Bacterial Community and Genomic Analysis of Carbapenem-Resistant Acinetobacter baumannii Isolates from the Environment of a Health Care Facility in the Western Region of Saudi Arabia
Abstract
:1. Introduction
2. Results
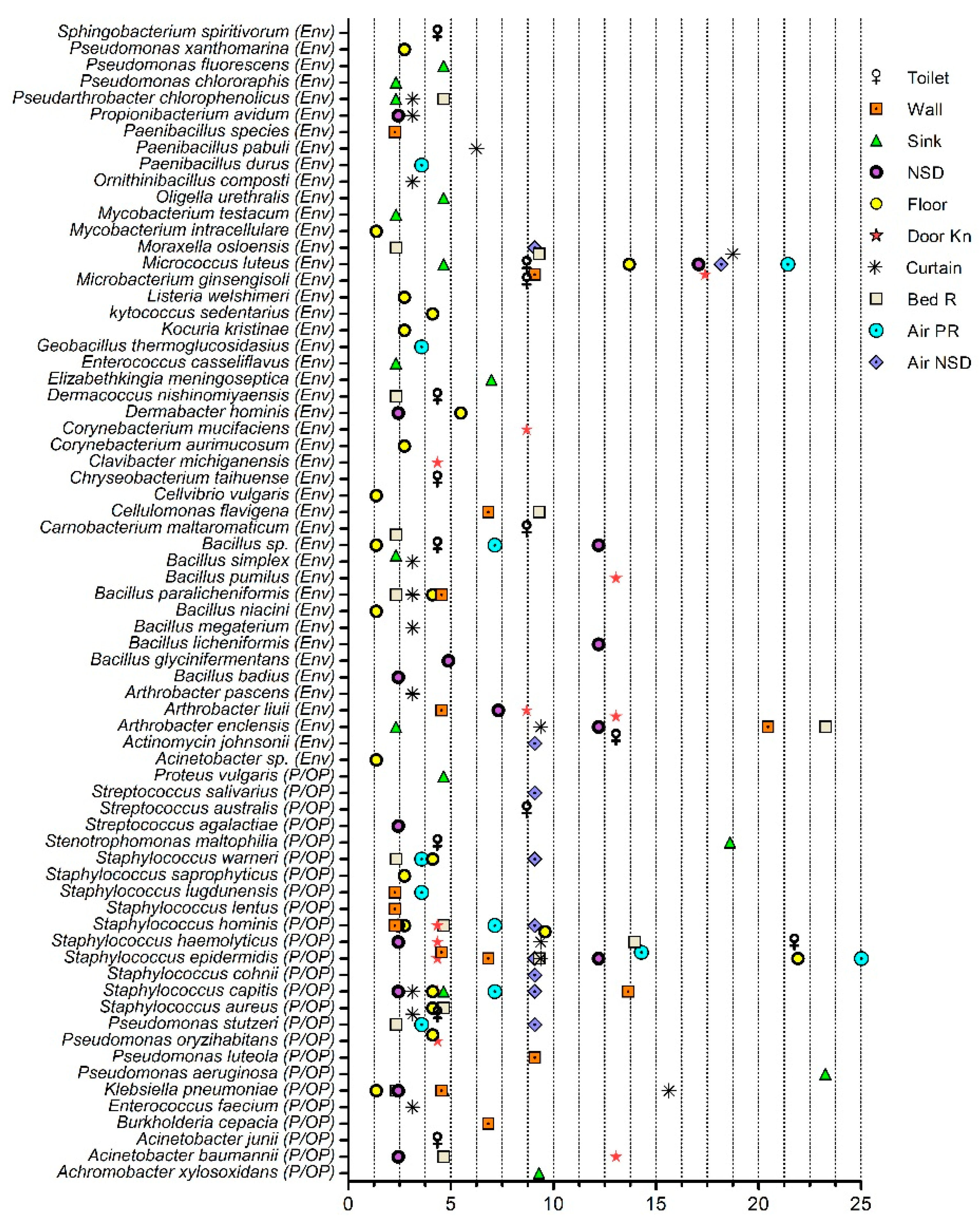
2.1. Bacterial Community Analysis
Pathogenic and Opportunistic Pathogenic Bacteria
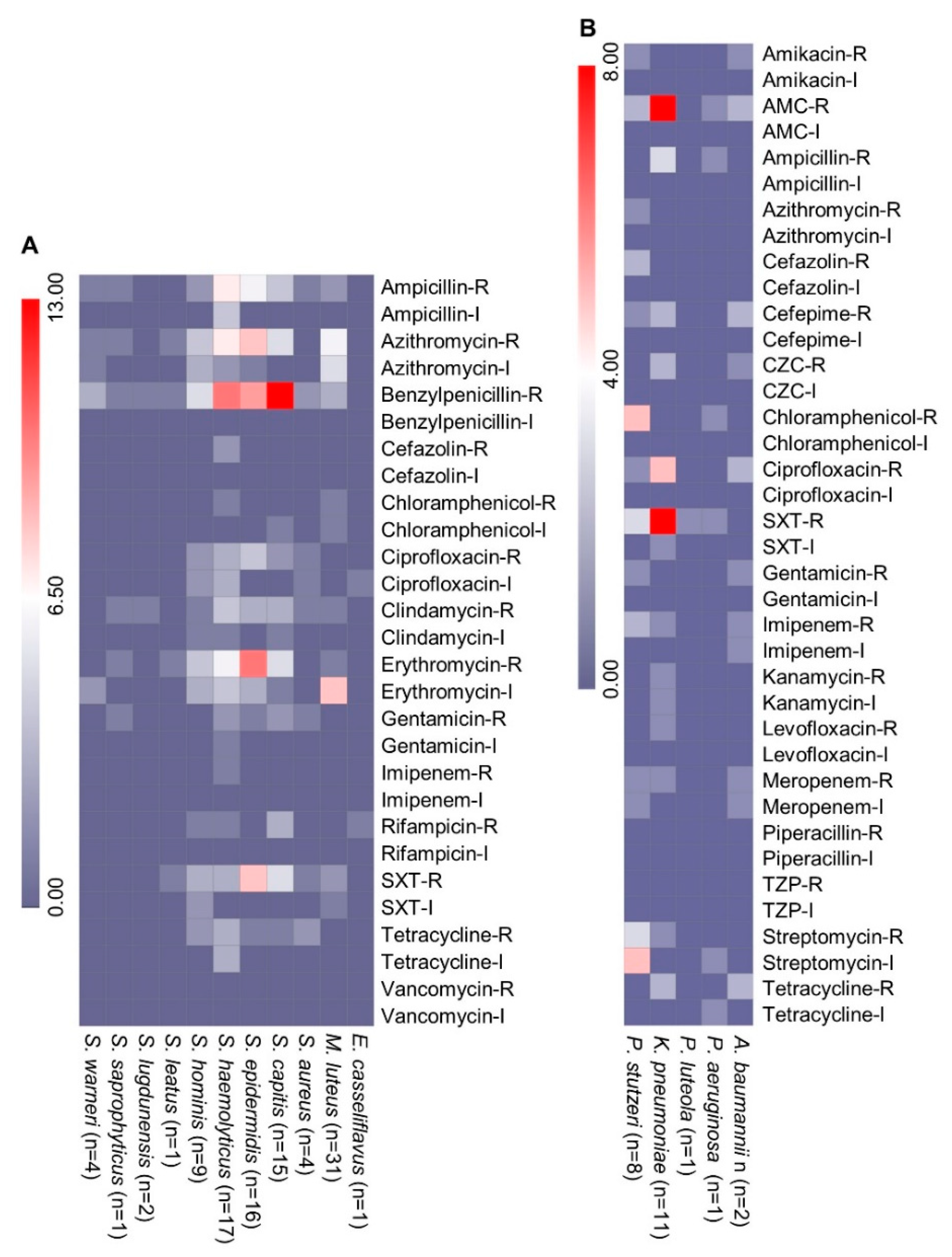
2.2. Phenotypic Antimicrobial Resistance
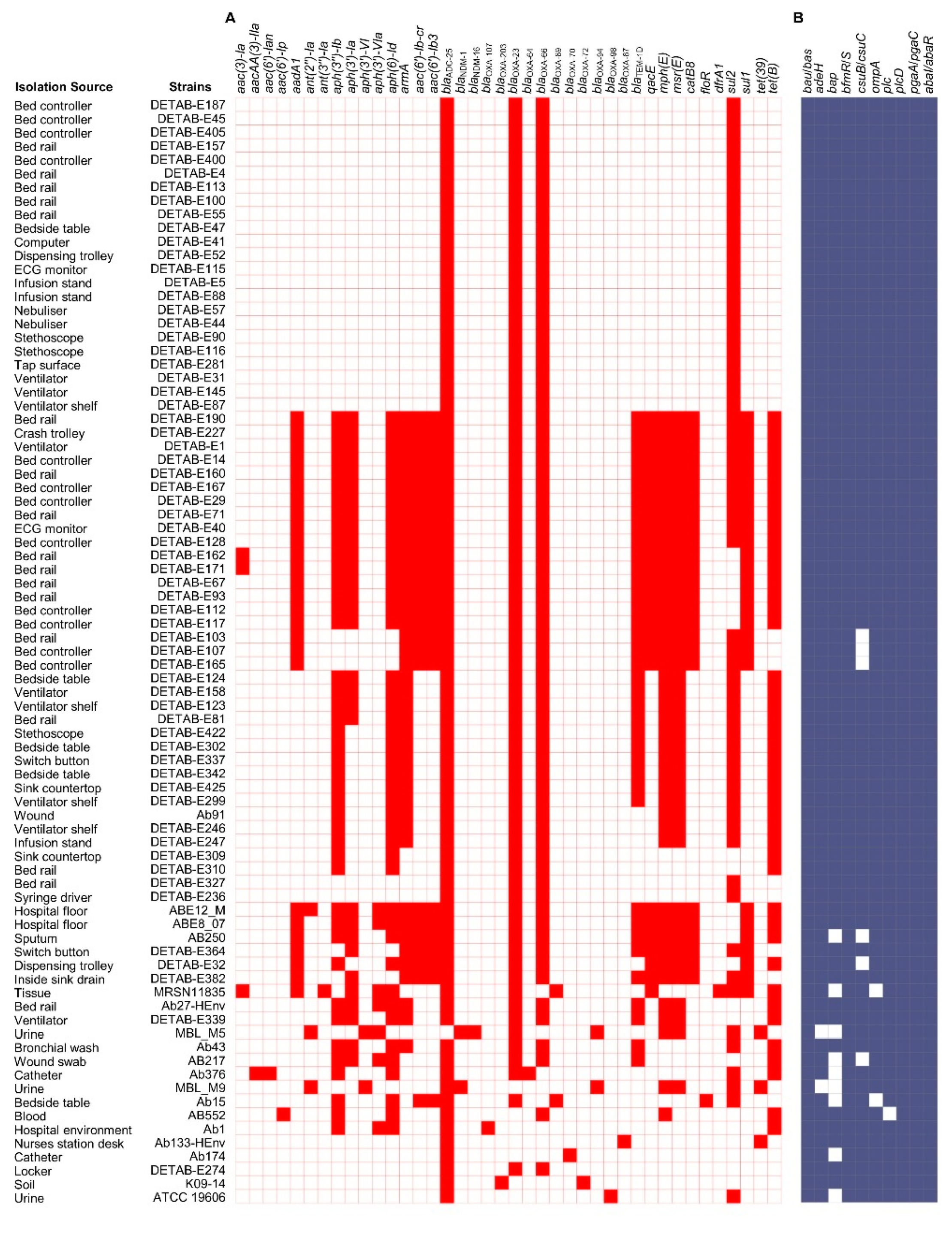
2.3. Genomic Analysis of A. baumannii Isolates
Antimicrobial Resistance Genes and Mobile Genetic Elements Analysis
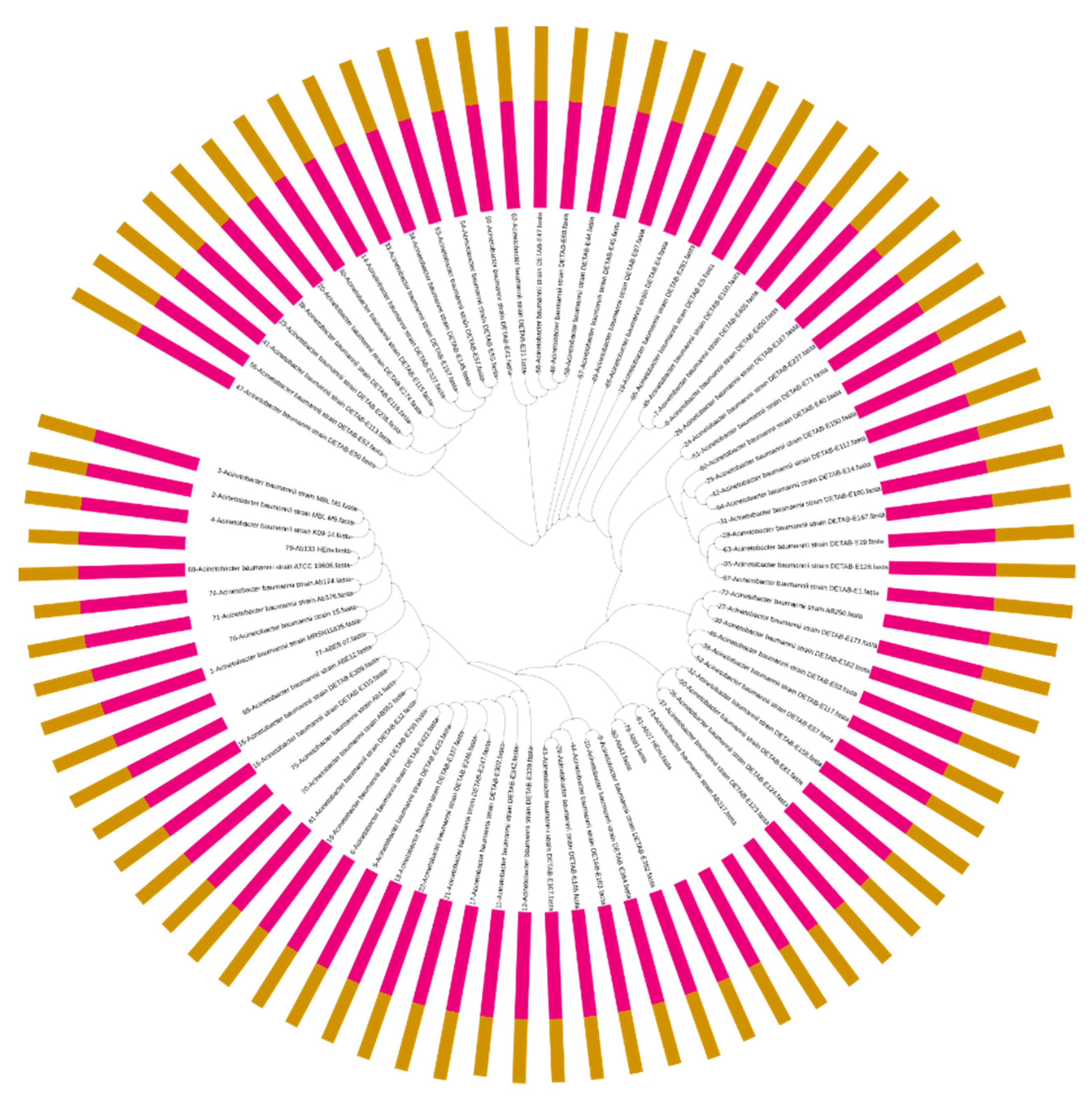
2.4. Comparative Analysis of A. baumannii Genomes from Genbank
2.4.1. Acquired AMR Genes Analysis
2.4.2. Virulence Factor Genes and Phylogenetic Analysis
3. Discussion
4. Materials and Methods
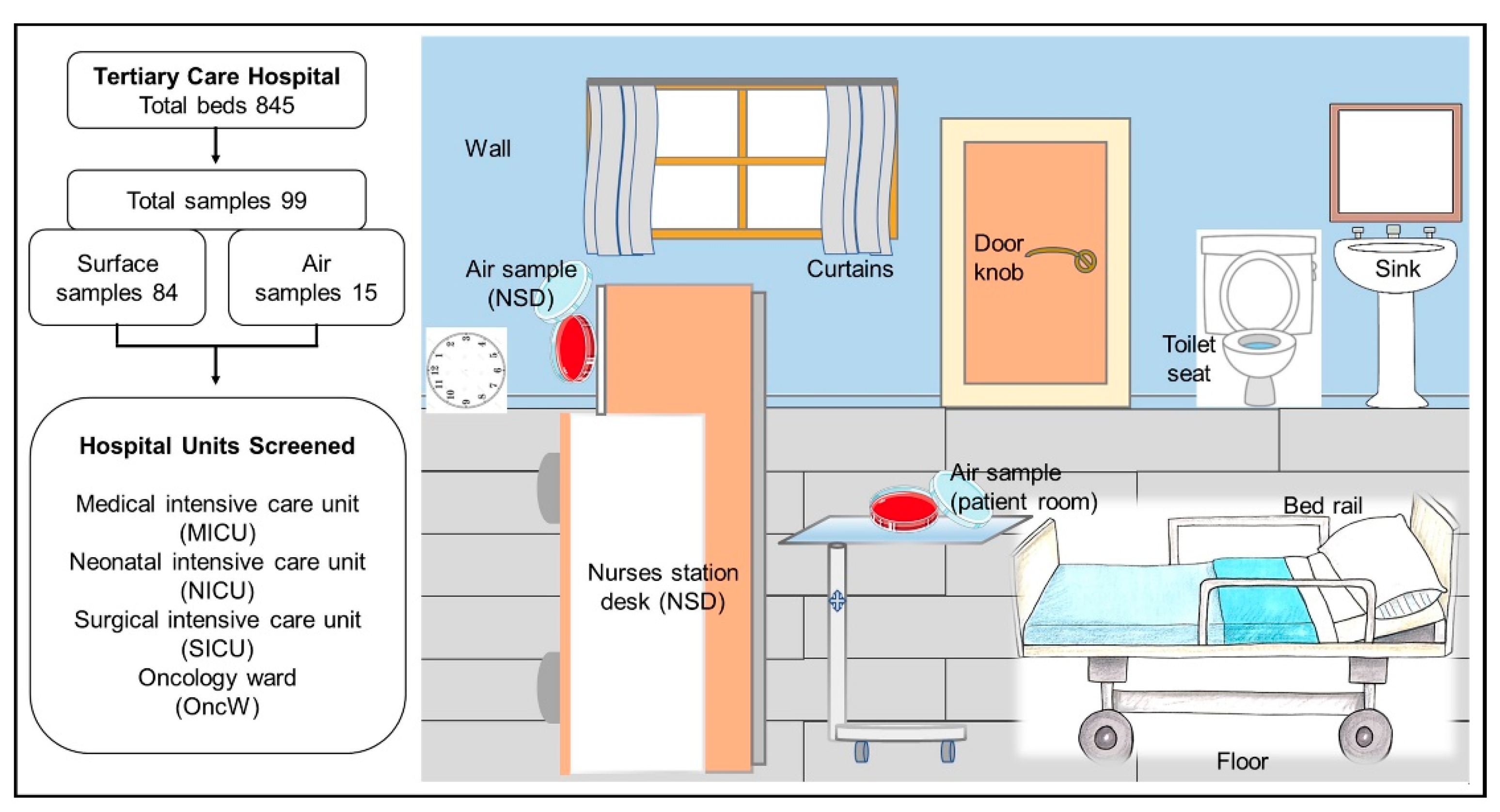
4.1. Sample Collection
4.2. Identification and Antimicrobial Susceptibility Screening
