Benzothiazole Derivatives Endowed with Antiproliferative Activity in Paraganglioma and Pancreatic Cancer Cells: Structure–Activity Relationship Studies and Target Prediction Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
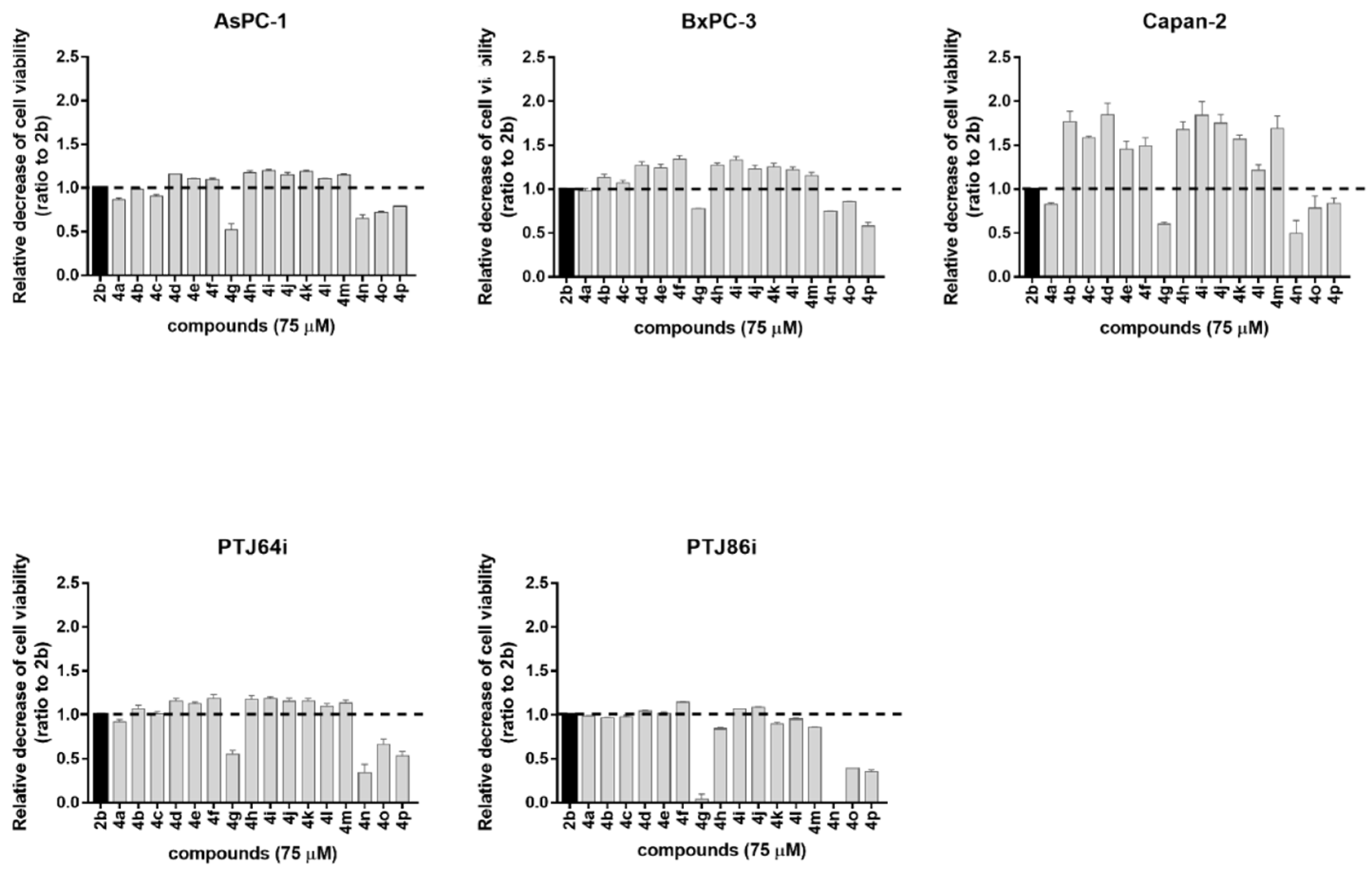
2.2. Antiproliferative Activity
2.3. Target Prediction Studies
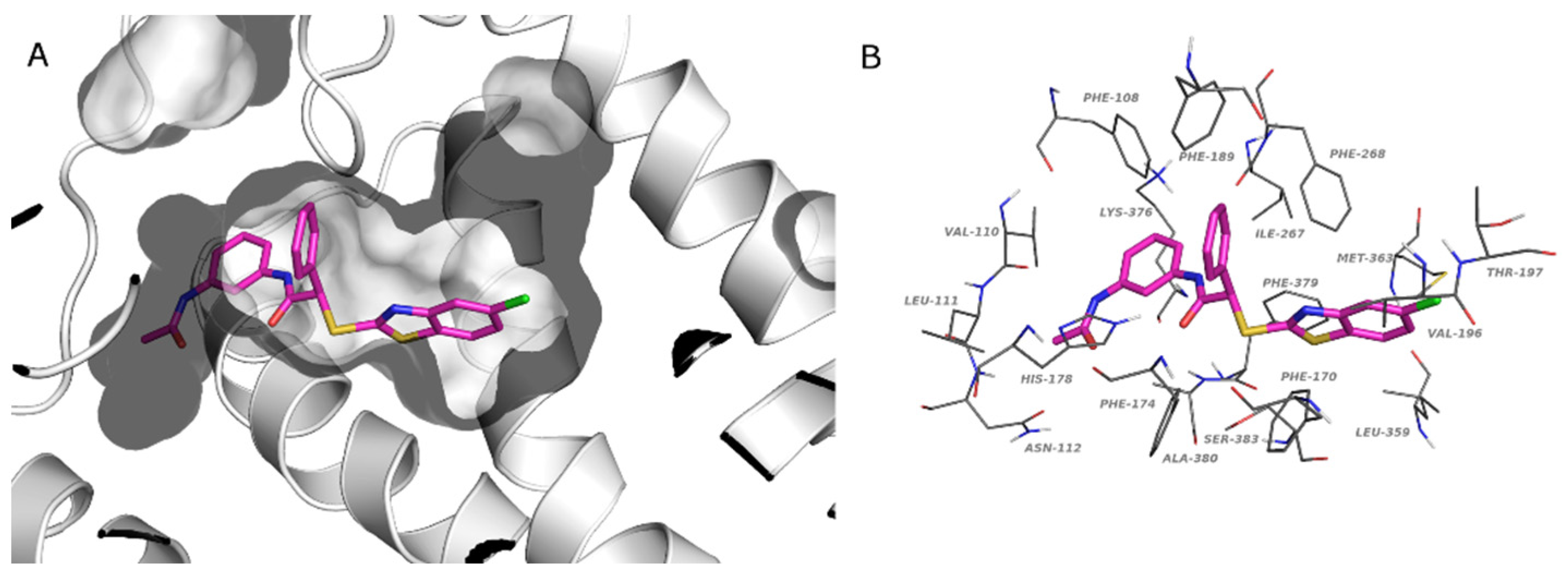
2.4. Docking Studies
2.5. Physicochemical and Pharmacokinetic Properties Calculation
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of Ethyl 2-[(Methylsulfonyl)oxy]-2-phenylacetate 1
3.1.2. Synthesis of Ethyl 2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-2-phenylacetate 2
3.1.3. Synthesis of 2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-2-phenylacetic Acid 3
3.1.4. General Procedure for the Synthesis of Amides 4a–p
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(4-methoxyphenyl)-2-phenylacetamide 4a
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(4-chlorophenyl)-2-phenylacetamide 4b
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(4-fluorophenyl)-2-phenylacetamide 4c
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-2-phenyl-N-[4-(trifluoromethyl)phenyl] Acetamide 4d
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(4-nitrophenyl)-2-phenylacetamide 4e
N-(4-Acetamidophenyl)-2-[(5-chlorobenzo[d]thiazol-2-yl)thio]-2-phenylacetamide 4f
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(3-methoxyphenyl)-2-phenylacetamide 4g
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(3-chlorophenyl)-2-phenylacetamide 4h
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(3-fluorophenyl)-2-phenylacetamide 4i
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-2-phenyl-N-[3-(trifluoromethyl) phenyl]acetamide 4j
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(3-nitrophenyl)-2-phenylacetamide 4k
N-(3-Acetamidophenyl)-2-[(5-chlorobenzo[d]thiazol-2-yl)thio]-2-phenylacetamide 4l
2-[(5-Chlorobenzo[d]thiazol-2-yl)thio]-N-(3,4-dichlorophenyl)-2-phenyl Acetamide 4m
N-(2-Bromo-5-nitrophenyl)-2-[(5-chlorobenzo[d]thiazol-2-yl)thio]-2-phenyl Acetamide 4n
N-[2-Bromo-4-(trifluoromethyl)phenyl]-2-[(5-chlorobenzo[d]thiazol-2-yl)thio]-2-phenylacetamide 4o
N-[2-Bromo-5-(trifluoromethyl)phenyl]-2-[(5-chlorobenzo[d]thiazol-2-yl)thio]-2-phenylacetamide 4p
3.2. Cell Lines, Treatments, and Cell Viability Assay
3.3. Calculation of Half Maximal Inhibitory Concentration (IC50), Selectivity Index (SI), Combination Index (CI) Values and Statistical Analysis
