruplexes and Their Ligands: Biophysical Methods to Unravel G-Quadruplex/Ligand Interactions. Pharmaceuticals 2021, 14, 769. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Wang, X.N.; Cheng, S.Q.; Su, X.X.; Ou, T.M. Developing novel G-quadruplex ligands: From interaction with nucleic acids to interfering with nucleic acid–protein interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naasani, I.; Seimiya, H.; Yamori, T.; Tsuruo, T. FJ5002: A potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res. 1999, 59, 4004–4011. [Google Scholar] [PubMed]
- Franceschin, M.; Rossetti, L.; D’Ambrosio, A.; Schirripa, S.; Bianco, A.; Ortaggi, G.; Savino, M.; Schultes, C.; Neidle, S. Natural and synthetic G-quadruplex interactive berberine derivatives. Bioorg. Med. Chem. Lett. 2006, 16, 1707–1711. [Google Scholar] [CrossRef]
- Zhang, W.J.; Ou, T.M.; Lu, Y.J.; Huang, Y.Y.; Wu, W.B.; Huang, Z.S.; Zhou, J.L.; Wong, K.Y.; Gu, L.Q. 9-Substituted berberine derivatives as G-quadruplex stabilizing ligands in telomeric DNA. Bioorg. Med. Chem. 2007, 15, 5493–5501. [Google Scholar] [CrossRef]
- Ma, Y.; Ou, T.M.; Hou, J.Q.; Lu, Y.J.; Tan, J.H.; Gu, L.Q.; Huang, Z.S. 9-N-Substituted berberine derivatives: Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc. Bioorg. Med. Chem. 2008, 16, 7582–7591. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.X.; Su, H.F.; Lv, P.; Ma, Y.; Wang, S.K.; Miao, H.; Liu, H.Y.; Tan, J.H.; Ou, T.M.; Gu, L.Q.; et al. A newly identified berberine derivative induces cancer cell senescence by stabilizing endogenous G-quadruplexes and sparking a DNA damage response at the telomere region. Oncotarget 2015, 6, 35625–35635. [Google Scholar] [CrossRef] [Green Version]
- Becher, J.; Berdnikova, D.V.; Ihmels, H.; Stremmel, C. Synthesis and investigation of quadruplex-DNA-binding, 9-O-substituted berberine derivatives. Beilstein J. Org. Chem. 2020, 16, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- Moraca, F.; Amato, J.; Ortuso, F.; Artese, A.; Pagano, B.; Novellino, E.; Alcaro, S.; Parrinello, M.; Limongelli, V. Ligand binding to telomeric G-quadruplex DNA investigated by funnel-metadynamics simulations. Proc. Natl. Acad. Sci. USA 2017, 114, E2136–E2145. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Shao, Y.; Ma, K.; Cui, Q.; Liu, G.; Xu, S. Simultaneous fluorescence light-up and selective multicolor nucleobase recognition based on sequence-dependent strong binding of berberine to DNA abasic site. Org. Biomol. Chem. 2012, 10, 3300–3307. [Google Scholar] [CrossRef]
- Xu, L.; Hong, S.; Sun, N.; Wang, K.; Zhou, L.; Ji, L.; Pei, R. Berberine as a novel light-up i-motif fluorescence ligand and its application in designing molecular logic systems. Chem. Commun. 2016, 52, 179–182. [Google Scholar] [CrossRef]