4.3. Genome Sequencing and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Babady, N.E. Hospital-Associated Infections. Microbiol. Spectr. 2016, 4, 735–758. [Google Scholar] [CrossRef] [PubMed]
- Gaynes, R.; Edwards, J.R.; National Nosocomial Infections Surveillance System. Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 2005, 41, 848–854. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.; Al-Saryi, N.; Al-Kadmy, I.M.S.; Aziz, S.N. Multidrug-resistant Acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 2021, 48, 6987–6998. [Google Scholar] [CrossRef]
- Mayon-White, R.T.; Ducel, G.; Kereselidze, T.; Tikomirov, E. An international survey of the prevalence of hospital-acquired infection. J. Hosp. Infect. 1988, 11 (Suppl. A), 43–48. [Google Scholar] [CrossRef]
- Allegranzi, B.; Nejad, S.B.; Chraiti, M.-N.; Combescure, C.; Shorbaji, N.A.; Sudan, R. Report on the Burden of Endemic Health Care-Associated Infection Worldwide; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Vincent, J.L. Nosocomial infections in adult intensive-care units. Lancet 2003, 361, 2068–2077. [Google Scholar] [CrossRef]
- Avershina, E.; Shapovalova, V.; Shipulin, G. Fighting Antibiotic Resistance in Hospital-Acquired Infections: Current State and Emerging Technologies in Disease Prevention, Diagnostics and Therapy. Front. Microbiol. 2021, 12, 707330. [Google Scholar] [CrossRef]
- Lemmen, S.W.; Hafner, H.; Zolldann, D.; Stanzel, S.; Lutticken, R. Distribution of multi-resistant Gram-negative versus Gram-positive bacteria in the hospital inanimate environment. J. Hosp. Infect. 2004, 56, 191–197. [Google Scholar] [CrossRef]
- Al-Hamad, A.; Pal, T.; Leskafi, H.; Abbas, H.; Hejles, H.; Alsubikhy, F.; Darwish, D.; Ghazawi, A.; Sonnevend, A. Molecular characterization of clinical and environmental carbapenem resistant Acinetobacter baumannii isolates in a hospital of the Eastern Region of Saudi Arabia. J. Infect. Public Health 2020, 13, 632–636. