Network Pharmacology and Bioinformatics Approach Reveals the Multi-Target Pharmacological Mechanism of Fumaria indica in the Treatment of Liver Cancer
Abstract
:1. Introduction
2. Results
2.1. Screening of Active Compounds and Targets
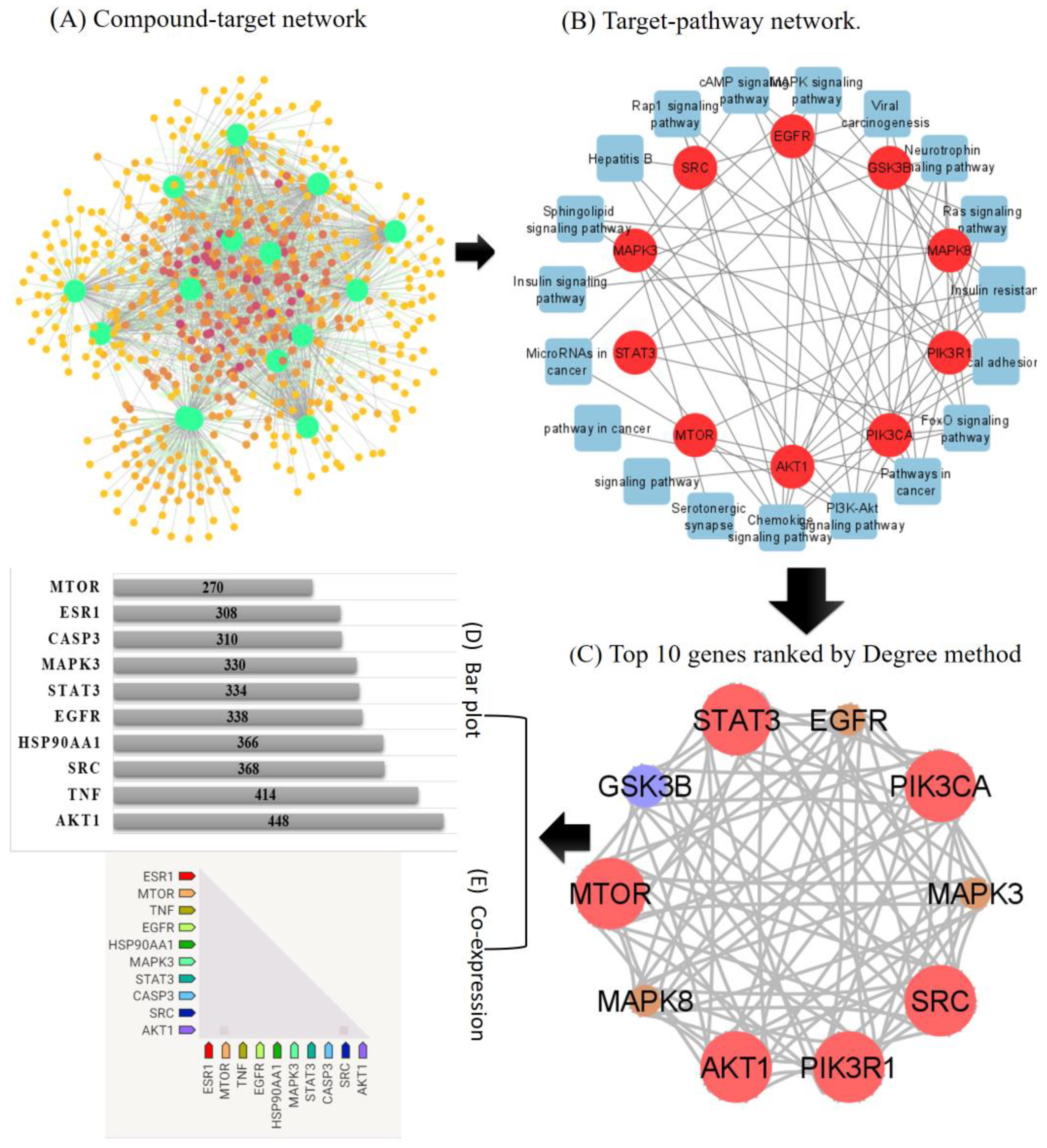
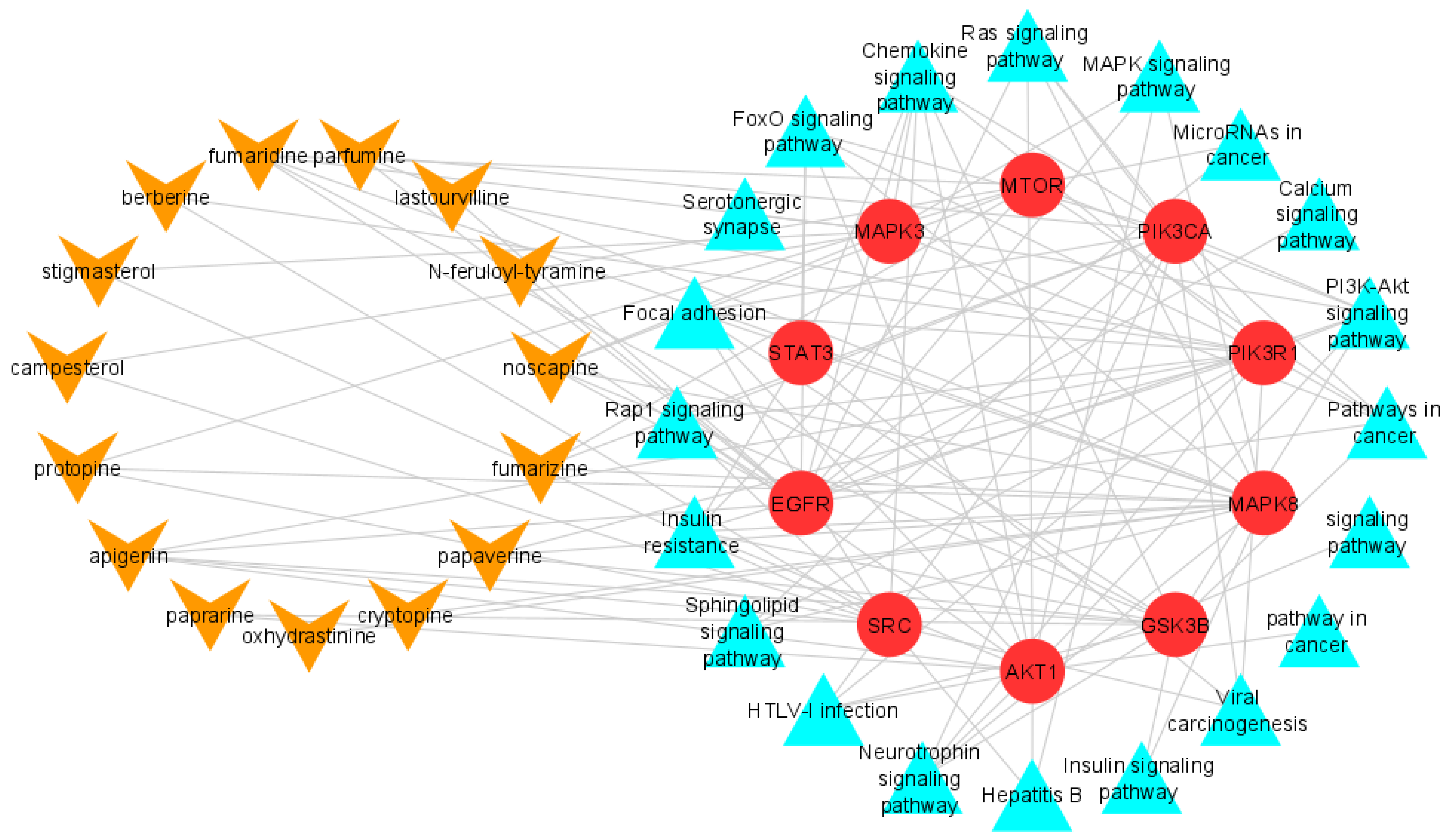
2.2. Compounds-Target Network Construction
2.3. PPI Network Construction
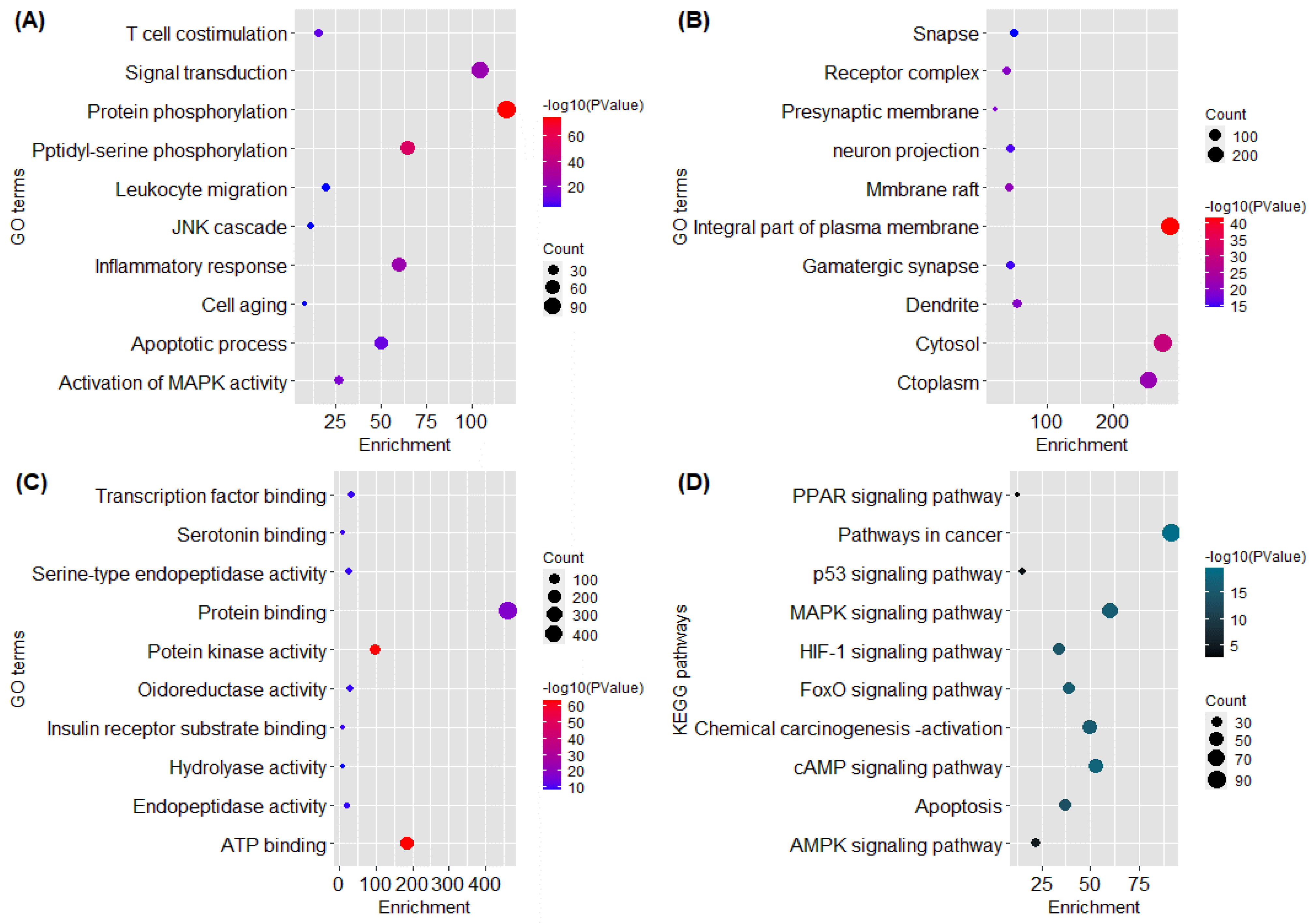
2.4. GO and KEGG Pathway Analysis
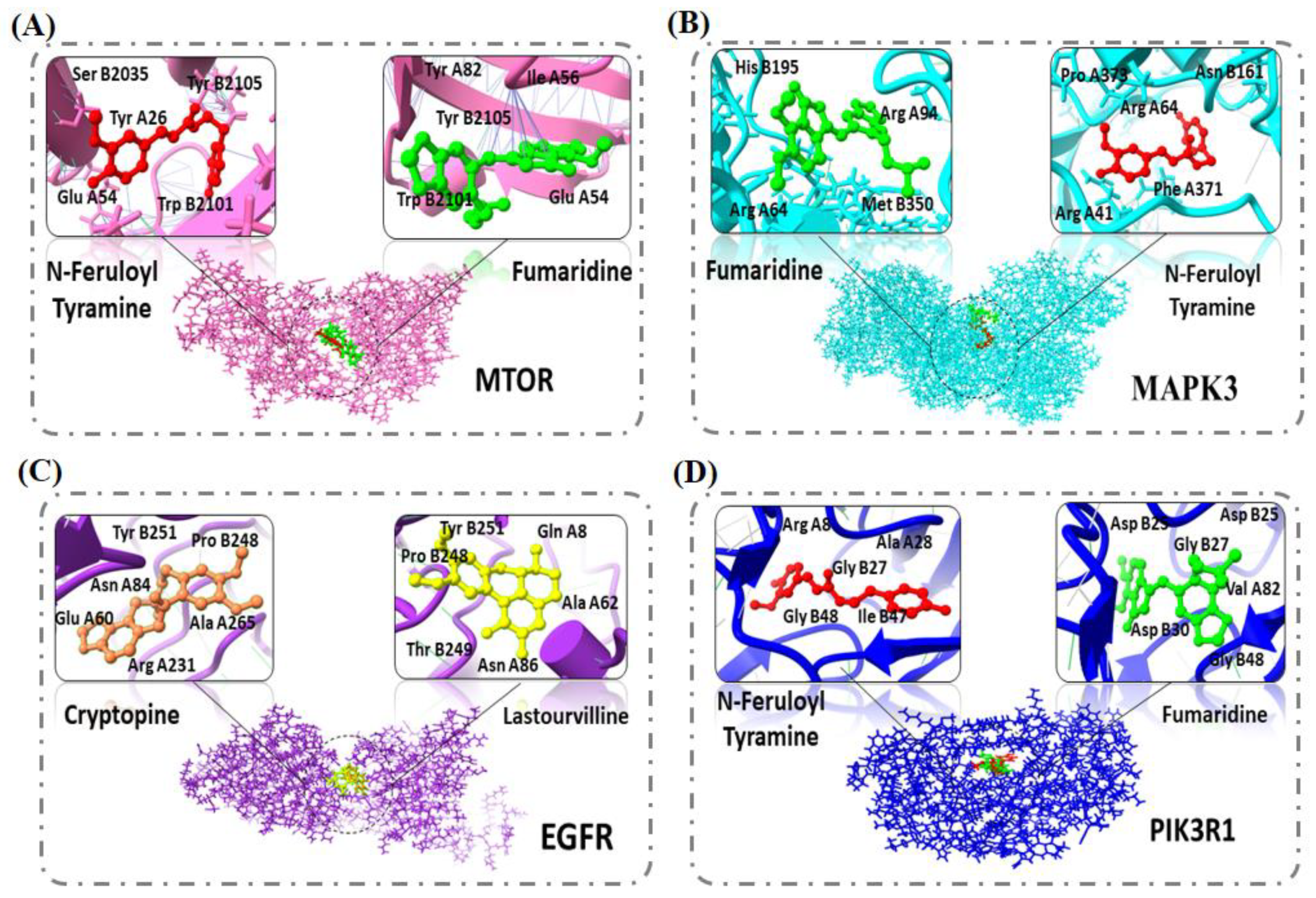
2.5. Molecular Docking
2.6. ADMET Profiling
3. Discussion
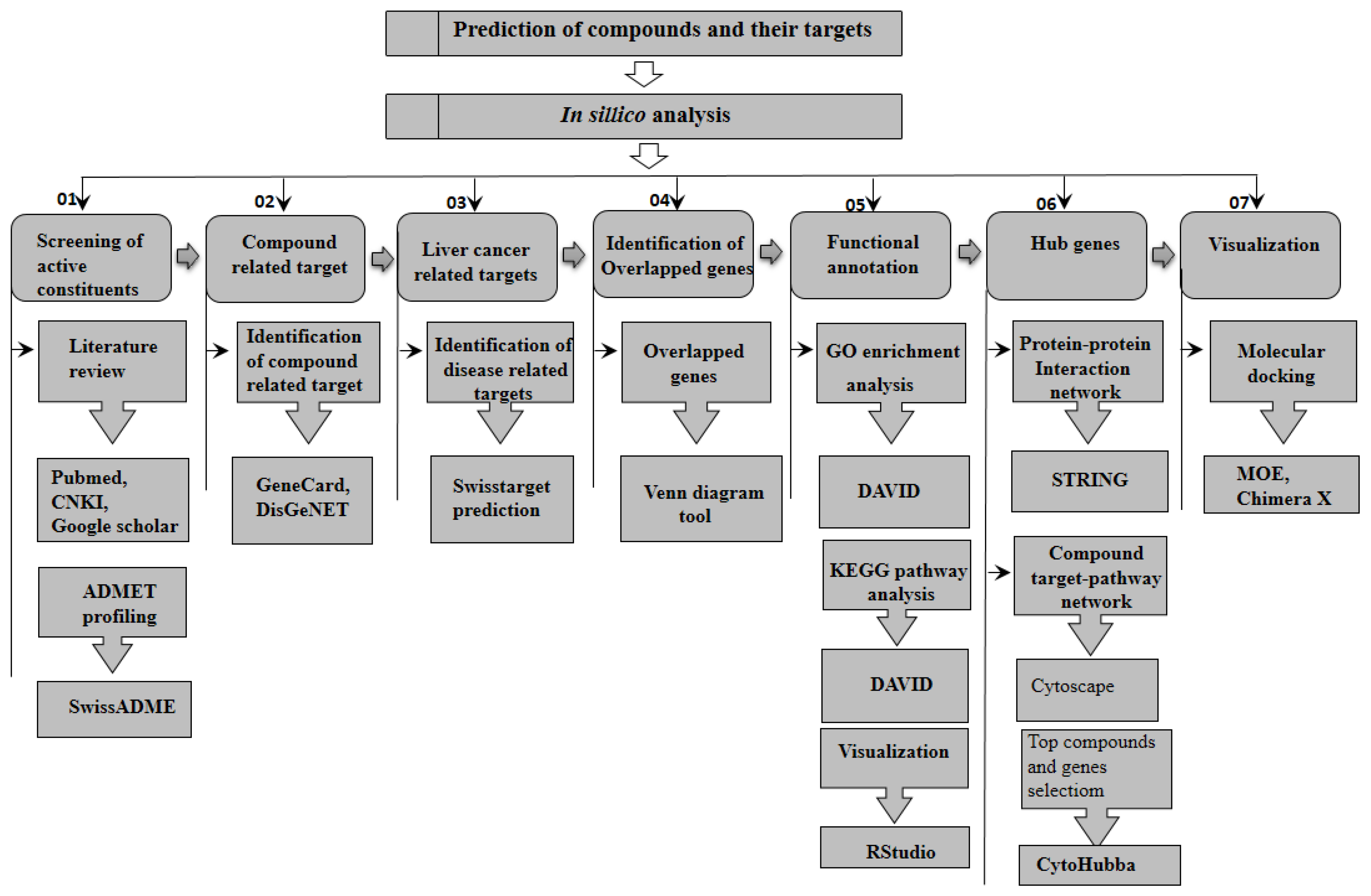
4. Materials and Methods
4.1. Virtual Screening of Active Constituents
4.2. Target Genes Screening
4.3. Pathway and Functional Enrichment Analysis
4.4. Network Construction
4.5. PPI Network Construction and Molecular Docking Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.; Zhu, J.; Wang, G.; Groopman, J.D.; Kensler, T.W. Qidong: A crucible for studies on liver cancer etiology and prevention. Cancer Biol. Med. 2019, 16, 24. [Google Scholar] [PubMed] [Green Version]
- Chan, L.-K.; Tsui, Y.-M.; Ho, D.W.-H.; Ng, I.O.-L. Cellular heterogeneity and plasticity in liver cancer. Semin. Cancer Biol. 2022, 82, 134–149. [Google Scholar] [CrossRef]
- Singh, A.K.; Kumar, R.; Pandey, A.K. Hepatocellular carcinoma: Causes, mechanism of progression and biomarkers. Curr. Chem. Genom. Transl. Med. 2018, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Z.; Zhao, X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J. Hematol. Oncol. 2019, 12, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakya, A.; Chatterjee, S.S.; Kumar, V. Role of fumarates in adaptogenics like efficacies of traditionally used Fumaria indica extracts. CELLMED 2015, 5, 6.1–6.10. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, Q.; Kennelly, E.J.; Long, C.; Chai, X. Diverse alkaloids and biological activities of Fumaria (Papaveraceae): An ethnomedicinal group. Fitoterapia 2020, 146, 104697. [Google Scholar] [CrossRef]
- Ataei, A.; Gholamalipour Alamdari, E.; Avarseji, Z.; Rahemi Karizaki, A. Study of allelopathic effect of aqueous extract of various organs of Fumaria parviflora on morphological, physiological and biochemical charateractics of Lolium rigidum. Appl. Biol. 2022, 34, 94–112. [Google Scholar]
- Hussain, T.; Siddiqui, H.H.; Fareed, S.; Sweety, K.; Vijayakumar, M.; Rao, C.V. Chemopreventive effect of Fumaria indica that modulates the oxidant-antioxidant imbalance during N-nitrosodiethylamine and CC14-induced hepatocarcinogenesis in Wistar rats. Asian Pac. J. Trop. Biomed. 2012, 2, S995–S1001. [Google Scholar] [CrossRef]