- Wickhorst, P.J.; Ihmels, H.; Memoriam Professor, I.; Hünig, S. Selective, pH-Dependent Colorimetric and Fluorimetric Detection of Quadruplex DNA with 4-Dimethylamino(phenyl)-Substituted Berberine Derivatives. Chem. Eur. J. 2021, 27, 8580–8589. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kendrick, S.; Hecht, S.M.; Hurley, L.H. The Dynamic Character of the BCL2 Promoter i-Motif Provides a Mechanism for Modulation of Gene Expression by Compounds That Bind Selectively to the Alternative DNA Hairpin Structure. J. Am. Chem. Soc. 2014, 136, 4161–4171. [Google Scholar] [CrossRef]
- Gargallo, R.; Aviñó, A.; Eritja, R.; Jarosova, P.; Mazzini, S.; Scaglioni, L.; Taborsky, P. Study of alkaloid berberine and its interaction with the human telomeric i-motif DNA structure. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2021, 248, 119185. [Google Scholar] [CrossRef] [PubMed]
- Pagano, A.; Iaccarino, N.; Abdelhamid, M.A.S.; Brancaccio, D.; Garzarella, E.U.; Di Porzio, A.; Novellino, E.; Waller, Z.A.E.; Pagano, B.; Amato, J.; et al. Common G-quadruplex binding agents found to interact with i-motif-forming DNA: Unexpected multi-target-directed compounds. Front. Chem. 2018, 6, 281. [Google Scholar] [CrossRef] [PubMed]
- Cristofari, C.; Rigo, R.; Greco, M.L.; Ghezzo, M.; Sissi, C. pH-driven conformational switch between non-canonical DNA structures in a C-rich domain of EGFR promoter. Sci. Rep. 2019, 9, 1210. [Google Scholar] [CrossRef] [PubMed]
- Lavrado, J.; Moreira, R.; Paulo, A. Indoloquinolines as Scaffolds for Drug Discovery. Curr. Med. Chem. 2010, 17, 2348–2370. [Google Scholar] [CrossRef] [PubMed]
- Che, T.; Wang, Y.Q.; Huang, Z.L.; Tan, J.H.; Huang, Z.S.; Chen, S. Bin Natural alkaloids and heterocycles as G-quadruplex ligands and potential anticancer agents. Molecules 2018, 23, 493. [Google Scholar] [CrossRef] [Green Version]
- Riechert-Krause, F.; Weisz, K. Indoloquinolines as DNA binding ligands. Heterocycl. Commun. 2013, 19, 145–166. [Google Scholar] [CrossRef]
- Alberti, P.; Schmitt, P.; Nguyen, C.H.; Rivalle, C.; Hoarau, M.; Grierson, D.S.; Mergny, J.L. Benzoindoloquinolines interact with DNA tetraplexes and inhibit telomerase. Bioorg. Med. Chem. Lett. 2002, 12, 1071–1074. [Google Scholar] [CrossRef]
- Caprio, V.; Guyen, B.; Opoku-Boahen, Y.; Mann, J.; Gowan, S.M.; Kelland, L.M.; Read, M.A.; Neidle, S. A novel inhibitor of human telomerase derived from 10h-indolo[3,2-b]quinoline. Bioorg. Med. Chem. Lett. 2000, 10, 2063–2066. [Google Scholar] [CrossRef]