3.4. In Silico Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, P.C.; Sinhmar, A.; Sharma, A.; Rajak, H.; Pathak, D.P. Medicinal significance of benzothiazole scaffold: An insight view. J. Enzyme Inhib. Med. Chem. 2013, 28, 240–266. [Google Scholar] [CrossRef]
- Weekes, A.A.; Westwell, A.D. 2-Arylbenzothiazole as a privileged scaffold in drug discovery. Curr. Med. Chem. 2009, 16, 2430–2440. [Google Scholar] [CrossRef]
- Zhilitskaya, L.V.; Shainyan, B.A.; Yarosh, N.O. Modern approaches to the synthesis and transformations of practically valuable benzothiazole derivatives. Molecules 2021, 26, 2190. [Google Scholar] [CrossRef]
- Keri, R.S.; Patil, M.R.; Patil, S.A.; Budagumpi, S. A comprehensive review in current developments of benzothiazole-based molecules in medicinal chemistry. Eur. J. Med. Chem. 2015, 89, 207–251. [Google Scholar] [CrossRef]
- Irfan, A.; Batool, F.; Zahra Naqvi, S.A.; Islam, A.; Osman, S.M.; Nocentini, A.; Alissa, S.A.; Supuran, C.T. Benzothiazole derivatives as anticancer agents. J. Enzyme Inhib. Med. Chem. 2020, 35, 265–279. [Google Scholar] [CrossRef] [Green Version]
- Ammazzalorso, A.; Carradori, S.; Amoroso, R.; Fernández, I.F. 2-substituted benzothiazoles as antiproliferative agents: Novel insights on structure-activity relationships. Eur. J. Med. Chem. 2020, 207, 112762. [Google Scholar] [CrossRef]
- Bradshaw, T.D.; Stevens, M.F.; Westwell, A.D. The discovery of the potent and selective antitumour agent 2-(4-amino-3-methylphenyl)benzothiazole (DF 203) and related compounds. Curr. Med. Chem. 2001, 8, 203–210. [Google Scholar] [CrossRef]
- Bradshaw, T.D.; Westwell, A.D. The development of the antitumour benzothiazole prodrug, Phortress, as a clinical candidate. Curr. Med. Chem. 2004, 11, 1009–1021. [Google Scholar] [CrossRef]
- Strachan, D.C.; Ruffell, B.; Oei, Y.; Bissell, M.J.; Coussens, L.M.; Pryer, N.; Daniel, D. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8+ T cells. Oncoimmunology 2013, 2, e26968. [Google Scholar] [CrossRef] [Green Version]
- Krauser, J.A.; Jin, Y.; Walles, M.; Pfaar, U.; Sutton, J.; Wiesmann, M.; Graf, D.; Pflimlin-Fritschy, V.; Wolf, T.; Camenisch, G.; et al. Phenotypic and metabolic investigation of a CSF-1R kinase receptor inhibitor (BLZ945) and its pharmacologically active metabolite. Xenobiotica 2015, 45, 107–123. [Google Scholar] [CrossRef]
