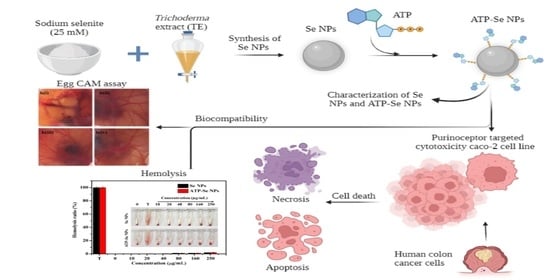
Purinoceptor Targeted Cytotoxicity of Adenosine Triphosphate-Conjugated Biogenic Selenium Nanoparticles in Human Colon Cancer Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of Nanoparticles
2.1.1. UV–Vis Spectrophotometer Analysis
2.1.2. X-ray Diffraction (XRD) Pattern Analysis
2.1.3. Morphological Characteristics
2.1.4. Hydrodynamic Size and Zeta Potential
2.1.5. Functional Group Analysis
2.2. Cytotoxicity
2.2.1. WST-1 Assay and Cellular Uptake
2.2.2. Staining Assay and FACS
2.2.3. Hemolysis and Egg CAM Toxicity
3. Materials and Methods
3.1. Chemicals, Microorganisms, and Cell Lines
3.2. Preparation of Trichoderma Extract and Synthesis of SeNPs and ATP-SeNPs
3.3. Characterization of SeNPs and ATP-SeNPs
3.4. Cytotoxicity Assay
3.5. Hemolysis Assay
3.6. Chick Egg CAM Assay
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brar, B.; Ranjan, K.; Palria, A.; Kumar, R.; Ghosh, M.; Sihag, S.; Minakshi, P. Nanotechnology in Colorectal Cancer for Precision Diagnosis and Therapy. Front. Nanotechnol. 2021, 3, 699266. [Google Scholar] [CrossRef]
- Huyghe, N.; Baldin, P.; Van den Eynde, M. Immunotherapy with immune checkpoint inhibitors in colorectal cancer: What is the future beyond deficient mismatch-repair tumours? Gastroenterol. Rep. 2019, 8, 11–24. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Huang, Z.; Zheng, W.; Fan, C.; Chen, T. Enhancement of cell permeabilization apoptosis-inducing activity of selenium nanoparticles by ATP surface decoration. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Amorim, C.D.M.; Couto, A.G.; Netz, D.J.A.; de Freitas, R.A.; Bresolin, T.M.B. Antioxidant idebenone-loaded nanoparticles based on chitosan and N-carboxymethylchitosan. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Greco, F.; Vicent, M.J. Combination therapy: Opportunities and challenges for polymer–drug conjugates as anticancer nanomedicines. Adv. Drug Deliv. Rev. 2009, 61, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Sathiyaseelan, A.; Mariadoss, A.V.A.; Jeevithan, E.; Hu, X.; Shin, S.; Wang, M.-H. Dual stimuli-responsive release of aptamer AS1411 decorated erlotinib loaded chitosan nanoparticles for non-small-cell lung carcinoma therapy. Carbohydr. Polym. 2020, 245, 116407. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Yang, J.; Zhang, Y.; Fang, Y.; Nishinari, K.; Phillips, G.O. Synthesis and antioxidant properties of gum arabic-stabilized selenium nanoparticles. Int. J. Biol. Macromol. 2014, 65, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.; Ks, S.D.; Santhiya, R.; Rajeshkumar, S.; Kumar, V. Selenium nanoparticles: A potent chemotherapeutic agent and an elucidation of its mechanism. Colloids Surf. B Biointerf. 2018, 170, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef]
- Sanmartín, C.; Plano, D.; Sharma, A.K.; Palop, J.A. Selenium Compounds, Apoptosis and Other Types of Cell Death: An Overview for Cancer Therapy. Int. J. Mol. Sci. 2012, 13, 9649–9672. [Google Scholar] [CrossRef]