- Guyen, B.; Schultes, C.M.; Hazel, P.; Mann, J.; Neidle, S. Synthesis and evaluation of analogues of 10H-indolo[3,2-b]-quinoline as G-quadruplex stabilising ligands and potential inhibitors of the enzyme telomerase. Org. Biomol. Chem. 2004, 2, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.L.; Lu, Y.J.; Ou, T.M.; Zhou, J.M.; Huang, Z.S.; Zhu, X.F.; Du, C.J.; Bu, X.Z.; Ma, L.; Gu, L.Q.; et al. Synthesis and evaluation of quindoline derivatives as G-quadruplex inducing and stabilizing ligands and potential inhibitors of telomerase. J. Med. Chem. 2005, 48, 7315–7321. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.M.; Lu, Y.J.; Zhang, C.; Huang, Z.S.; Wang, X.D.; Tan, J.H.; Chen, Y.; Ma, D.L.; Wong, K.Y.; Tang, J.C.O.; et al. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J. Med. Chem. 2007, 50, 1465–1474. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.M.; Lin, J.; Lu, Y.J.; Hou, J.Q.; Tan, J.H.; Chen, S.H.; Li, Z.; Li, Y.P.; Li, D.; Gu, L.Q.; et al. Inhibition of cell proliferation by quindoline derivative (SYUIQ-05) through its preferential interaction with c-myc promoter G-quadruplex. J. Med. Chem. 2011, 54, 5671–5679. [Google Scholar] [CrossRef] [PubMed]
- Boddupally, P.V.L.; Hahn, S.; Beman, C.; De, B.; Brooks, T.A.; Gokhale, V.; Hurley, L.H. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J. Med. Chem. 2012, 55, 6076–6086. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.J.; Ou, T.M.; Tan, J.H.; Hou, J.Q.; Shao, W.Y.; Peng, D.; Sun, N.; Wang, X.D.; Wu, W.B.; Bu, X.Z.; et al. 5-N-methylated quindoline derivatives as telomeric G-quadruplex stabilizing ligands: Effects of 5-N positive charge on quadruplex binding affinity and cell proliferation. J. Med. Chem. 2008, 51, 6381–6392. [Google Scholar] [CrossRef]
- Lavrado, J.; Reszka, A.P.; Moreira, R.; Neidle, S.; Paulo, A. C-11 diamino cryptolepine derivatives NSC748392, NSC748393, and NSC748394: Anticancer profile and G-quadruplex stabilization. Bioorg. Med. Chem. Lett. 2010, 20, 7042–7045. [Google Scholar] [CrossRef]
- Lavrado, J.; Borralho, P.M.; Ohnmacht, S.A.; Castro, R.E.; Rodrigues, C.M.P.; Moreira, R.; Dos Santos, D.J.V.A.; Neidle, S.; Paulo, A. Synthesis, G-quadruplex stabilisation, docking studies, and effect on cancer cells of indolo[3,2-b]quinolines with one, two, or three basic side chains. ChemMedChem 2013, 8, 1648–1661. [Google Scholar] [CrossRef] [Green Version]
- Lavrado, J.; Brito, H.; Borralho, P.M.; Ohnmacht, S.A.; Kim, N.S.; Leitão, C.; Pisco, S.; Gunaratnam, M.; Rodrigues, C.M.P.; Moreira, R.; et al. KRAS oncogene repression in colon cancer cell lines by G-quadruplex binding indolo[3,2-c]quinolines. Sci. Rep. 2015, 5, 9696. [Google Scholar] [CrossRef] [Green Version]