- Lin, C.-C.; Gil-Martin, M.; Bauer, T.M.; Naing, A.; Wan-Teck Lim, D.; Sarantopoulos, J.; Geva, R.; Ando, Y.; Fan, L.; Choudhury, S.; et al. Abstract nr CT171: Phase I study of BLZ945 alone and with spartalizumab (PDR001) in patients (pts) with advanced solid tumors [abstract]. Cancer Res. 2020, 80 (Suppl. 16), CT171. [Google Scholar] [CrossRef]
- El-Damasy, A.K.; Cho, N.C.; Nam, G.; Pae, A.N.; Keum, G. Discovery of a nanomolar multikinase inhibitor (KST016366): A new benzothiazole derivative with remarkable broad-spectrum antiproliferative activity. ChemMedChem 2016, 11, 1587–1595. [Google Scholar] [CrossRef]
- Ammazzalorso, A.; D’Angelo, A.; Giancristofaro, A.; De Filippis, B.; Di Matteo, M.; Fantacuzzi, M.; Giampietro, L.; Linciano, P.; Maccallini, C.; Amoroso, R. Fibrate-derived N-(methylsulfonyl)amides with antagonistic properties on PPARα. Eur. J. Med. Chem. 2012, 58, 317–322. [Google Scholar] [CrossRef]
- Ammazzalorso, A.; De Lellis, L.; Florio, R.; Bruno, I.; De Filippis, B.; Fantacuzzi, M.; Giampietro, L.; Maccallini, C.; Perconti, S.; Verginelli, F.; et al. Cytotoxic effect of a family of peroxisome proliferator-activated receptor antagonists in colorectal and pancreatic cancer cell lines. Chem. Biol. Drug Des. 2017, 90, 1029–1035. [Google Scholar] [CrossRef]
- Benedetti, E.; d’Angelo, M.; Ammazzalorso, A.; Gravina, G.L.; Laezza, C.; Antonosante, A.; Panella, G.; Cinque, B.; Cristiano, L.; Dhez, A.C.; et al. PPARα antagonist AA452 triggers metabolic reprogramming and increases sensitivity to radiation therapy in human glioblastoma primary cells. J. Cell Physiol. 2017, 232, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Ammazzalorso, A.; De Lellis, L.; Florio, R.; Laghezza, A.; De Filippis, B.; Fantacuzzi, M.; Giampietro, L.; Maccallini, C.; Tortorella, P.; Veschi, S.; et al. Synthesis of novel benzothiazole amides: Evaluation of PPAR activity and anti-proliferative effects in paraganglioma, pancreatic and colorectal cancer cell lines. Bioorg. Med. Chem. Lett. 2019, 29, 2302–2306. [Google Scholar] [CrossRef]
- Florio, R.; De Lellis, L.; di Giacomo, V.; Di Marcantonio, M.C.; Cristiano, L.; Basile, M.; Verginelli, F.; Verzilli, D.; Ammazzalorso, A.; Prasad, S.C.; et al. Effects of PPARα inhibition in head and neck paraganglioma cells. PLoS ONE 2017, 12, e0178995. [Google Scholar] [CrossRef] [Green Version]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Pujadas, G.; Garcia-Vallve, S. Tools for in silico target fishing. Methods 2015, 71, 981–983. [Google Scholar] [CrossRef] [PubMed]