- Noor, F.; Rehman, A.; Ashfaq, U.A.; Saleem, M.H.; Okla, M.K.; Al-Hashimi, A.; AbdElgawad, H.; Aslam, S.J.P. Integrating Network Pharmacology and Molecular Docking Approaches to Decipher the Multi-Target Pharmacological Mechanism of Abrus precatorius L. Acting on Diabetes. Pharmaceuticals 2022, 15, 414. [Google Scholar] [CrossRef]
- Zhai, Z.; Tao, X.; Alami, M.M.; Shu, S.; Wang, X. Network Pharmacology and Molecular Docking Combined to Analyze the Molecular and Pharmacological Mechanism of Pinellia ternata in the Treatment of Hypertension. Curr. Issues Mol. Biol. 2021, 43, 65–78. [Google Scholar] [CrossRef]
- Noor, F.; Ahmad, S.; Saleem, M.; Alshaya, H.; Qasim, M.; Rehman, A.; Ehsan, H.; Talib, N.; Saleem, H.; Bin Jardan, Y.A.; et al. Designing a multi-epitope vaccine against Chlamydia pneumoniae by integrating the core proteomics, subtractive proteomics and reverse vaccinology-based immunoinformatics approaches. Comput. Biol. Med. 2022, 145, 105507. [Google Scholar] [CrossRef]
- Noor, F.; Ashfaq, U.A.; Javed, M.R.; Saleem, M.H.; Ahmad, A.; Aslam, M.F.; Aslam, S. Comprehensive computational analysis reveals human respiratory syncytial virus encoded microRNA and host specific target genes associated with antiviral immune responses and protein binding. J. King Saud Univ.-Sci. 2021, 33, 101562. [Google Scholar] [CrossRef]
- Noor, F.; Noor, A.; Ishaq, A.R.; Farzeen, I.; Saleem, M.H.; Ghaffar, K.; Aslam, M.F.; Aslam, S.; Chen, J.-T. Recent Advances in Diagnostic and Therapeutic Approaches for Breast Cancer: A Comprehensive Review. Curr. Pharm. Des. 2021, 27, 2344–2365. [Google Scholar] [CrossRef]
- Noor, F.; Saleem, M.H.; Aslam, M.F.; Ahmad, A.; Aslam, S. Construction of miRNA-mRNA network for the identification of key biological markers and their associated pathways in IgA nephropathy by employing the integrated bioinformatics analysis. Saudi J. Biol. Sci. 2021, 28, 4938–4945. [Google Scholar] [CrossRef]
- Noor, F.; Saleem, M.H.; Chen, J.-T.; Javed, M.R.; Al-Megrin, W.A.; Aslam, S. Integrative bioinformatics approaches to map key biological markers and therapeutic drugs in Extramammary Paget’s disease of the scrotum. PLoS ONE 2021, 16, e0254678. [Google Scholar]
- Li, L.; Qiu, H.; Liu, M.; Cai, Y. A network pharmacology-based study of the molecular mechanisms of shaoyao-gancao decoction in treating Parkinson’s disease. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 131–144. [Google Scholar] [CrossRef]