- Lavrado, J.; Ohnmacht, S.A.; Correia, I.; Leitão, C.; Pisco, S.; Gunaratnam, M.; Moreira, R.; Neidle, S.; dos Santos, D.J.V.A.; Paulo, A. Indolo[3,2-c]quinoline G-quadruplex stabilizers: A structural analysis of binding to the human telomeric G-quadruplex. ChemMedChem 2015, 10, 836–849. [Google Scholar] [CrossRef]
- Zhang, B.; Li, X.; Li, B.; Gao, C.; Jiang, Y. Acridine and its derivatives: A patent review (2009–2013). Expert Opin. Ther. Pat. 2014, 24, 647–664. [Google Scholar] [CrossRef]
- Sullivan, H.J.; Readmond, C.; Radicella, C.; Persad, V.; Fasano, T.J.; Wu, C. Binding of Telomestatin, TMPyP4, BSU6037, and BRACO19 to a Telomeric G-Quadruplex-Duplex Hybrid Probed by All-Atom Molecular Dynamics Simulations with Explicit Solvent. ACS Omega 2018, 3, 14788–14806. [Google Scholar] [CrossRef]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rache, A.; Mergny, J.L. Assessment of selectivity of G-quadruplex ligands via an optimised FRET melting assay. Biochimie 2015, 115, 194–202. [Google Scholar] [CrossRef]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Nakajima, A.; Shimamoto, T.; Ohyashiki, J.H.; Ohyashiki, K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene 2003, 22, 5338–5347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monchaud, D.; Granzhan, A.; Saettel, N.; Guédin, A.; Mergny, J.L.; Teulade-Fichou, M.P. “One ring to bind them all”—Part I: The efficiency of the macrocyclic scaffold for G-quadruplex DNA recognition. J. Nucleic Acids 2010, 525862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tera, M.; Ishizuka, H.; Takagi, M.; Suganuma, M.; Shin-Ya, K.; Nagasawa, K. Macrocyclic hexaoxazoles as sequence- and mode-selective G-quadruplex binders. Angew. Chem. Int. Ed. 2008, 47, 5557–5560. [Google Scholar] [CrossRef]
- Iida, K.; Tera, M.; Hirokawa, T.; Shin-Ya, K.; Nagasawa, K. G-quadruplex recognition by macrocyclic hexaoxazole (6OTD) dimer: Greater selectivity than monomer. Chem. Commun. 2009, 6481–6483. [Google Scholar] [CrossRef]
- Iida, K.; Nagasawa, K. Macrocyclic polyoxazoles as G-quadruplex ligands. Chem. Rec. 2013, 13, 539–548. [Google Scholar] [CrossRef]
- Wheelhouse, R.T.; Sun, D.; Han, H.; Han, F.X.; Hurley, L.H. Cationic porphyrins as telomerase inhibitors: The interaction of tetra- (N-methyl-4-pyridyl)porphine with quadruplex DNA. J. Am. Chem. Soc. 1998, 120, 3261–3262. [Google Scholar] [CrossRef]
- Han, F.X.; Wheelhouse, R.T.; Hurley, L.H. Interactions of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc. 1999, 121, 3561–3570. [Google Scholar] [CrossRef]
- Han, H.; Langley, D.R.; Rangan, A.; Hurley, L.H. Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc. 2001, 123, 8902–8913. [Google Scholar] [CrossRef]