- Sydow, D.; Burggraaff, L.; Szengel, A.; Van Vlijmen, H.W.T.; Ijzerman, A.P.; Van Westen, G.J.P.; Volkamer, A. Advances and challenges in computational target prediction. J. Chem. Inf. Model. 2019, 59, 1728–1742. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. Swiss Target Prediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciriaco, F.; Gambacorta, N.; Alberga, D.; Nicolotti, O. Quantitative polypharmacology profiling based on a multifingerprint similarity predictive approach. J. Chem. Inf. Model. 2021, 61, 4868–4876. [Google Scholar] [CrossRef]
- Alberga, D.; Trisciuzzi, D.; Montaruli, M.; Leonetti, F.; Mangiatordi, G.F.; Nicolotti, O. A new approach for drug target and bioactivity prediction: The Multifingerprint Similarity Search Algorithm (MuSSeL). J. Chem. Inf. Model. 2019, 59, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Montaruli, M.; Alberga, D.; Ciriaco, F.; Trisciuzzi, D.; Tondo, A.R.; Mangiatordi, G.F.; Nicolotti, O. Accelerating drug discovery by early protein drug target prediction based on a multi-fingerprint similarity search. Molecules 2019, 24, 2233. [Google Scholar] [CrossRef] [Green Version]
- Ciriaco, F.; Gambacorta, N.; Trisciuzzi, D.; Nicolotti, O. PLATO: A predictive drug discovery web platform for efficient target fishing and bioactivity profiling of small molecules. Int. J. Mol. Sci. 2022, 23, 5245. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awale, M.; Reymond, J.L. Polypharmacology browser PPB2: Target prediction combining nearest neighbors with machine learning. J. Chem. Inf. Model. 2019, 59, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunkel, M.; Günther, S.; Ahmed, J.; Wittig, B.; Preissner, R. SuperPred: Drug classification and target prediction. Nucleic Acids Res. 2008, 36, W55–W59. [Google Scholar] [CrossRef]
- Gong, J.; Cai, C.; Liu, X.; Ku, X.; Jiang, H.; Gao, D.; Li, H. ChemMapper: A versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics 2013, 29, 1827–1829. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Di Guan, Y.; Zhang, L.X.; Liu, S.; Lu, A.P.; Cheng, Y.; Cao, D.S. A combinatorial target screening strategy for deorphaning macromolecular targets of natural product. Eur. J. Med. Chem. 2020, 204, 112644. [Google Scholar] [CrossRef] [PubMed]