- Yu, S.; Gao, W.; Zeng, P.; Chen, C.; Zhang, Z.; Liu, Z.; Liu, J. Exploring the effect of Gupi Xiaoji Prescription on hepatitis B virus-related liver cancer through network pharmacology and in vitro experiments. Biomed. Pharmacother. 2021, 139, 111612. [Google Scholar] [CrossRef]
- Huang, J.; Cheung, F.; Tan, H.Y.; Hong, M.; Wang, N.; Yang, J.; Feng, Y.; Zheng, Q. Identification of the active compounds and significant pathways of yinchenhao decoction based on network pharmacology. Mol. Med. Rep. 2017, 16, 4583–4592. [Google Scholar] [CrossRef] [Green Version]
- Nikolaou, K.; Sarris, M.; Talianidis, I. Molecular pathways: The complex roles of inflammation pathways in the development and treatment of liver cancer. Clin. Cancer Res. 2013, 19, 2810–2816. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Shen, H.-M.; Lim, L.H.K. The role of autophagy in liver cancer: Crosstalk in signaling pathways and potential therapeutic targets. Pharmaceuticals 2020, 13, 432. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, X.; Li, Y.; Wang, Y.; Tang, K.; Wu, D.; Zhao, W.; Ma, Y.; Liu, P.; Cao, Z. Comparative network pharmacology analysis of classical TCM prescriptions for chronic liver disease. Front. Pharmacol. 2019, 10, 1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shawky, E. Prediction of potential cancer-related molecular targets of North African plants constituents using network pharmacology-based analysis. J. Ethnopharmacol. 2019, 238, 111826. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.-F.; Bian, Y.-Q.; Wu, R.; Sun, Y.; Chen, X.-L.; Yang, M.-D.; Zhang, Q.-R.; Hu, Y.; Sun, M.-Y.; Su, S.-B. Yinchenhao decoction suppresses rat liver fibrosis involved in an apoptosis regulation mechanism based on network pharmacology and transcriptomic analysis. Biomed. Pharmacother. 2019, 114, 108863. [Google Scholar] [CrossRef] [PubMed]
- Hungin, A.; Whorwell, P.; Tack, J.; Mearin, F. The prevalence, patterns and impact of irritable bowel syndrome: An international survey of 40,000 subjects. Aliment. Pharmacol. Ther. 2003, 17, 643–650. [Google Scholar] [CrossRef] [Green Version]
- Honorio, K.M.; Moda, T.L.; Andricopulo, A.D. Pharmacokinetic properties and in silico ADME modeling in drug discovery. Med. Chem. 2013, 9, 163–176. [Google Scholar] [CrossRef]