- Izbicka, E.; Wheelhouse, R.T.; Raymond, E.; Davidson, K.K.; Lawrence, R.A.; Sun, D.; Windle, B.E.; Hurley, L.H.; Von Hoff, D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999, 59, 639–644. [Google Scholar] [PubMed]
- Grand, C.L.; Han, H.; Mũnoz, R.M.; Weitman, S.; Von Hoff, D.D.; Hurley, L.H.; Bearss, D.J. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol. Cancer Ther. 2002, 1, 565–573. [Google Scholar] [PubMed]
- Fedoroff, O.Y.; Rangan, A.; Chemeris, V.V.; Hurley, L.H. Cationic porphyrins promote the formation of i-motif DNA and bind peripherally by a nonintercalative mechanism. Biochemistry 2000, 39, 15083–15090. [Google Scholar] [CrossRef]
- Nielsen, C.M.; Ulven, T. Macrocyclic G-Quadruplex Ligands. Curr. Med. Chem. 2012, 17, 3438–3448. [Google Scholar] [CrossRef]
- Neidle, S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017, 1, 41. [Google Scholar] [CrossRef]
- Ruan, T.L.; Davis, S.J.; Powell, B.M.; Harbeck, C.P.; Habdas, J.; Habdas, P.; Yatsunyk, L.A. Lowering the overall charge on TMPyP4 improves its selectivity for G-quadruplex DNA. Biochimie 2017, 132, 121–130. [Google Scholar] [CrossRef]
- Yaku, H.; Murashima, T.; Miyoshi, D.; Sugimoto, N. Specific binding of anionic porphyrin and phthalocyanine to the G-quadruplex with a variety of in Vitro and in Vivo applications. Molecules 2012, 17, 10586–10613. [Google Scholar] [CrossRef]
- Weisman-Shomer, P.; Cohen, E.; Hershco, I.; Khateb, S.; Wolfovitz-Barchad, O.; Hurley, L.H.; Fry, M. The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res. 2003, 31, 3963–3970. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.; Singh, A.; Kukreti, S. Porphyrin induced structural destabilization of a parallel DNA G-quadruplex in human MRP1 gene promoter. J. Mol. Recognit. 2022, 35, e2950. [Google Scholar] [CrossRef]
- Morris, M.J.; Wingate, K.L.; Silwal, J.; Leeper, T.C.; Basu, S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012, 40, 4137–4145. [Google Scholar] [CrossRef]
- Zamiri, B.; Reddy, K.; Macgregor, R.B.; Pearson, C.E. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNAbinding proteins. J. Biol. Chem. 2014, 289, 4653–4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Joshi, S.; Kukreti, S. Cationic porphyrins as destabilizer of a G-quadruplex located at the promoter of human MYH7 β gene. J. Biomol. Struct. Dyn. 2020, 38, 4801–4816. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.Z.; Zhang, M.J.; Liao, T.C.; Li, J.H.; Zou, M.; Wang, Z.M.; Zhou, C.Q. Dimers formed with the mixed-type G-quadruplex binder pyridostatin specifically recognize human telomere G-quadruplex dimers. Org. Biomol. Chem. 2020, 18, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Waller, Z.A.E.; Shirude, P.S.; Rodriguez, R.; Balasubramanian, S. Triarylpyridines: A versatile small molecule scaffold for G-quadruplex recognition. Chem. Commun. 2008, 44, 1467–1469. [Google Scholar] [CrossRef]