- Mackie, K. Cannabinoid receptors: Where they are and what they do. J. Neuroendocrinol. 2008, 20, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Cavic, M.; Krivokuca, A.; Casadó, V.; Canela, E. The endocannabinoid system as a target in cancer diseases: Are we there yet? Front. Pharmacol. 2019, 10, 339. [Google Scholar] [CrossRef] [Green Version]
- Michalski, C.W.; Oti, F.E.; Erkan, M.; Sauliunaite, D.; Bergmann, F.; Pacher, P.; Batkai, S.; Müller, M.W.; Giese, N.A.; Friess, H. Cannabinoids in pancreatic cancer: Correlation with survival and pain. Int. J. Cancer 2008, 122, 742–750. [Google Scholar] [CrossRef] [Green Version]
- Garmpis, N.; Damaskos, C.; Dimitroulis, D.; Garmpi, A.; Diamantis, E.; Sarantis, P.; Georgakopoulou, V.E.; Patsouras, A.; Prevezanos, D.; Syllaios, A.; et al. Targeting the endocannabinoid system: From the need for new therapies to the development of a promising strategy. What about pancreatic cancer? In Vivo 2022, 36, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Celen, A.B.; Sahin, U. Sumoylation on its 25th anniversary: Mechanisms, pathology, and emerging concepts. FEBS J. 2020, 287, 3110–3140. [Google Scholar] [CrossRef]
- Jia, Y.; Claessens, L.A.; Vertegaal, A.C.O.; Ovaa, H. Chemical tools and biochemical assays for SUMO specific proteases (SENPs). ACS Chem. Biol. 2019, 14, 2389–2395. [Google Scholar] [CrossRef] [Green Version]
- Tokarz, P.; Woźniak, K. SENP proteases as potential targets for cancer therapy. Cancers 2021, 13, 2059. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://cancergenome.nih.gov/ (accessed on 1 March 2022).
- Schneeweis, C.; Hassan, Z.; Schick, M.; Keller, U.; Schneider, G. The SUMO pathway in pancreatic cancer: Insights and inhibition. Br. J. Cancer 2021, 124, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Y.; Zhang, J.; Ullah, S.; Kang, N.; Zhao, Y.; Zhou, H. Benzothiophene-2-carboxamide derivatives as SENPs inhibitors with selectivity within SENPs family. Eur. J. Med. Chem. 2020, 204, 112553. [Google Scholar] [CrossRef]
- Hua, T.; Li, X.; Wu, L.; Iliopoulos-Tsoutsouvas, C.; Wang, Y.; Wu, M.; Shen, L.; Brust, C.A.; Nikas, S.P.; Song, F.; et al. Activation and signaling mechanism revealed by cannabinoid receptor-Gi complex structures. Cell 2020, 180, 655–665. [Google Scholar] [CrossRef]
- Alegre, K.O.; Reverter, D. Structural insights into the SENP6 Loop1 structure in complex with SUMO2. Protein Sci. 2014, 23, 433–441. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger Release 2021-4: Maestro, Glide, Protein Preparation Wizard, Epik, SiteMap, QikProp, MacroModel; Schrödinger, LLC.: New York, NY, USA, 2021.