- Bocci, G.; Carosati, E.; Vayer, P.; Arrault, A.; Lozano, S.; Cruciani, G. ADME-Space: A new tool for medicinal chemists to explore ADME properties. Sci. Rep. 2017, 7, 6359. [Google Scholar] [CrossRef]
- Melo, A.P.S.; França, E.B.; Malta, D.C.; Garcia, L.P.; Mooney, M.; Naghavi, M. Mortality due to cirrhosis, liver cancer, and disorders attributed to alcohol use: Global Burden of Disease in Brazil, 1990 and 2015. Rev. Bras. Epidemiol. 2017, 20, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Rush, B.; Walley, K.R.; Celi, L.A.; Rajoriya, N.; Brahmania, M. Palliative care access for hospitalized patients with end—Stage liver disease across the United States. Hepatology 2017, 66, 1585–1591. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA A Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Adnan, M.; Jeon, B.-B.; Chowdhury, M.H.U.; Oh, K.-K.; Das, T.; Chy, M.N.U.; Cho, D.-H.J.L. Network Pharmacology Study to Reveal the Potentiality of a Methanol Extract of Caesalpinia sappan L. Wood against Type-2 Diabetes Mellitus. Life 2022, 12, 277. [Google Scholar] [CrossRef]
- Oh, K.-K.; Adnan, M.; Cho, D.-H.J. Network pharmacology-based study to uncover potential pharmacological mechanisms of Korean thistle (Cirsium japonicum var. maackii (maxim.) Matsum.) flower against cancer. Molecules 2021, 26, 5904. [Google Scholar] [CrossRef]
- Shakya, A.; Chatterjee, S.S.; Kumar, V. Holistic psychopharmacology of Fumaria indica (Fumitory). Chin. Med. 2012, 3, 26043. [Google Scholar] [CrossRef] [Green Version]
- Hördegen, P.; Hertzberg, H.; Heilmann, J.; Langhans, W.; Maurer, V. The anthelmintic efficacy of five plant products against gastrointestinal trichostrongylids in artificially infected lambs. Vet. Parasitol. 2003, 117, 51–60. [Google Scholar] [CrossRef]
- Gilani, A.H.; Bashir, S.; Janbaz, K.H.; Khan, A. Pharmacological basis for the use of Fumaria indica in constipation and diarrhea. J. Ethnopharmacol. 2005, 96, 585–589. [Google Scholar] [CrossRef]
- Akhtar, M.; Khan, Q.; Khaliq, T. Effects of Euphorbia prostrata and Fumaria parviflora in normoglycaemic and alloxan-treated hyperglycaemic rabbits. Planta Med. 1984, 50, 138–142. [Google Scholar] [CrossRef]
- Liu, B.; Bai, C. Regulatory Mechanisms of Coicis Semen on Bionetwork of Liver Cancer Based on Network Pharmacology. BioMed Res. Int. 2020, 2020, 5860704. [Google Scholar] [CrossRef]