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Eze, E.C.; Chenia, H.Y.; El Zowalaty, M.E. Acinetobacter baumannii biofilms: Effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect. Drug Resist. 2018, 11, 2277–2299. [Google Scholar] [CrossRef] [Green Version]
- Cornejo-Juarez, P.; Cevallos, M.A.; Castro-Jaimes, S.; Castillo-Ramirez, S.; Velazquez-Acosta, C.; Martinez-Oliva, D.; Perez-Oseguera, A.; Rivera-Buendia, F.; Volkow-Fernandez, P. High mortality in an outbreak of multidrug resistant Acinetobacter baumannii infection introduced to an oncological hospital by a patient transferred from a general hospital. PLoS ONE 2020, 15, e0234684. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Quirino, A.; Giordano, M.; Marano, V.; Rizzo, C.; Liberto, M.C.; Foca, A.; Pavia, M. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect. Dis. 2016, 16, 747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ababneh, Q.; Abulaila, S.; Jaradat, Z. Isolation of extensively drug resistant Acinetobacter baumannii from environmental surfaces inside intensive care units. Am. J. Infect. Control. 2022, 50, 159–165. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [Green Version]
- Giammanco, A.; Cala, C.; Fasciana, T.; Dowzicky, M.J. Global Assessment of the Activity of Tigecycline against Multidrug-Resistant Gram-Negative Pathogens between 2004 and 2014 as Part of the Tigecycline Evaluation and Surveillance Trial. mSphere 2017, 2, e00310-16. [Google Scholar] [CrossRef] [Green Version]
- Spellberg, B.; Rex, J.H. The value of single-pathogen antibacterial agents. Nat. Rev. Drug Discov. 2013, 12, 963. [Google Scholar] [CrossRef] [Green Version]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 10 April 2022).