- Florio, R.; Veschi, S.; di Giacomo, V.; Pagotto, S.; Carradori, S.; Verginelli, F.; Cirilli, R.; Casulli, A.; Grassadonia, A.; Tinari, N.; et al. The benzimidazole-based anthelmintic parbendazole: A repurposed drug candidate that synergizes with gemcitabine in pancreatic cancer. Cancers 2019, 11, 2042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammazzalorso, A.; Bruno, I.; Florio, R.; De Lellis, L.; Laghezza, A.; Cerchia, C.; De Filippis, B.; Fantacuzzi, M.; Giampietro, L.; Maccallini, C.; et al. Sulfonimide and amide derivatives as novel PPARα antagonists: Synthesis, antiproliferative activity, and docking studies. ACS Med. Chem. Lett. 2020, 11, 624–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aid. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Halgren, T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shaw, D.E.; Shelley, M.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
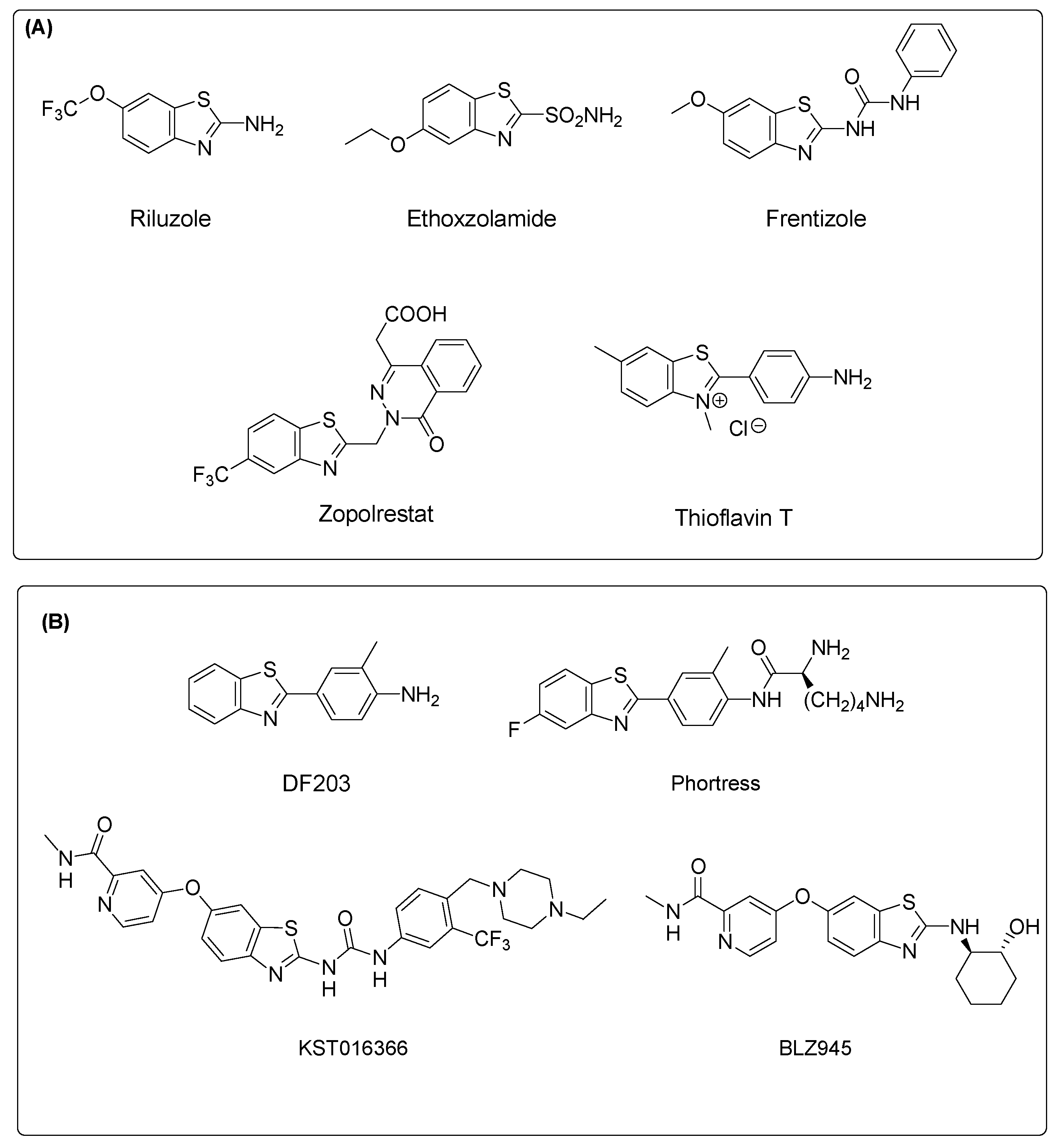




| Cpd | R | Purification Conditions | Yield % | m.p. |
|---|---|---|---|---|
| 4a | p-OCH3 | Silica gel, eluent chloroform | 58 | 190 °C (dec) |
| 4b | p-Cl | Silica gel, eluent chloroform | 63 | 178–180 °C |
| 4c | p-F | Silica gel, eluent dichloromethane | 52 | 168–170 °C |
| 4d | p-CF3 | Silica gel, eluent dichloromethane | 47 | 183–185 °C |
| 4e | p-NO2 | Silica gel, eluent dichloromethane | 43 | 191–193 °C |
| 4f | p-NHCOCH3 | Crystallization from ethyl acetate/methanol | 59 | 243 °C (dec) |
| 4g | m-OCH3 | Silica gel, eluent cyclohexane/ethyl acetate 7:1 | 44 | 160–162 °C |
| 4h | m-Cl | Silica gel, eluent dichloromethane | 76 | 175–177 °C |
| 4i | m-F | Silica gel, eluent dichloromethane | 51 | 151–153 °C |
| 4j | m-CF3 | Silica gel, eluent cyclohexane/diethyl ether 4:1 | 45 | 155–157 °C |