- Gao, L.; Wang, X.-D.; Niu, Y.-Y.; Duan, D.-D.; Yang, X.; Hao, J.; Zhu, C.-H.; Chen, D.; Wang, K.-X.; Qin, X.-M. Molecular targets of Chinese herbs: A clinical study of hepatoma based on network pharmacology. Sci. Rep. 2016, 6, 24944. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-J.; Hu, M.-H.; Xu, F.-G.; Xu, H.-J.; She, J.-J.; Xia, H.-P. Understanding the inflammation-cancer transformation in the development of primary liver cancer. Hepatoma Res. 2018, 4, 29. [Google Scholar] [CrossRef]
- Ferrín, G.; Guerrero, M.; Amado, V.; Rodríguez-Perálvarez, M.; De la Mata, M. Activation of mTOR signaling pathway in hepatocellular carcinoma. Int. J. Mol. Sci. 2020, 21, 1266. [Google Scholar] [CrossRef] [Green Version]
- Wahlang, B.; McClain, C.; Barve, S.; Gobejishvili, L. Role of cAMP and phosphodiesterase signaling in liver health and disease. Cell. Signal. 2018, 49, 105–115. [Google Scholar] [CrossRef]
- Gupta, P.; Naithani, S.; Preece, J.; Kim, S.; Cheng, T.; D’Eustachio, P.; Elser, J.; Bolton, E.E.; Jaiswal, P. Plant Reactome and PubChem: The Plant Pathway and (Bio) Chemical Entity Knowledgebases. In Plant Bioinformatics; Humana Press: New York, NY, USA, 2022; pp. 511–525. [Google Scholar]
- Masri, D.E.; Alsaayed, B.; Masri, J.E.; Zreika, B.; Chanbour, H.; Salameh, P. Contribution of Arab countries to familial Mediterranean fever research: A PubMed-based bibliometric analysis. Rheumatol. Int. 2022, 42, 95–100. [Google Scholar] [CrossRef]
- Williams, A.J. Chemspider: A platform for crowdsourced collaboration to curate data derived from public compound databases. In Collaborative Computational Technologies for Biomedical Research; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; pp. 363–386. [Google Scholar]
- James, T.; Hsieh, M.-L.; Knipling, L.; Hinton, D. Determining the Architecture of a Protein–DNA Complex by Combining FeBABE Cleavage Analyses, 3-D Printed Structures, and the ICM Molsoft Program. In DNA-Protein Interactions; Humana Press: New York, NY, USA, 2015; pp. 29–40. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [Green Version]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [Green Version]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
| Molecule Name | Molecular Weight (MW) | Drug Likeness (DL) | Oral Bioavailability (OB) | Structure | PubChem ID |
|---|---|---|---|---|---|
| Protopine | 353.37 | 0.29 | 0.55 |  | 4970 |
| Fumaridine | 396.44 | 0.55 | 0.55 | 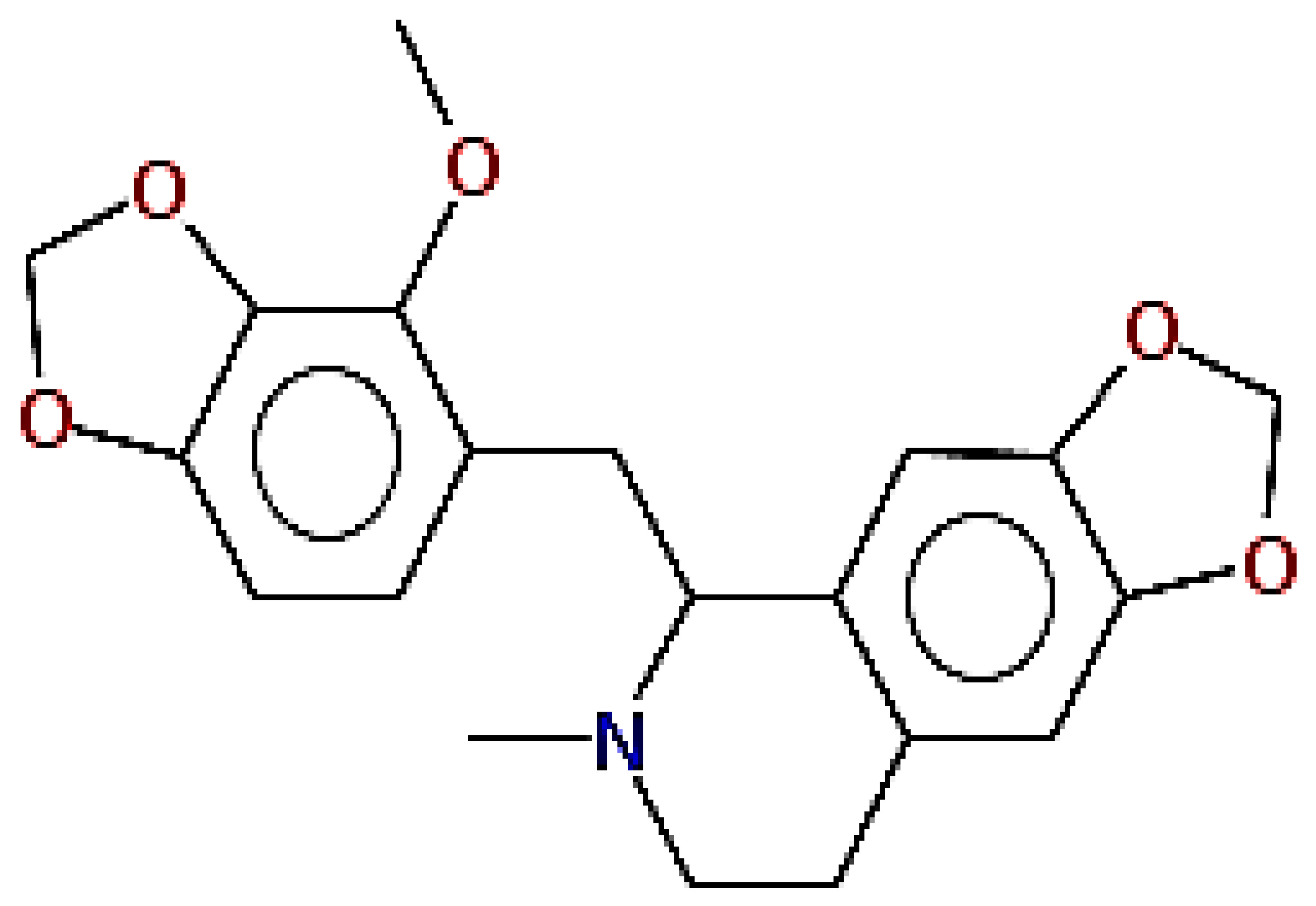 | 6537302 |
| Parfumine | 353.37 | 0.33 | 0.55 |  | 185623 |
| Lastourvilline | 327.37 | 0.74 | 0.55 | 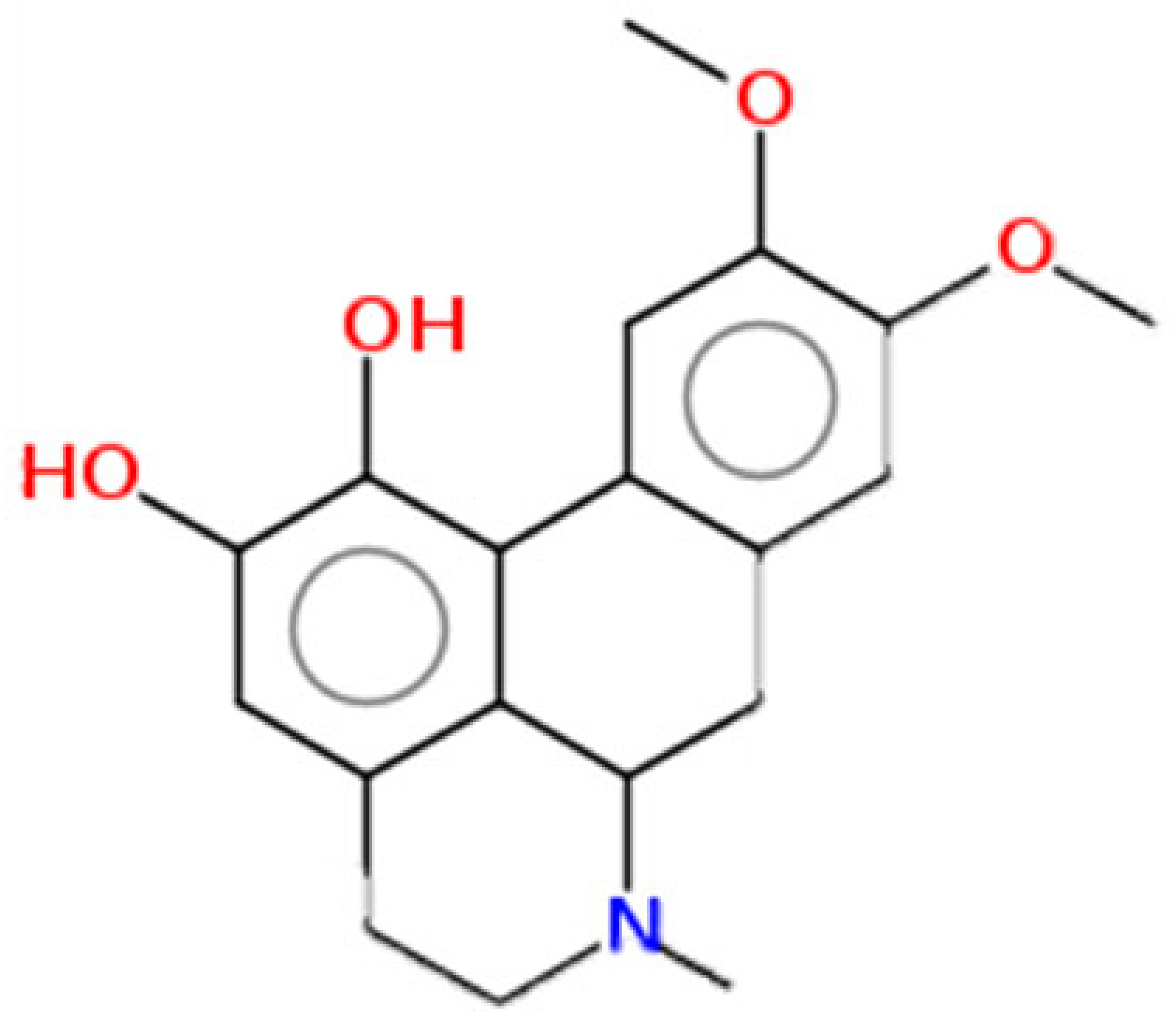 | 155514 |
| N-feruloyl tyramine | 313.35 | 0.21 | 0.55 | 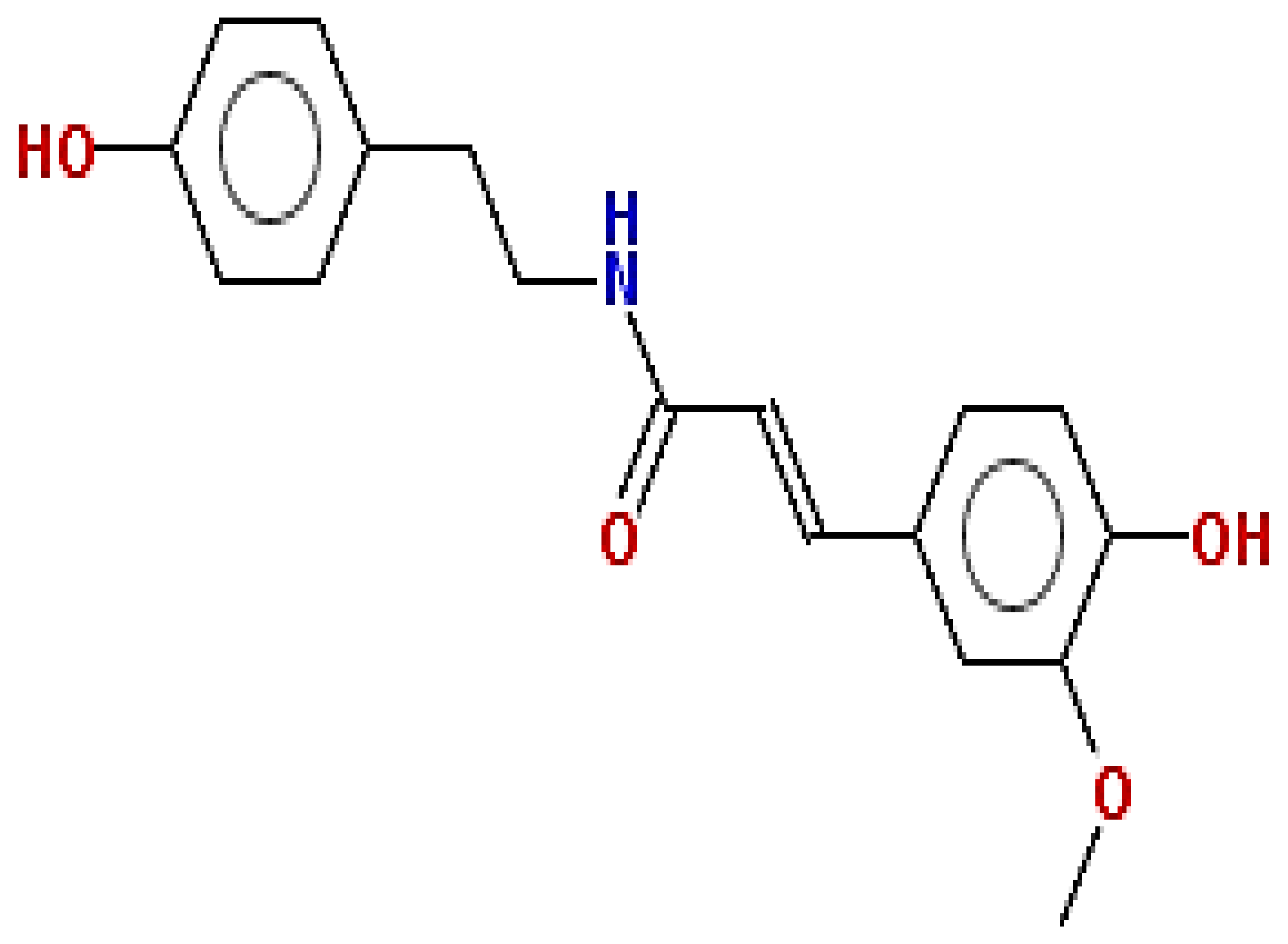 | 45257345 |