- Wright, M.S.; Iovleva, A.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med. 2016, 8, 26. [Google Scholar] [CrossRef] [Green Version]
- Price, J.R.; Cole, K.; Bexley, A.; Kostiou, V.; Eyre, D.W.; Golubchik, T.; Wilson, D.J.; Crook, D.W.; Walker, A.S.; Peto, T.E.A.; et al. Transmission of Staphylococcus aureus between health-care workers, the environment, and patients in an intensive care unit: A longitudinal cohort study based on whole-genome sequencing. Lancet Infect. Dis. 2017, 17, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alzahrani, A.J.; Tobaiqy, M.; Alresasi, A.M.; Bu-Shehab, I.; Al-Hadary, I.; Alhmeed, N.; Alismail, M.; et al. Antimicrobial susceptibility of gram-positive and gram-negative bacteria: A 5-year retrospective analysis at a multi-hospital healthcare system in Saudi Arabia. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 43. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.W.; Yasir, M.; Farman, M.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Alawi, M.; El-Hossary, D.; Azhar, E.I. Antimicrobial Susceptibility and Molecular Characterization of Clinical Strains of Acinetobacter baumannii in Western Saudi Arabia. Microb. Drug Resist. 2019, 25, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, A.A.; Al-Azraqi, T.A.; Abbag, F.I.; Al-Gamal, M.N.; Seef, S.; Bello, C.S. Nosocomial infections in a neonatal intensive care unit in south-western Saudi Arabia. East. Mediterr. Health J. 2010, 16, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Balkhy, H.H.; Walsh, T.R.; Paterson, D.L. beta-Lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clin. Microbiol. Rev. 2013, 26, 361–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.Y.; Dickter, J.K. Nosocomial Infections: A History of Hospital-Acquired Infections. Gastrointest. Endosc. Clin. N. Am. 2020, 30, 637–652. [Google Scholar] [CrossRef]
- Vandijck, D.M.; Labeau, S.O.; Vogelaers, D.P.; Blot, S.I. Prevention of nosocomial infections in intensive care patients. Nurs. Crit. Care 2010, 15, 251–256. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [Green Version]
- Johani, K.; Abualsaud, D.; Costa, D.M.; Hu, H.; Whiteley, G.; Deva, A.; Vickery, K. Characterization of microbial community composition, antimicrobial resistance and biofilm on intensive care surfaces. J. Infect. Public Health 2018, 11, 418–424. [Google Scholar] [CrossRef]
- Markwart, R.; Saito, H.; Harder, T.; Tomczyk, S.; Cassini, A.; Fleischmann-Struzek, C.; Reichert, F.; Eckmanns, T.; Allegranzi, B. Epidemiology and burden of sepsis acquired in hospitals and intensive care units: A systematic review and meta-analysis. Intensive Care Med. 2020, 46, 1536–1551. [Google Scholar] [CrossRef]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Yang, C.; Zhang, J.; Hu, K.; Zou, J.; Li, J.; Wang, J.; Huang, W.; Yin, L.; Zhang, X. An Outbreak of Carbapenem-Resistant Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Chongqing, China. Front. Cell. Infect. Microbiol. 2021, 11, 656070. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Fonseca, T.A.P.; Pessoa, R.; Felix, A.C.; Sanabani, S.S. Diversity of Bacterial Communities on Four Frequently Used Surfaces in a Large Brazilian Teaching Hospital. Int. J. Environ. Res. Public Health 2016, 13, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenza, G. Multidrug-resistant gram-negative rods in the intensive care unit: Epidemiology, prevention and treatment options. Med. Klin. Intensivmed. Notf. 2019, 114, 263–275. [Google Scholar] [CrossRef]
- Mehrad, B.; Clark, N.M.; Zhanel, G.G.; Lynch, J.P., 3rd. Antimicrobial resistance in hospital-acquired gram-negative bacterial infections. Chest 2015, 147, 1413–1421. [Google Scholar] [CrossRef] [Green Version]
- Yezli, S.; Shibl, A.M.; Livermore, D.M.; Memish, Z.A. Prevalence and antimicrobial resistance among Gram-negative pathogens in Saudi Arabia. J. Chemother. 2014, 26, 257–272. [Google Scholar] [CrossRef]