- Sinha, R.; El-Bayoumy, K. Apoptosis is a Critical Cellular Event in Cancer Chemoprevention and Chemotherapy by Selenium Compounds. Curr. Cancer Drug Targets 2004, 4, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Wrobel, J.K.; Power, R.; Toborek, M. Biological activity of selenium: Revisited. IUBMB Life 2016, 68, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Björnstedt, M.; Fernandes, A.P. Selenium in the prevention of human cancers. EPMA J. 2010, 1, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Abdulah, R.; Miyazaki, K.; Nakazawa, M.; Koyama, H. Chemical forms of selenium for cancer prevention. J. Trace Elem. Med. Biol. 2005, 19, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.X.; Gutterman, D.D. Mitochondrial reactive oxygen species-mediated signaling in endothelial cells. Am. J. Physiol. -Heart Circ. Physiol. 2007, 292, H2023–H2031. [Google Scholar] [CrossRef] [PubMed]
- Khurana, A.; Tekula, S.; Saifi, M.A.; Venkatesh, P.; Godugu, C. Therapeutic applications of selenium nanoparticles. Biomed. Pharmacother. 2019, 111, 802–812. [Google Scholar] [CrossRef]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.; Yu, H. Elemental selenium at nano size possesses lower toxicity without compromising the fundamental effect on selenoenzymes: Comparison with selenomethionine in mice. Free Radic. Biol. Med. 2007, 42, 1524–1533. [Google Scholar] [CrossRef]
- Chen, T.; Wong, Y.S.; Zheng, W.; Bai, Y.; Huang, L. Selenium nanoparticles fabricated in Undaria pinnatifida polysaccharide solutions induce mitochondria-mediated apoptosis in A375 human melanoma cells. Colloids Surf. B Biointerf. 2008, 67, 26–31. [Google Scholar] [CrossRef]
- Wan, H.X.; Hu, J.H.; Xie, R.; Yang, S.M.; Dong, H. Important roles of P2Y receptors in the inflammation and cancer of digestive system. Oncotarget 2016, 7, 28736–28747. [Google Scholar] [CrossRef]
- White, N.; Burnstock, G. P2 receptors and cancer. Trends Pharmacol. Sci. 2006, 27, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Hertel, C.; Coulter, S.J.; Perkins, J.P. The involvement of cellular ATP receptor-mediated internalization of epidermal growth factor and hormone-induced internalization of β-adrenergic receptors. J. Biol. Chem. 1986, 261, 5974–5980. [Google Scholar] [CrossRef]
- Yaguchi, T.; Saito, M.; Yasuda, Y.; Kanno, T.; Nakano, T.; Nishizaki, T. Higher Concentrations of Extracellular ATP Suppress Proliferation of Caco-2 Human Colonic Cancer Cells via an Unknown Receptor Involving PKC Inhibition. Cell. Physiol. Biochem. 2010, 26, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Menon, S.; Ks, S.D.; Agarwal, H.; Shanmugam, V.K. Efficacy of Biogenic Selenium Nanoparticles from an Extract of Ginger towards Evaluation on Anti-Microbial and Anti-Oxidant Activities. Colloid Interface Sci. Commun. 2019, 29, 1–8. [Google Scholar] [CrossRef]
- Menon, S.; Rajeshkumar, S.; Kumar, S.V. A review on biogenic synthesis of gold nanoparticles, characterization, and its applications. Resour. Effic. Technol. 2017, 3, 516–527. [Google Scholar] [CrossRef]
- Husen, A.; Siddiqi, K.S. Plants and microbes assisted selenium nanoparticles: Characterization and application. J. Nanobiotechnol. 2014, 12, 28. [Google Scholar] [CrossRef]
- Kessi, J.; Ramuz, M.; Wehrli, E.; Spycher, M.; Bachofen, R. Reduction of Selenite and Detoxification of Elemental Selenium by the Phototrophic Bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 1999, 65, 4734–4740. [Google Scholar] [CrossRef]
- Tejo Prakash, N.; Sharma, N.; Prakash, R.; Raina, K.K.; Fellowes, J.; Pearce, C.I.; Lloyd, J.R.; Pattrick, R.A.D. Aerobic microbial manufacture of nanoscale selenium: Exploiting nature’s bio-nanomineralization potential. Biotechnol. Lett. 2009, 31, 1857. [Google Scholar] [CrossRef]