| Fumarizine | 355.38 | 1.06 | 0.55 |  | 11131999 |
| Paprarine | 397.38 | 0.3 | 0.55 | 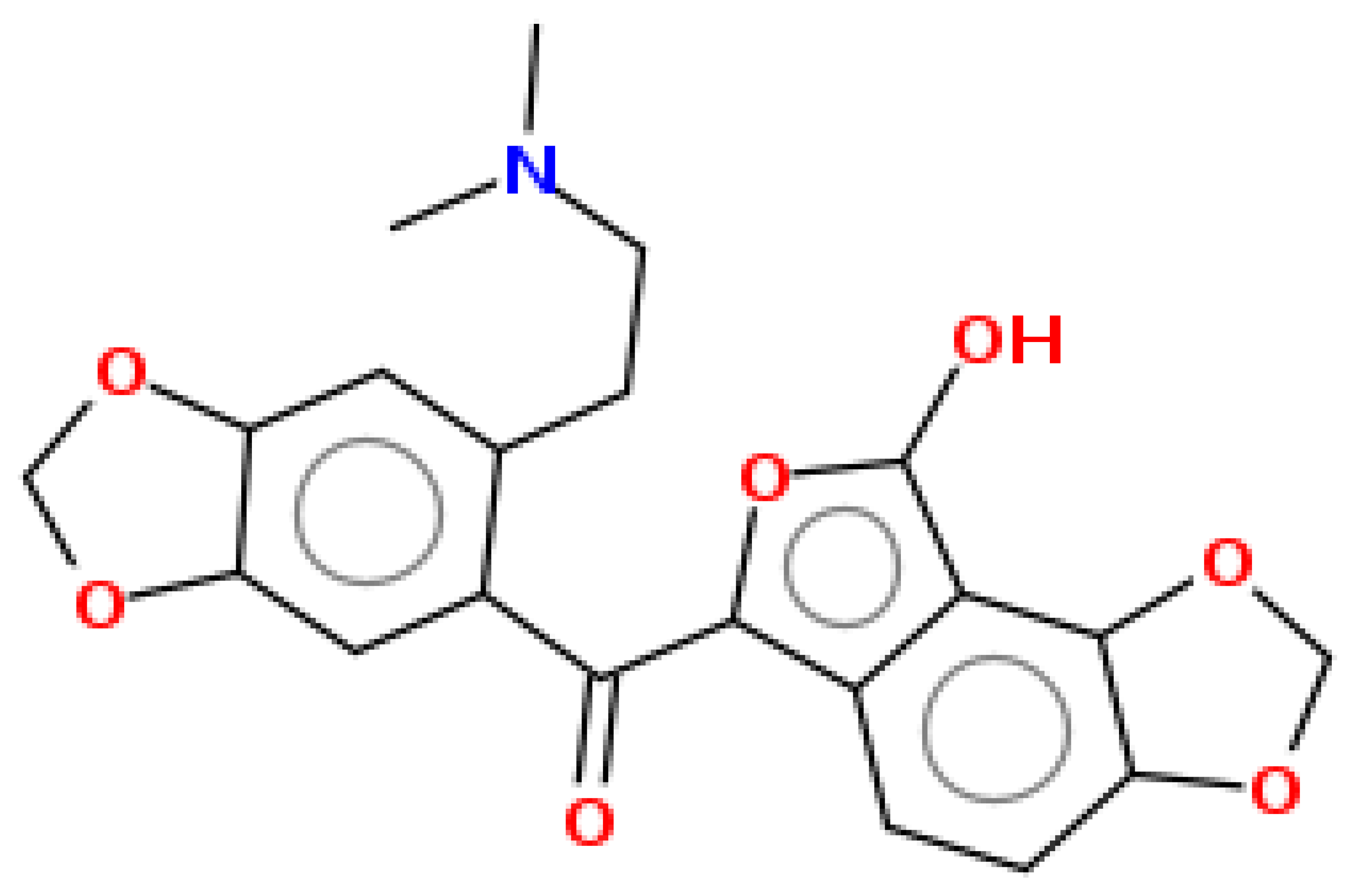 | 15226320 |
| Cryptopine | 369.41 | 0.35 | 0.55 | 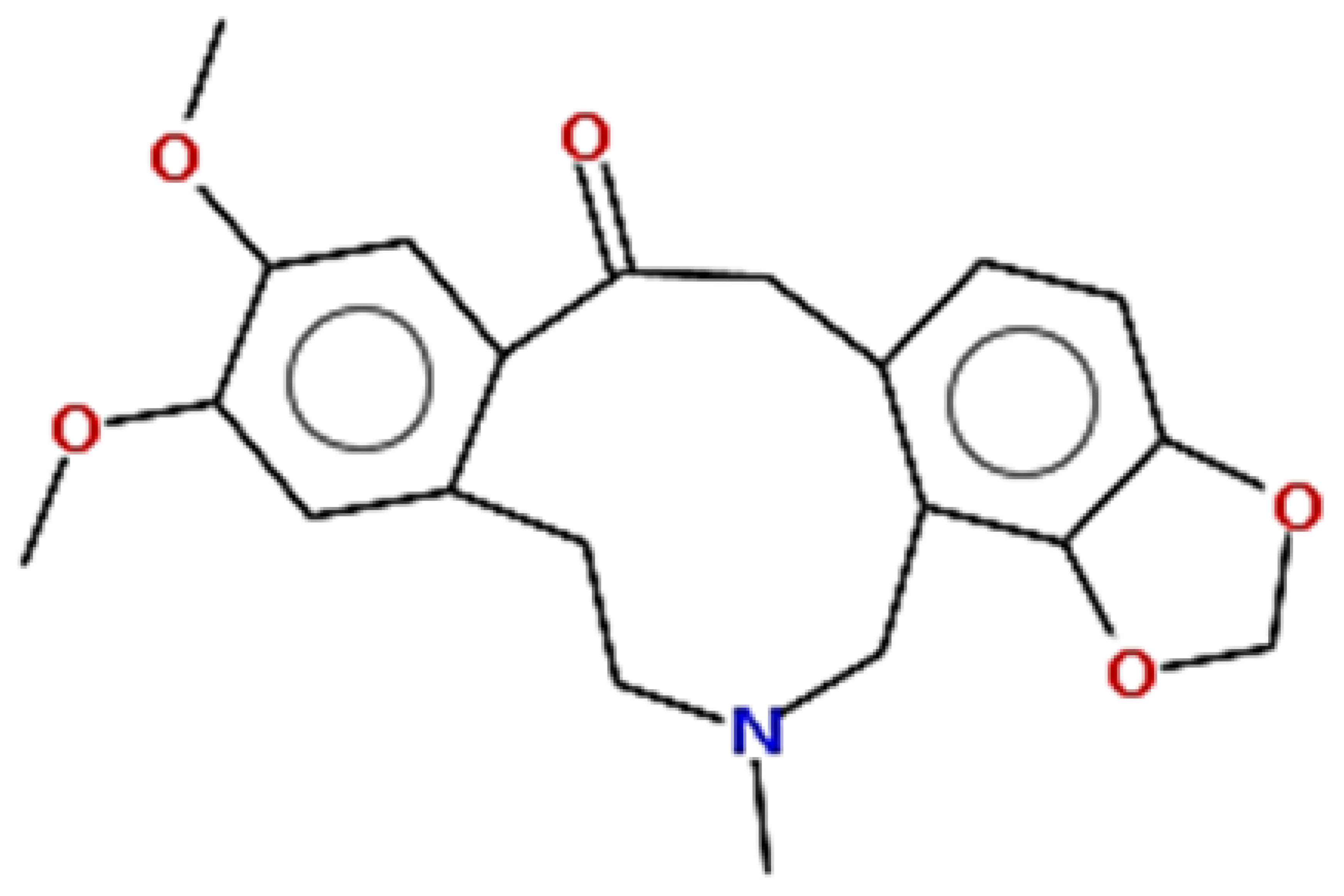 | 72616 |
| Berberine | 336.36 | 0.77 | 0.55 |  | 2353 |
| Stigmesterol | 412.69 | 0.62 | 0.55 | 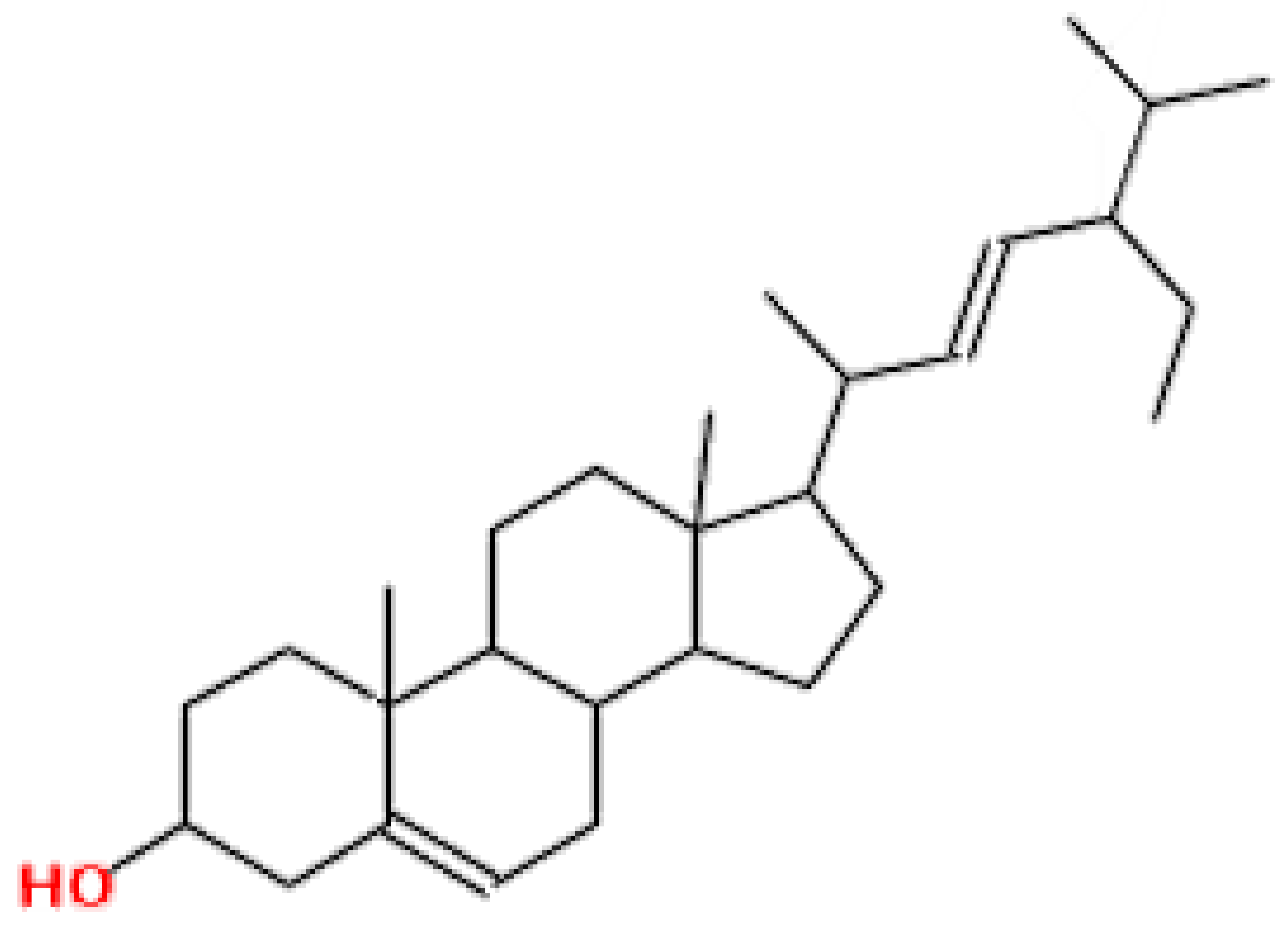 | 5280794 |
| Campesterol | 400.68 | 0.59 | 0.55 |  | 173183 |
| Papaverine | 339.39 | 0.75 | 0.55 | 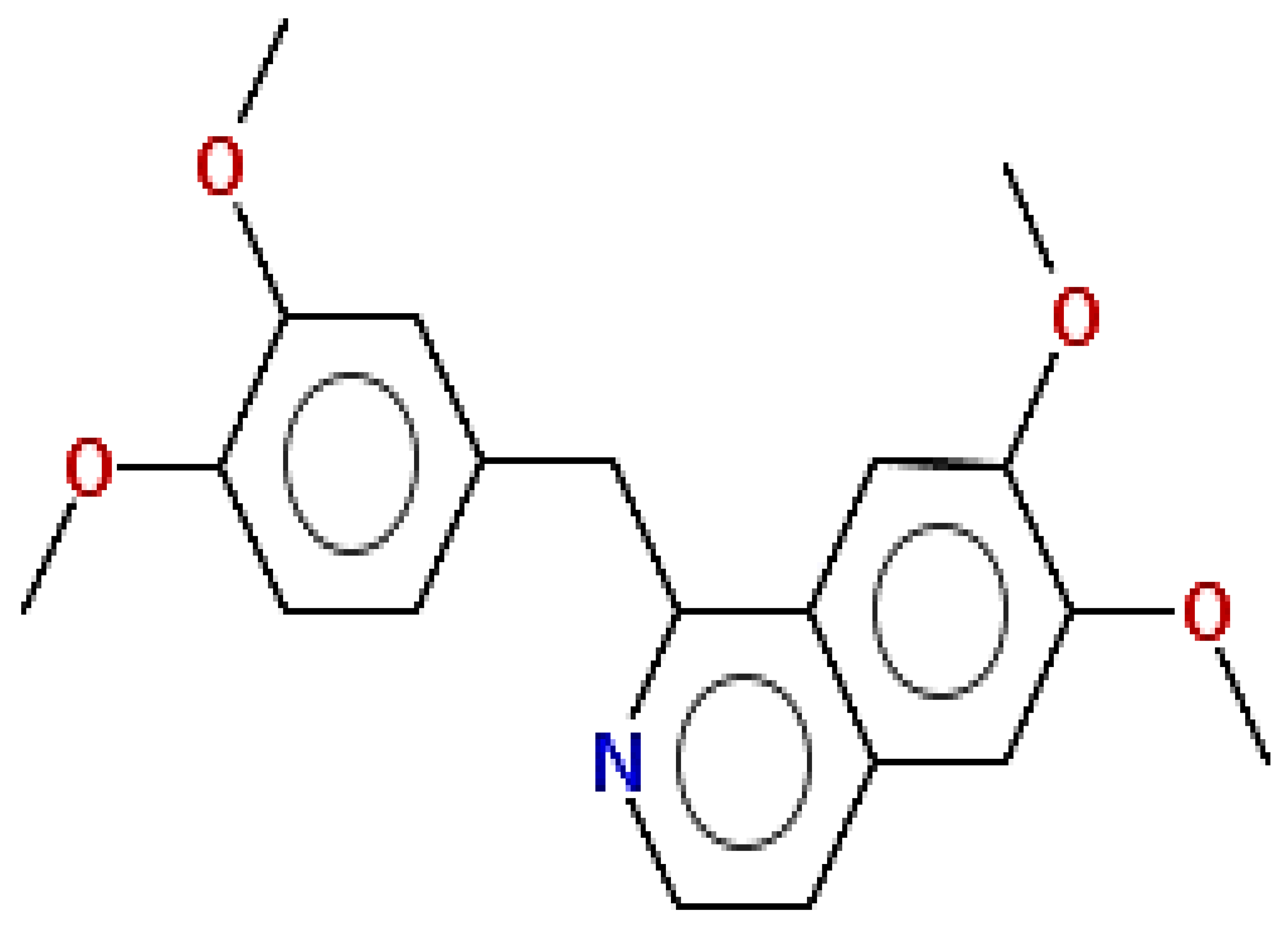 | 4680 |
| Oxyhydrastinine | 205.21 | 0.18 | 0.55 | 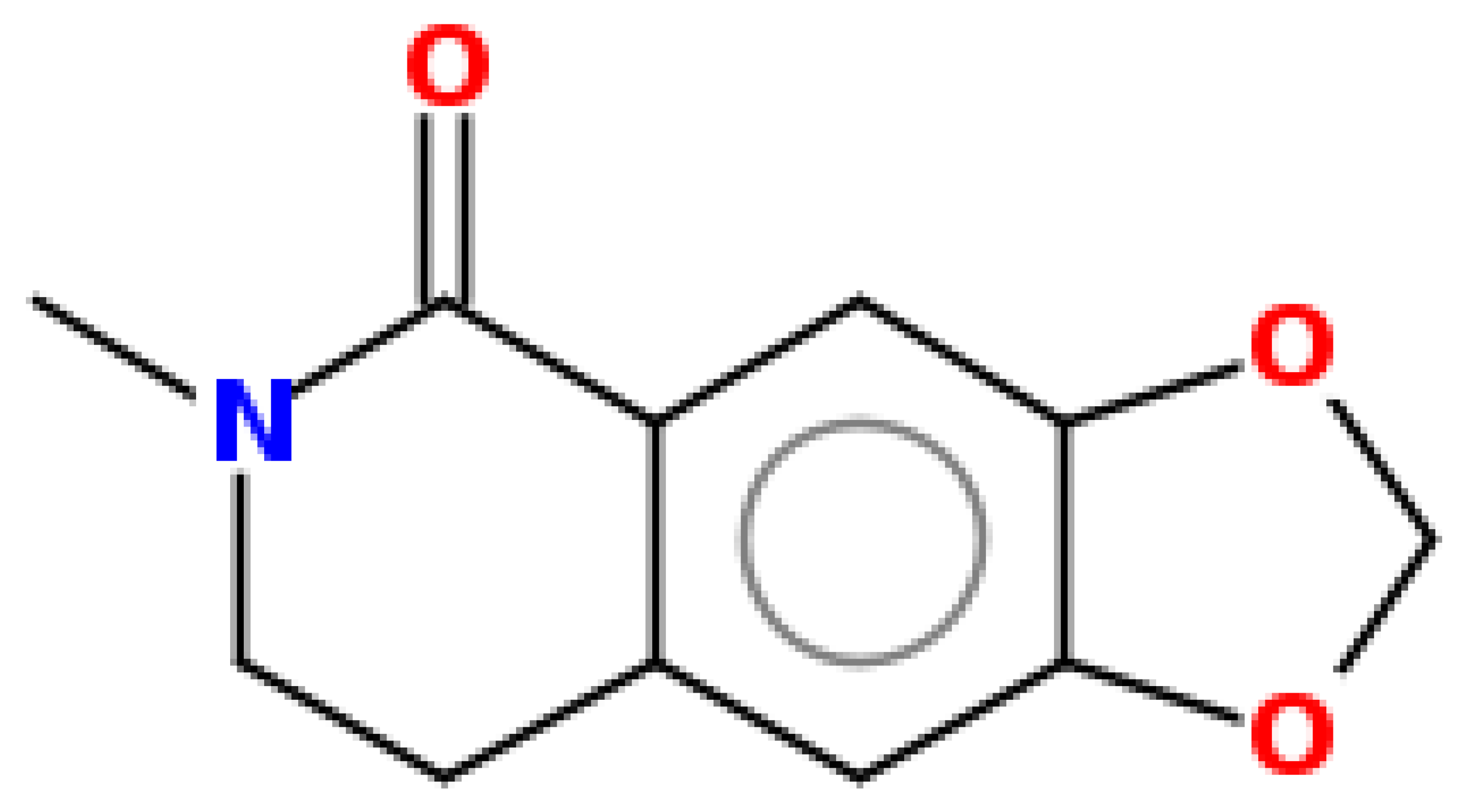 | 160522 |
| Noscapine | 413.42 | 0.54 | 0.55 |  | 275196 |
| Apigenin | 270.24 | 0.39 | 0.55 |  | 5280443 |
| Molecule Name | Class | Degree |
|---|---|---|
| Protopine | Alkaloids | 4 |
| Fumaridine | Alkaloids | 8 |
| Parfumine | Alkaloids | 3 |
| Lastourvilline | Alkylamides | 1 |
| N-Feruloyltyramine | Tyramines | 4 |
| Cryptopine | Alkaloids | 7 |
| Berberine | Alkaloids | 3 |
| Stigmasterol | Steroid | 3 |
| Campesterol | Steroid | 2 |
| Papaverine | Alkaloids | 2 |
| Oxhydrastinine | Alkaloids | 2 |
| Noscapine | Alkaloids | 6 |
| Gene Name | Compounds | Score | Pathways |
|---|---|---|---|
| AKT1 | Fumaridine/Paprarine/Apigenin | 224 | Neuroactive ligand–receptor interaction, pathways in cancer, cAMP signaling pathway, |
| TNF | N-feruloyl tyramine | 207 | Proteoglycans in cancer, MAPK signaling pathway, insulin resistance |
| SRC | Protopine/Stigmasterol/Fumaridine/Berberine Campesterol/Cryptopine/Apigenin | 184 | Chemokine signaling pathway, viral carcinogenesis |
| EGFR | Fumaridine/Parfumine/Lastourvilline/N-feruloyl tyramine/Noscapine/Apigenin | 169 | Focal adhesion, Rap1 signaling pathway, serotonergic synapse |
| STAT3 | Cryptopine | 167 | Pathway in cancer, proteoglycans in cancer, FoxO signaling pathway |
| MAPK3 | Fumaridine/Cryptopine/Stigmasterol/Campesterol/, Noscapine | 165 | Viral carcinogenesis, focal adhesion, Rap1 signaling pathway |
| CASP3 | Oxyhydrastinine | 155 | Pathways in cancer, proteoglycans in cancer, MAPK signaling pathway, Hepatitis B |
| MTOR | Protopine/Fumaridine/N-feruloyl tyramine/Fumarizine, Cryptopine/ Noscapine | 135 | MicroRNAS in cancer, insulin resistance |
| MAPK1 | Protopine/Fumaridine/Cryptopine/Noscapine | 134 | Neurotrophin signaling pathway, serotonergic synapse |
| PIK3R1 | Apigenin/Lastourvilline/Cryptopine/Berberine/Papaverine | 94 | Sphingolipid signaling pathway, Hepatitis B |