- Yasir, M.; Ajlan, A.M.; Shakil, S.; Jiman-Fatani, A.A.; Almasaudi, S.B.; Farman, M.; Baazeem, Z.M.; Baabdullah, R.; Alawi, M.; Al-Abdullah, N.; et al. Molecular characterization, antimicrobial resistance and clinico-bioinformatics approaches to address the problem of extended-spectrum beta-lactamase-producing Escherichia coli in western Saudi Arabia. Sci. Rep. 2018, 8, 14847. [Google Scholar] [CrossRef] [Green Version]
- Karaiskos, I.; Giamarellou, H. Carbapenem-Sparing Strategies for ESBL Producers: When and How. Antibiotics 2020, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Avent, M.L.; Rogers, B.A.; Cheng, A.C.; Paterson, D.L. Current use of aminoglycosides: Indications, pharmacokinetics and monitoring for toxicity. Intern. Med. J. 2011, 41, 441–449. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Donhofer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Rahnama’I, M.S.; Wagenvoort, J.H.; van der Linden, C.J. Amoxicillin/clavulanate (Augmentin) resistant Escherichia coli in bacterial peritonitis after abdominal surgery--clinical outcome in ICU patients. Neth. J. Med. 2009, 67, 173–176. [Google Scholar] [PubMed]
- Yasir, M.; Farman, M.; Shah, M.W.; Jiman-Fatani, A.A.; Othman, N.A.; Almasaudi, S.B.; Alawi, M.; Shakil, S.; Al-Abdullah, N.; Ismaeel, N.A.; et al. Genomic and antimicrobial resistance genes diversity in multidrug-resistant CTX-M-positive isolates of Escherichia coli at a health care facility in Jeddah. J. Infect. Public Health 2020, 13, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Singh, A.; Chowdhary, P.; Pandey, A.; Gupta, P. Occurrence of emerging sulfonamide resistance (sul1 and sul2) associated with mobile integrons-integrase (intI1 and intI2) in riverine systems. Sci. Total Environ. 2021, 751, 142217. [Google Scholar] [CrossRef] [PubMed]
- Mamishi, S.; Mohammadian, M.; Pourakbari, B.; Hosseinpour Sadeghi, R.; Haghi Ashtiani, M.T.; Abdosalehi, M.R.; Rahmani, M.; Mahmoudi, S. Antibiotic Resistance And Genotyping Of Gram-Positive Bacteria Causing Hospital-Acquired Infection In Patients Referring To Children’s Medical Center. Infect. Drug Resist. 2019, 12, 3719–3726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-Resistant Bacterial Infections in U.S. Hospitalized Patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Tajeddin, E.; Rashidan, M.; Razaghi, M.; Javadi, S.S.; Sherafat, S.J.; Alebouyeh, M.; Sarbazi, M.R.; Mansouri, N.; Zali, M.R. The role of the intensive care unit environment and health-care workers in the transmission of bacteria associated with hospital acquired infections. J. Infect. Public Health 2016, 9, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Sonnevend, A.; Ghazawi, A.; Al Munthari, N.; Pitout, M.; Hamadeh, M.B.; Hashmey, R.; Girgis, S.K.; Sheikh, F.A.; Al Haj, M.; Nagelkerke, N.; et al. Characteristics of epidemic and sporadic strains of Acinetobacter baumannii isolated in Abu Dhabi hospitals. J. Med. Microbiol. 2013, 62, 582–590. [Google Scholar] [CrossRef] [Green Version]
- Lopes, B.S.; Al-Agamy, M.H.; Ismail, M.A.; Shibl, A.M.; Al-Qahtani, A.A.; Al-Ahdal, M.N.; Forbes, K.J. The transferability of blaOXA-23 gene in multidrug-resistant Acinetobacter baumannii isolates from Saudi Arabia and Egypt. Int. J. Med. Microbiol. 2015, 305, 581–588. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Zhang, X.; Li, F.; Awan, F.; Jiang, H.; Zeng, Z.; Lv, W. Molecular Epidemiology and Clone Transmission of Carbapenem-Resistant Acinetobacter baumannii in ICU Rooms. Front. Cell. Infect. Microbiol. 