- Waller, Z.A.E.; Sewitz, S.A.; Hsu, S.D. A Small Molecule That Disrupts G-Quadruplex DNA Structure and Enhances Gene Expression. J. Am. Chem. Soc. 2009, 131, 12628–12633. [Google Scholar] [CrossRef] [Green Version]
- Smith, N.M.; Labrunie, G.; Corry, B.; Tran, P.L.T.; Norret, M.; Djavaheri-Mergny, M.; Raston, C.L.; Mergny, J.L. Unraveling the relationship between structure and stabilization of triarylpyridines as G-quadruplex binding ligands. Org. Biomol. Chem. 2011, 9, 6154–6162. [Google Scholar] [CrossRef]
- O’Hagan, M.P.; Haldar, S.; Duchi, M.; Oliver, T.A.A.; Mulholland, A.J.; Morales, J.C.; Galan, M.C. A Photoresponsive Stiff-Stilbene Ligand Fuels the Reversible Unfolding of G-Quadruplex DNA. Angew. Chem. Int. Ed. 2019, 58, 4334–4338. [Google Scholar] [CrossRef] [Green Version]
- O’Hagan, M.P.; Peñalver, P.; Gibson, R.S.L.; Morales, J.C.; Galan, M.C. Stiff-Stilbene Ligands Target G-Quadruplex DNA and Exhibit Selective Anticancer and Antiparasitic Activity. Chem. Eur. J. 2020, 26, 6224–6233. [Google Scholar] [CrossRef]
- Mitteaux, J.; Lejault, P.; Wojciechowski, F.; Joubert, A.; Boudon, J.; Desbois, N.; Gros, C.P.; Hudson, R.H.E.; Boulé, J.B.; Granzhan, A.; et al. Identifying G-Quadruplex-DNA-Disrupting Small Molecules. J. Am. Chem. Soc. 2021, 143, 12567–12577. [Google Scholar] [CrossRef] [PubMed]
- D’Urso, A.; Randazzo, R.; Rizzo, V.; Gangemi, C.M.A.; Romanucci, V.; Zarrelli, A.; Tomaselli, G.A.; Milardi, D.; Borbone, N.; Purrello, R.; et al. Stabilization: Vs. destabilization of G-quadruplex superstructures: The role of the porphyrin derivative having spermine arms. Phys. Chem. Chem. Phys. 2017, 19, 17404–17410. [Google Scholar] [CrossRef]
- Ma, Y.; Iida, K.; Nagasawa, K. Topologies of G-quadruplex: Biological functions and regulation by ligands. Biochem. Biophys. Res. Commun. 2020, 531, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tsushima, Y.; Sakuma, M.; Sasaki, S.; Iida, K.; Okabe, S.; Seimiya, H.; Hirokawa, T.; Nagasawa, K. Development of G-quadruplex ligands for selective induction of a parallel-type topology. Org. Biomol. Chem. 2018, 16, 7375–7382. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Panto, G.D.; Gonçalves, D.P.N.; Sanders, J.K.M. Ligand-Driven G-Quadruplex Conformational Switching By Using an Unusual Mode of Interaction. Angew. Chem. Int. Ed. 2008, 46, 5405–5407. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Q.; Zhuo, S.T.; Tan, J.H.; Ou, T.M.; Li, D.; Gu, L.Q.; Huang, Z.S. Facile syntheses of disubstituted bis(vinylquinolinium)benzene derivatives as G-quadruplex DNA binders. Tetrahedron 2013, 69, 4922–4932. [Google Scholar] [CrossRef]