| MTOR | ||||
|---|---|---|---|---|
| Compound ID | Compound Name | Docking Score (kcal/mol) | RMSD | Hydrogen Bond and Other Interacting Residues |
| 6537302 | Fumaridine | −13.86 | 1.71 | Tyr A82, Tyr B2105, Phe B2108, Phe B2039, Ile A56, Phe A46, Glu A54, Trp B2101 |
| 5280537 | N-feruloyl tyramine | −12.94 | 0.93 | Ser B2035, Glu A52, Tyr A26, Phe A46, Asp A37, Arg A42, Thr B2098, Asp B2102, Lys B2095, Trp B2101, Phe B2039, Tyr B2105, Phe B2108, Leu B2031 |
| 72616 | Cryptopine | −10.95 | 1.4349 | Phe B2039, Ile A56, Tyr A82, Thr B2098, Arg A42, Asp A37, Phe A46, Asp B2102 |
| 155514 | Lastourvilline | −9.77 | 2.9765 | Phe B2039, Tyr A82, Glu A54, Ser B2035, Trp B2101, Ser B203 |
| Standard Drug | ||||
| 54675783 | Minocycline | −7.79 | 0.79 | ASP A37 PHE B2039 |
| MAPK3 | ||||
| 6537302 | Fumaridine | −12.32 | 1.17 | His B195, Arg A64 Met B350, Arg A94 |
| 5280537 | N-feruloyl tyramine | −12.07 | 1.77 | Asn B161, Phe A371, Arg A64, Pro A373, Arg A41, Thr B347, Glu B194, Arg A104 |
| 72616 | Cryptopine | −10.1110 | 1.01 | Arg A64, Arg A41, Arg A104, Asp A105, Phe A371, Pro B193, Asn B161, Asp B192, IIe B190 |
| 155514 | Lastourvilline | −11.0807 | 1.31 | Arg A41, Arg A64, Thr B347, Glu B194, Pro B193, Asn B161, Phe A371, Arg A370, Pro A373, Asp A105 |
| Standard Drug | ||||
| 54675783 | Minocycline | −7.75 | 2.9235 | Arq A41,Asp B192 |
| EGFR | ||||
| 155514 | Lastourvilline | −12.6598 | 1.8080 | Tyr B251, Gln A8, Leu A38, Ala A62, Asn A86, Thr B249, Pro B248, Lys A407 |
| 72616 | Cryptopine | −10.2961 | 0.7945 | Tyr B251, Arg A84 Ala A62, Thr B249, Pro B248, Asn A86, Glu A60, Arg A231, Ala A265, Leu A38, Gly A264 |
| 6537302 | Fumaridine | −10.12 | 1.5566 | Tyr B251, Lys A322, Asn A86, Thr B249, Ala A62, Pro B248, Leu A38, Lys A407, Met A87 |
| 5280537 | N-feruloyl tyramine | −10.27 | 2.4809 | Asn A12, His A409, Ser A11, Gly A410, Thr A10, Arg A285, Lys A407, Arg A405, Tyr B251, Leo A38, Gln A8, Gly C39 |
| Standard Drug | ||||
| 176870 | Erlotinib | −8.06 | 2.3660 | Arg A231 |
| PIK3R1 | ||||
| 6537302 | Fumaridine | −12.10 | 3.0608 | Agr A8, Asp B30, Val A82, Ile A84, Asp B25, Gly B27, Gly B48, Ala B28, Asp B29, Asp B30 |
| 5280537 | N-feruloyl tyramine | −10.69 | 3.4899 | Arg A8, Ala A28 Gly B27, Asp A25 Ile A84, Asp B25, Gly B48, Ile B47, Asp B29, Ile A50 Gly A49, Ile B50 |
| 72616 | Cryptopine | −10.45 | 1.2321 | Arg A8, Ala A28, Asp A25, Asp B25, Ala B28, Gly B27, Leu A23, Arg A8, Val A82, Gly B48, Pro A81, Ile B50, Gly A49 |