2021, 11, 633817. [Google Scholar] [CrossRef]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napoli, C.; Marcotrigiano, V.; Montagna, M.T. Air sampling procedures to evaluate microbial contamination: A comparison between active and passive methods in operating theatres. BMC Public Health 2012, 12, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farman, M.; Yasir, M.; Al-Hindi, R.R.; Farraj, S.A.; Jiman-Fatani, A.A.; Alawi, M.; Azhar, E.I. Genomic analysis of multidrug-resistant clinical Enterococcus faecalis isolates for antimicrobial resistance genes and virulence factors from the western region of Saudi Arabia. Antimicrob. Resist. Infect. Control 2019, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Aslam, Z.; Kim, S.W.; Lee, S.W.; Jeon, C.O.; Chung, Y.R. Bacterial community composition and chitinase gene diversity of vermicompost with antifungal activity. Bioresour. Technol. 2009, 100, 4396–4403. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Rogers, M.R.C.; Braat, J.C.; Verschuuren, T.D.; Top, J.; Corander, J.; Willems, R.J.L.; Schurch, A.C. mlplasmids: A user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 2018, 4, e000224. [Google Scholar] [CrossRef] [Green Version]

| Features | Ab27-HEnv | Ab133-HEnv | Ab43 | Ab91 |
|---|---|---|---|---|
| Phenotypic Resistance | ||||
| Ciprofloxacin | R | R | R | R |
| Gentamicin | R | S | R | R |
| Tobramycin | R | S | R | R |
| Ampicillin | R | S | R | R |
| Cefepime | R | R | R | R |
| Ceftazidime | R | S | R | R |
| Ceftriaxone | S | S | S | I |
| Imipenem | R | S | R | R |
| Meropenem | R | S | R | R |
| TZP | R | S | R | R |
| Tigecycline | S | S | I | I |
| SXT | S | S | R | R |
| Minocycline | R | S | R | I |
| Genomic features | ||||
| MLST (Oxford) | ST218 | ST2528 | ST218 | ST218 |
| MLST (Pasture) | ST2 | ST2089 | ST2 | ST2 |
| Contigs | 75 | 21 | 150 | 65 |
| GC Content | 38.87 | 39.03 | 39.01 | 38.92 |
| Contig L50 | 9 | 2 | 22 | 9 |
| Contig N50 | 131,103 | 838,947 | 64,059 | 165,021 |
| CDS | 3904 | 3439 | 3822 | 3783 |
| tRNA | 64 | 63 | 59 | 57 |
| rRNA | 4 | 4 | 5 | 3 |
| Hypothetical proteins | 1007 | 671 | 938 | 930 |
| Proteins with functional assignments | 2897 | 2768 | 2884 | 2853 |
| Proteins with EC number assignments | 938 | 931 | 939 | 937 |
| Proteins with GO assignments | 806 | 799 | 808 | 806 |
| Proteins with pathway assignments | 726 | 728 | 728 | 728 |
| Proteins with PATRIC genus-specific family (PLfam) assignments | 3720 | 3352 | 3651 | 3646 |
| Proteins with PATRIC cross-genus family (PGfam) assignments | 3841 | 3385 | 3765 | 3730 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasir, M.; Subahi, A.M.; Shukri, H.A.; Bibi, F.; Sohrab, S.S.; Alawi, M.; Sindi, A.A.; Jiman-Fatani, A.A.; Azhar, E.I. Bacterial Community and Genomic Analysis of Carbapenem-Resistant Acinetobacter baumannii Isolates from the Environment of a Health Care Facility in the Western Region of Saudi Arabia. Pharmaceuticals 2022, 15, 611. https://doi.org/10.3390/ph15050611
Yasir M, Subahi AM, Shukri HA, Bibi F, Sohrab SS, Alawi M, Sindi AA, Jiman-Fatani AA, Azhar EI. Bacterial Community and Genomic Analysis of Carbapenem-Resistant Acinetobacter baumannii Isolates from the Environment of a Health Care Facility in the Western Region of Saudi Arabia. Pharmaceuticals. 2022; 15(5):611. https://doi.org/10.3390/ph15050611
Chicago/Turabian StyleYasir, Muhammad, Abdullah Mohammad Subahi, Hani A. Shukri, Fehmida Bibi, Sayed Sartaj Sohrab, Maha Alawi, Anees A. Sindi, Asif A. Jiman-Fatani, and Esam I. Azhar. 2022. "Bacterial Community and Genomic Analysis of Carbapenem-Resistant Acinetobacter baumannii Isolates from the Environment of a Health Care Facility in the Western Region of Saudi Arabia" Pharmaceuticals 15, no. 5: 611. https://doi.org/10.3390/ph15050611
APA StyleYasir, M., Subahi, A. M., Shukri, H. A., Bibi, F., Sohrab, S. S., Alawi, M., Sindi, A. A., Jiman-Fatani, A. A., & Azhar, E. I. (2022). Bacterial Community and Genomic Analysis of Carbapenem-Resistant Acinetobacter baumannii Isolates from the Environment of a Health Care Facility in the Western Region of Saudi Arabia. Pharmaceuticals, 15(5), 611. https://doi.org/10.3390/ph15050611