- He, J.H.; Liu, H.Y.; Li, Z.; Tan, J.H.; Ou, T.M.; Huang, S.L.; An, L.K.; Li, D.; Gu, L.Q.; Huang, Z.S. New quinazoline derivatives for telomeric G-quadruplex DNA: Effects of an added phenyl group on quadruplex binding ability. Eur. J. Med. Chem. 2013, 63, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Zhang, X.; Zheng, W.; Liu, X.; Qi, C.; Wang, F.; Shangguan, D. Fluorescence light-up probe for parallel G-quadruplexes. Anal. Chem. 2014, 86, 943–952. [Google Scholar] [CrossRef]
- Chen, S.B.; Liu, G.C.; Gu, L.Q.; Huang, Z.S.; Tan, J.H. Identification of small molecules capable of regulating conformational changes of telomeric G-quadruplex. J. Mol. Struct. 2018, 1154, 1–7. [Google Scholar] [CrossRef]
- Hao, X.; Wang, C.; Wang, Y.; Li, C.; Hou, J.; Zhang, F.; Kang, C.; Gao, L. Topological conversion of human telomeric G-quadruplexes from hybrid to parallel form induced by naphthalene diimide ligands. Int. J. Biol. Macromol. 2021, 167, 1048–1058. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Hao, X.; Wang, L.; Ma, X.; Jin, R.; Kang, C.; Gao, L. A naphthyridine-indole ligand for selective stabilization of G-quadruplexes and conformational conversion of hybrid topology. Bioorg. Med. Chem. 2021, 48, 116416. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Reznichenko, O.; Chaput, L.; Martin, P.; Teulade-Fichou, M.P.; Granzhan, A. Topology-Selective, Fluorescent “Light-Up” Probes for G-Quadruplex DNA Based on Photoinduced Electron Transfer. Chem. Eur. J. 2018, 24, 12638–12651. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Perry, M.D.J.; Moreira, R.; Mendes, E. Alkylating Agents. Anticancer. Ther. 2008, 133–158. [Google Scholar] [CrossRef]
- Di Antonio, M.; McLuckie, K.I.E.; Balasubramanian, S. Reprogramming the mechanism of action of chlorambucil by coupling to a G-quadruplex ligand. J. Am. Chem. Soc. 2014, 136, 5860–5863. [Google Scholar] [CrossRef] [PubMed]
- Verga, D.; Hamon, F.; Poyer, F.; Bombard, S.; Teulade-Fichou, M.P. Photo-cross-linking probes for trapping G-quadruplex DNA. Angew. Chem. Int. Ed. 2014, 53, 994–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, Y.; Zhang, L.; He, H.; Zhou, X. Quinone Methide Derivatives: Important Intermediates to DNA Alkylating and DNA Cross-linking Actions. Curr. Med. Chem. 2005, 12, 2893–2913. [Google Scholar] [CrossRef]