| 155514 | Lastourvilline | −10.02 | 0.7393 | Asp B30, Ala B28, Arg A8, Asp A25, Pro A31, Ile B50, Val A32, Gly B49, Gly B48, Asp B29 |
| Standard Drug | ||||
| 49867926 | XL-765 | −9.80 | 1.73 | Asp B25 |
| Standard Parameters | Fumaridine | N-Feruloyl Tyramine | Cryptopine | Lastourvilline |
|---|---|---|---|---|
| GI absorption | High | High | High | High |
| BBB | Yes | No | Yes | Yes |
| P-gp substrate | Yes | No | Yes | Yes |
| CYP1A2 inhibitor | No | No | Yes | Yes |
| CYP2C19 inhibitors | Yes | No | No | No |
| CYP2C9 inhibitors | Yes | No | Yes | No |
| CYP2D6 inhibitors | Yes | Yes | Yes | Yes |
| CYP3A4 inhibitors | Yes | Yes | Yes | Yes |
| Log Kp (skin permeation) | −6.62 cm/s | −6.72 cm/s | −6.48 cm/s | −6.71 cm/s |
| Toxicity | ||||
| Carcinogens | Non-carcinogenic | Non-carcinogenic | Non-carcinogenic | Non-carcinogenic |
| Cytotoxicity | Non-toxic | Non-toxic | Non-toxic | Non-toxic |
| Mutagenicity | Nil | Nil | Nil | Nil |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batool, S.; Javed, M.R.; Aslam, S.; Noor, F.; Javed, H.M.F.; Seemab, R.; Rehman, A.; Aslam, M.F.; Paray, B.A.; Gulnaz, A. Network Pharmacology and Bioinformatics Approach Reveals the Multi-Target Pharmacological Mechanism of Fumaria indica in the Treatment of Liver Cancer. Pharmaceuticals 2022, 15, 654. https://doi.org/10.3390/ph15060654
Batool S, Javed MR, Aslam S, Noor F, Javed HMF, Seemab R, Rehman A, Aslam MF, Paray BA, Gulnaz A. Network Pharmacology and Bioinformatics Approach Reveals the Multi-Target Pharmacological Mechanism of Fumaria indica in the Treatment of Liver Cancer. Pharmaceuticals. 2022; 15(6):654. https://doi.org/10.3390/ph15060654
Chicago/Turabian StyleBatool, Sara, Muhammad Rizwan Javed, Sidra Aslam, Fatima Noor, Hafiz Muhammad Faizan Javed, Riffat Seemab, Abdur Rehman, Muhammad Farhan Aslam, Bilal Ahamad Paray, and Aneela Gulnaz. 2022. "Network Pharmacology and Bioinformatics Approach Reveals the Multi-Target Pharmacological Mechanism of Fumaria indica in the Treatment of Liver Cancer" Pharmaceuticals 15, no. 6: 654. https://doi.org/10.3390/ph15060654
APA StyleBatool, S., Javed, M. R., Aslam, S., Noor, F., Javed, H. M. F., Seemab, R., Rehman, A., Aslam, M. F., Paray, B. A., & Gulnaz, A. (2022). Network Pharmacology and Bioinformatics Approach Reveals the Multi-Target Pharmacological Mechanism of Fumaria indica in the Treatment of Liver Cancer. Pharmaceuticals, 15(6), 654. https://doi.org/10.3390/ph15060654