- Stolz, J.F.; Basu, P.; Santini, J.M.; Oremland, R.S. Arsenic and Selenium in Microbial Metabolism. Annu. Rev. Microbiol. 2006, 60, 107–130. [Google Scholar] [CrossRef]
- Spyridopoulou, K.; Tryfonopoulou, E.; Aindelis, G.; Ypsilantis, P.; Sarafidis, C.; Kalogirou, O.; Chlichlia, K. Biogenic selenium nanoparticles produced by Lactobacillus casei ATCC 393 inhibit colon cancer cell growth in vitro and in vivo. Nanoscale Adv. 2021, 3, 2516–2528. [Google Scholar] [CrossRef]
- Nandini, B.; Hariprasad, P.; Prakash, H.S.; Shetty, H.S.; Geetha, N. Trichogenic-selenium nanoparticles enhance disease suppressive ability of Trichoderma against downy mildew disease caused by Sclerospora graminicola in pearl millet. Sci. Rep. 2017, 7, 2612. [Google Scholar] [CrossRef] [PubMed]
- Arunthirumeni, M.; Veerammal, V.; Shivakumar, M.S. Biocontrol Efficacy of Mycosynthesized Selenium Nanoparticle Using Trichoderma sp. on Insect Pest Spodoptera litura. J. Clust. Sci. 2021, 1–9. [Google Scholar] [CrossRef]
- Anu, K.; Devanesan, S.; Prasanth, R.; AlSalhi, M.S.; Ajithkumar, S.; Singaravelu, G. Biogenesis of selenium nanoparticles and their anti-leukemia activity. J. King Saud Univ. Sci. 2020, 32, 2520–2526. [Google Scholar] [CrossRef]
- Srivastava, N.; Mukhopadhyay, M. Green synthesis and structural characterization of selenium nanoparticles and assessment of their antimicrobial property. Bioprocess Biosyst. Eng. 2015, 38, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Kirupagaran, R.; Saritha, A.; Bhuvaneswari, S. Green synthesis of selenium nanoparticles from leaf and stem extract of Leucas lavandulifolia Sm. and their application. J. Nanosci. Technol. 2016, 2, 224–226. [Google Scholar]
- Kumaran, C.K.S.; Agilan, S.; Velauthapillai, D.; Muthukumarasamy, N.; Thambidurai, M.; Senthil, T.S.; Balasundaraprabhu, R. Synthesis and Characterization of Selenium Nanowires. ISRN Nanotechnol. 2011, 2011, 589073. [Google Scholar] [CrossRef][Green Version]
- Cittrarasu, V.; Kaliannan, D.; Dharman, K.; Maluventhen, V.; Easwaran, M.; Liu, W.C.; Balasubramanian, B.; Arumugam, M. Green synthesis of selenium nanoparticles mediated from Ceropegia bulbosa Roxb extract and its cytotoxicity, antimicrobial, mosquitocidal and photocatalytic activities. Sci. Rep. 2021, 11, 1032. [Google Scholar] [CrossRef]
- Diko, C.S.; Zhang, H.; Lian, S.; Fan, S.; Li, Z.; Qu, Y. Optimal synthesis conditions and characterization of selenium nanoparticles in Trichoderma sp. WL-Go culture broth. Mater. Chem. Phys. 2020, 246, 122583. [Google Scholar] [CrossRef]
- Ramamurthy, C.H.; Sampath, K.S.; Arunkumar, P.; Kumar, M.S.; Sujatha, V.; Premkumar, K.; Thirunavukkarasu, C. Green synthesis and characterization of selenium nanoparticles and its augmented cytotoxicity with doxorubicin on cancer cells. Bioprocess Biosyst. Eng. 2013, 36, 1131–1139. [Google Scholar] [CrossRef]
- Lin, Z.H.; Chris Wang, C.R. Evidence on the size-dependent absorption spectral evolution of selenium nanoparticles. Mater. Chem. Phys. 2005, 92, 591–594. [Google Scholar] [CrossRef]
- Tang, H.Y.; Huang, Q.; Wang, Y.L.; Yang, X.Q.; Su, D.X.; He, S.; Tan, J.C.; Zeng, Q.Z.; Yuan, Y. Development, structure characterization and stability of food grade selenium nanoparticles stabilized by Tilapia polypeptides. J. Food Eng. 2020, 275, 109878. [Google Scholar] [CrossRef]