- Percivalle, C.; Doria, F.; Freccero, M. Quinone Methides as DNA Alkylating Agents: An Overview on Efficient Activation Protocols for Enhanced Target Selectivity. Curr. Org. Chem. 2014, 18, 19–43. [Google Scholar] [CrossRef]
- Doria, F.; Nadai, M.; Folini, M.; Di Antonio, M.; Germani, L.; Percivalle, C.; Sissi, C.; Zaffaroni, N.; Alcaro, S.; Artese, A.; et al. Hybrid ligand-alkylating agents targeting telomeric G-quadruplex structures. Org. Biomol. Chem. 2012, 10, 2798–2806. [Google Scholar] [CrossRef] [Green Version]
- Minard, A.; Liano, D.; Wang, X.; Di Antonio, M. The unexplored potential of quinone methides in chemical biology. Bioorg. Med. Chem. 2019, 27, 2298–2305. [Google Scholar] [CrossRef]
- Cadoni, E.; Manicardi, A.; Fossépré, M.; Heirwegh, K.; Surin, M.; Madder, A. Teaching photosensitizers a new trick: Red light-triggered G-quadruplex alkylation by ligand co-localization. Chem. Commun. 2021, 57, 1010–1013. [Google Scholar] [CrossRef]
- Zuffo, M.; Gu, A.; Leriche, E.; Doria, F.; Pirota, V. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018, 46, e115. [Google Scholar] [CrossRef] [PubMed]
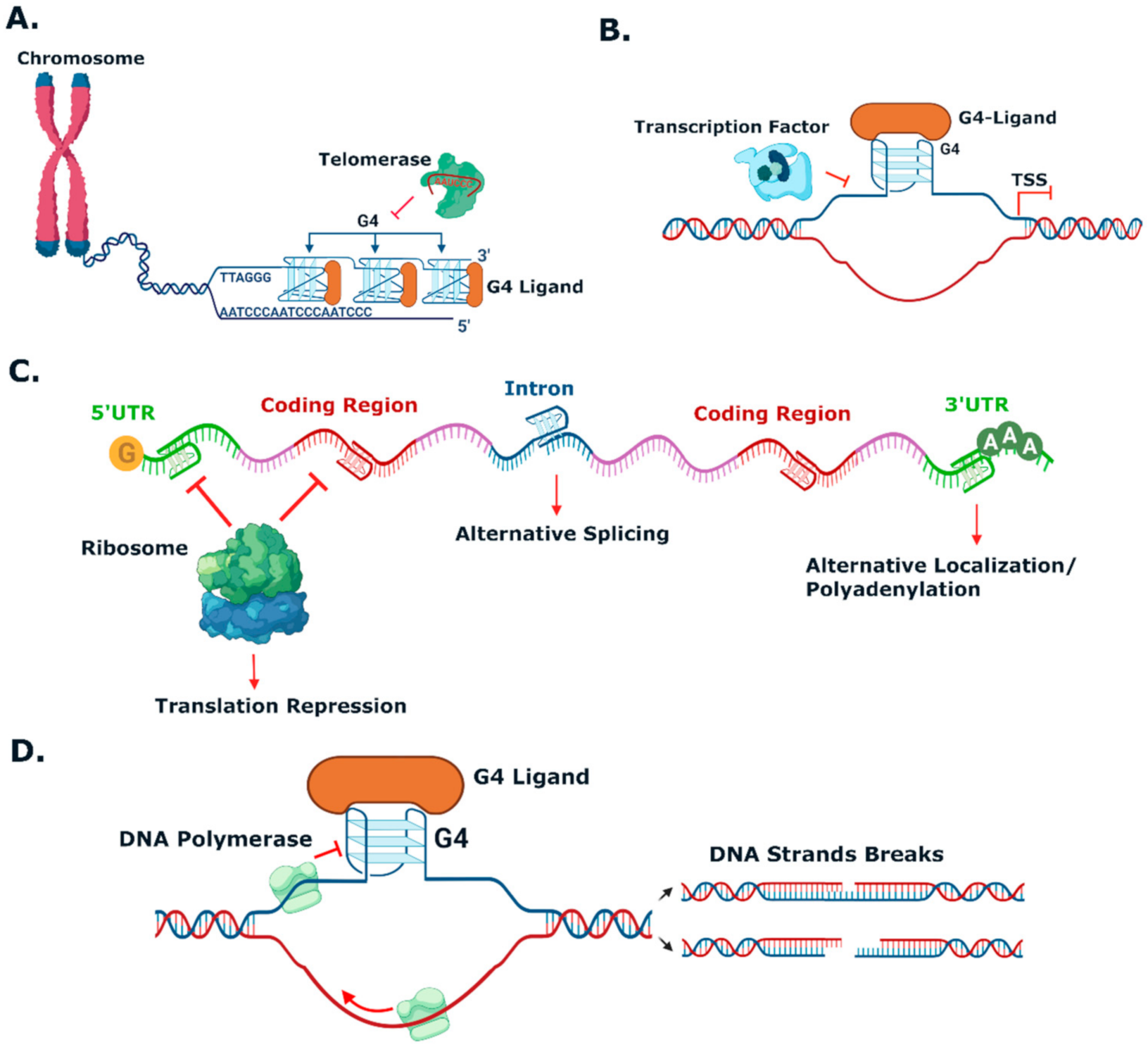




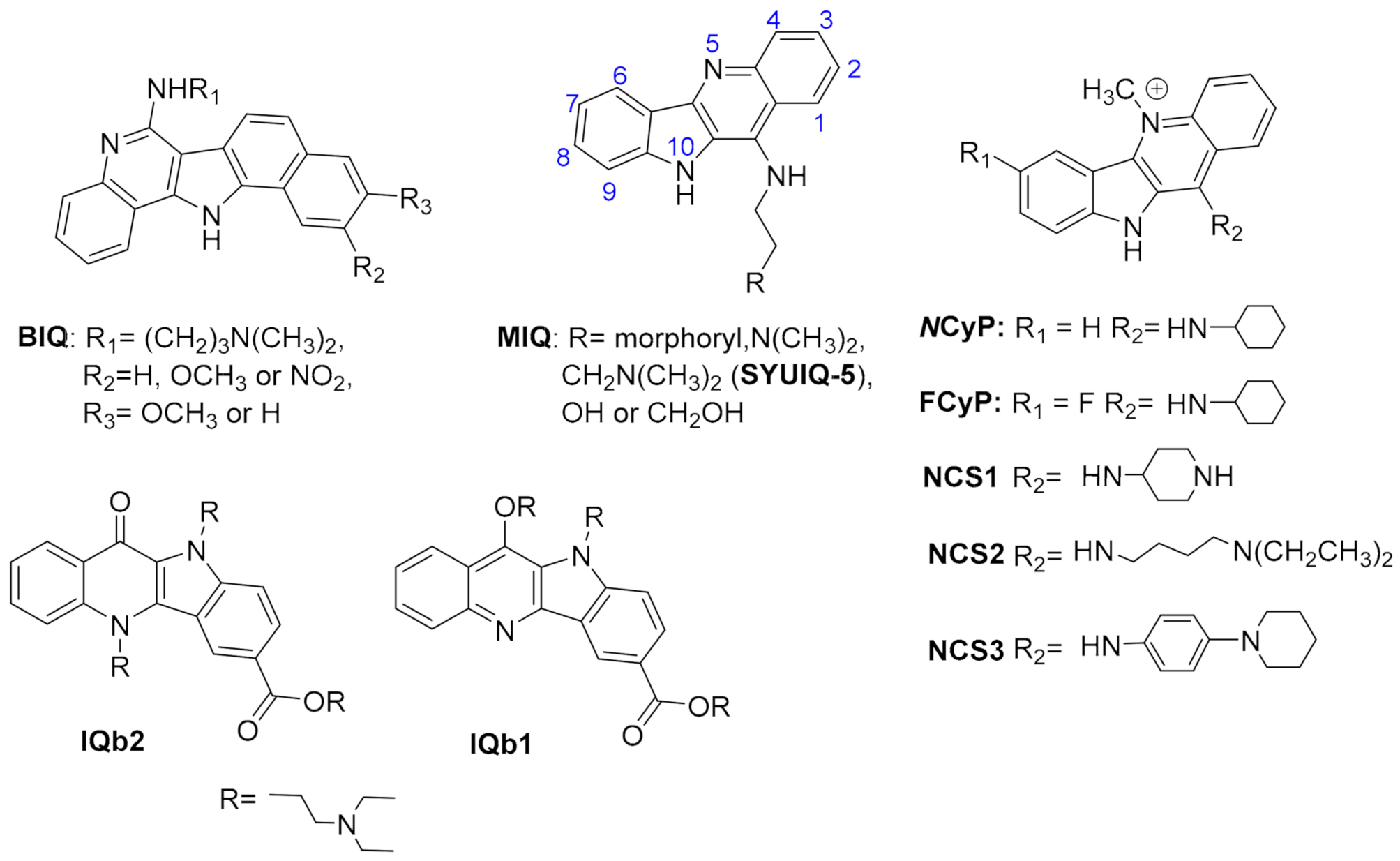



| Ligand (Chemotype) | Structure | Topology Switch | Refs. | ||
|---|---|---|---|---|---|
| 3,3-L2H2-6OTD (Oxazole-based) | Figure 15 | ssDNA | ➔ | antiparallel (ion free) | [257] |
| RHPS4 (acridinium) | Figure 4 | hybrid | ➔ | antiparallel (in K+) | [282] |
| Berberine | Figure 13 | hybrid | ➔ | antiparallel (in K+) | [282] |
| BRACO-19 (acridine) | Figure 4 | hybrid | ➔ | antiparallel (in K+) | [282] |
| pyridostatin | Figure 6 | hybrid | ➔ | antiparallel (in K+) | [253] |
| PhenDC3 (phenanthroline) | Figure 6 | hybrid | ➔ | antiparallel (in K+) | [253,282] |
| PDC-360A (pyridine-dicarboxamide) | Figure 6 | hybrid | ➔ | antiparallel (in 10 mM K+) | [253] |
| hybrid | ➔ | parallel (in 60 mM K+) | [183] | ||
| PDCd (pyridine-dicarboxamide) | Figure 17 | hybrid | ➔ | antiparallel (in K+) | [183] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, E.; Aljnadi, I.M.; Bahls, B.; Victor, B.L.; Paulo, A. Major Achievements in the Design of Quadruplex-Interactive Small Molecules. Pharmaceuticals 2022, 15, 300. https://doi.org/10.3390/ph15030300
Mendes E, Aljnadi IM, Bahls B, Victor BL, Paulo A. Major Achievements in the Design of Quadruplex-Interactive Small Molecules. Pharmaceuticals. 2022; 15(3):300. https://doi.org/10.3390/ph15030300
Chicago/Turabian StyleMendes, Eduarda, Israa M. Aljnadi, Bárbara Bahls, Bruno L. Victor, and Alexandra Paulo. 2022. "Major Achievements in the Design of Quadruplex-Interactive Small Molecules" Pharmaceuticals 15, no. 3: 300. https://doi.org/10.3390/ph15030300