- Jiang, W.; He, S.; Su, D.; Ye, M.; Zeng, Q.; Yuan, Y. Synthesis, characterization of tuna polypeptide selenium nanoparticle, and its immunomodulatory and antioxidant effects in vivo. Food Chem. 2022, 383, 132405. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Dua, K.; Dureja, H. Erlotinib loaded chitosan nanoparticles: Formulation, physicochemical characterization and cytotoxic potential. Int. J. Biol. Macromol. 2019, 139, 1304–1316. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Yu, S.; Yu, D.; Liu, N.; Tang, Y.; Fan, Y.; Wang, C.; Wu, A. Biogenic Trichoderma harzianum-derived selenium nanoparticles with control functionalities originating from diverse recognition metabolites against phytopathogens and mycotoxins. Food Control. 2019, 106, 106748. [Google Scholar] [CrossRef]
- Chen, L.F.; Liang, H.W.; Lu, Y.; Cui, C.H.; Yu, S.H. Synthesis of an Attapulgite Clay@Carbon Nanocomposite Adsorbent by a Hydrothermal Carbonization Process and Their Application in the Removal of Toxic Metal Ions from Water. Langmuir 2011, 27, 8998–9004. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Sathiyaseelan, A.; Park, S.; Kim, S.-R.; Priya, V.V.; Wang, M.-H. Monoclonal Antibody Functionalized, and L-lysine α-Oxidase Loaded PEGylated-Chitosan Nanoparticle for HER2/Neu Targeted Breast Cancer Therapy. Pharmaceutics 2022, 14, 927. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Mandava, S.; Chellia, R.; Jeevithan, E.; Babu Yelamanchi, R.S.; Mandava, D.; Wen-Hui, W.; Lee, J.; Oh, D.H.; Kathiresan, K.; et al. Novel metabolites from Trichoderma atroviride against human prostate cancer cells and their inhibitory effect on Helicobacter pylori and Shigella toxin producing Escherichia coli. Microb. Pathog. 2019, 126, 19–26. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical-Chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef]
- Sayes, C.M.; Reed, K.L.; Warheit, D.B. Assessing toxicology of fine and nanoparticles: Comparing in vitro measurements to in vivo pulmonary toxicity profiles. Toxicol. Sci. 2007, 97, 163–180. [Google Scholar] [CrossRef]
- Lin, Y.S.; Haynes, C.L. Impacts of mesoporous silica nanoparticle size, pore ordering, and pore integrity on hemolytic activity. J. Am. Chem. Soc. 2010, 132, 4834–4842. [Google Scholar] [CrossRef]
- Liao, K.H.; Lin, Y.S.; Macosko, C.W.; Haynes, C.L. Cytotoxicity of Graphene Oxide and Graphene in Human Erythrocytes and Skin Fibroblasts. ACS Appl. Mater. Interfaces 2011, 3, 2607–2615. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Reipa, V.; Hitchins, V.M.; Goering, P.L.; Malinauskas, R.A. Physicochemical Characterization and In Vitro Hemolysis Evaluation of Silver Nanoparticles. Toxicol. Sci. 2011, 123, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Arbab, A.S.; Bashaw, L.A.; Miller, B.R.; Jordan, E.K.; Lewis, B.K.; Kalish, H.; Frank, J.A. Characterization of Biophysical and Metabolic Properties of Cells Labeled with Superparamagnetic Iron Oxide Nanoparticles and Transfection Agent for Cellular MR Imaging. Radiology 2003, 229, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Jain, P.K.; El-Sayed, I.H.; El-Sayed, M.A. Gold nanoparticles: Interesting optical properties and recent applications in cancer diagnostics and therapy. Nanomedicine 2007, 2, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Aseichev, A.V.; Azizova, O.A.; Beckman, E.M.; Skotnikova, O.I.; Dudnik, L.B.; Shcheglovitova, O.N.; Sergienko, V.I. Effects of gold nanoparticles on erythrocyte hemolysis. Bull. Exp. Biol. Med. 2014, 156, 495–498. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Wang, M.H. Isolation and molecular identification of Trichoderma species from wetland soil and their antagonistic activity against phytopathogens. Physiol. Mol. Plant Pathol. 2020, 109, 101458. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Vivek, R.; Sithranga Boopathy, N.; Yaqian, L.; Kathiresan, K.; Chen, J. Anticancer potential of bioactive 16-methylheptadecanoic acid methyl ester derived from marine Trichoderma. J. Appl. Biomed. 2015, 13, 199–212. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Mariadoss, A.V.A.; Sathiyaseelan, A.; Wang, M.H. Synthesis and characterization of nano-chitosan capped gold nanoparticles with multifunctional bioactive properties. Int. J. Biol. Macromol. 2020, 165, 747–757. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Sriram, B.; Sathiyaseelan, A.; Hu, X.; Mariadoss, A.V.A.; MubarakAli, D.; Wang, M.H. Molecular identification, volatile metabolites profiling, and bioactivities of an indigenous endophytic fungus (Diaporthe sp.). Process Biochem. 2021, 102, 72–81. [Google Scholar] [CrossRef]
- Wu, S.; Xu, C.; Zhu, Y.; Zheng, L.; Zhang, L.; Hu, Y.; Yu, B.; Wang, Y.; Xu, F.J. Biofilm-Sensitive Photodynamic Nanoparticles for Enhanced Penetration and Antibacterial Efficiency. Adv. Funct. Mater. 2021, 31, 2103591. [Google Scholar] [CrossRef]
- Rajendran, I.; Dhandapani, H.; Anantanarayanan, R.; Rajaram, R. Apigenin mediated gold nanoparticle synthesis and their anti-cancer effect on human epidermoid carcinoma (A431) cells. RSC Adv. 2015, 5, 51055–51066. [Google Scholar] [CrossRef]
- Park, S.; Saravanakumar, K.; Sathiyaseelan, A.; Park, S.; Hu, X.; Wang, M.-H. Cellular antioxidant properties of nontoxic exopolysaccharide extracted from Lactobacillales (Weissella cibaria) isolated from Korean kimchi. LWT 2022, 154, 112727. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saravanakumar, K.; Sathiyaseelan, A.; Zhang, X.; Park, S.; Wang, M.-H. Purinoceptor Targeted Cytotoxicity of Adenosine Triphosphate-Conjugated Biogenic Selenium Nanoparticles in Human Colon Cancer Cells. Pharmaceuticals 2022, 15, 582. https://doi.org/10.3390/ph15050582
Saravanakumar K, Sathiyaseelan A, Zhang X, Park S, Wang M-H. Purinoceptor Targeted Cytotoxicity of Adenosine Triphosphate-Conjugated Biogenic Selenium Nanoparticles in Human Colon Cancer Cells. Pharmaceuticals. 2022; 15(5):582. https://doi.org/10.3390/ph15050582
Chicago/Turabian StyleSaravanakumar, Kandasamy, Anbazhagan Sathiyaseelan, Xin Zhang, Soyoung Park, and Myeong-Hyeon Wang. 2022. "Purinoceptor Targeted Cytotoxicity of Adenosine Triphosphate-Conjugated Biogenic Selenium Nanoparticles in Human Colon Cancer Cells" Pharmaceuticals 15, no. 5: 582. https://doi.org/10.3390/ph15050582
APA StyleSaravanakumar, K., Sathiyaseelan, A., Zhang, X., Park, S., & Wang, M.-H. (2022). Purinoceptor Targeted Cytotoxicity of Adenosine Triphosphate-Conjugated Biogenic Selenium Nanoparticles in Human Colon Cancer Cells. Pharmaceuticals, 15(5), 582. https://doi.org/10.3390/ph15050582