| 4k | m-NO2 | Silica gel, eluent dichloromethane | 48 | 193–195 °C |
| 4l | m-NHCOCH3 | Crystallization from chloroform | 41 | 197–199 °C |
| 4m | 3,4-diCl | Silica gel, eluent dichloromethane | 44 | 203–204 °C |
| 4n | 2-Br, 5-NO2 | Crystallization from cyclohexane/methanol | 48 | 176–178 °C |
| 4o | 2-Br, 4-CF3 | Crystallization from petroleum ether/methanol | 51 | 179–180 °C |
| 4p | 2-Br, 5-CF3 | Crystallization from petroleum ether | 46 | 157–159 °C |
| IC50 (µM) | ||||||
|---|---|---|---|---|---|---|
| Pancreatic Cancer | Paraganglioma | Normal Cells | ||||
| AsPC-1 | BxPC-3 | Capan-2 | PTJ64i | PTJ86i | HFF-1 | |
| 2b | 12.44 | 14.99 | 19.65 | 8.49 | 16.70 | 21.37 |
| 4d | 7.66 | 3.99 | 8.97 | 6.79 | 12.39 | 9.23 |
| 4e | 12.77 | 10.69 | 14.11 | 9.81 | 18.87 | 16.69 |
| 4f | 10.04 | 18.85 | 20.10 | 12.34 | 12.82 | 6.54 |
| 4h | 12.16 | 11.99 | 17.67 | 7.27 | 16.58 | 11.55 |
| 4i | 14.80 | 18.60 | 28.50 | 8.60 | 11.70 | 15.00 |
| 4j | 9.53 | 13.96 | 24.18 | 11.20 | 17.46 | 18.10 |
| 4k | 10.08 | 11.92 | 16.87 | 7.47 | 13.51 | 23.33 |
| 4l | 14.78 | 13.67 | 33.76 | 10.13 | 19.88 | 67.07 |
| 4m | 8.49 | 9.81 | 13.33 | 7.84 | 19.92 | 10.32 |
| Selectivity Index (SI) Values | |||||
|---|---|---|---|---|---|
| Pancreatic Cancer | Paraganglioma | ||||
| AsPC-1 | BxPC-3 | Capan-2 | PTJ64i | PTJ86i | |
| 4k | 2.31 | 1.96 | 1.38 | 3.12 | 1.73 |
| 4l | 4.54 | 4.91 | 1.99 | 6.62 | 3.37 |
| Inhibition Rate of Cell Viability, % | ||||
|---|---|---|---|---|
| Pancreatic Cancer | Normal Cells | |||
| AsPC-1 | BxPC-3 | Capan-2 | HFF-1 | |
| 4l–0.5 µM | 13.40 | 6.00 | 9.34 | 0.00 |
| 4l–5 µM | 18.87 | 9.54 | 30.25 | 1.00 |
| 4l–50 µM | 42.97 | 70.50 | 43.30 | 17.69 |
| GEM—0.1 µM | 54.21 | 65.02 | 26.87 | 27.39 |
| GEM—1 µM | 63.25 | 64.65 | 27.22 | 25.43 |
| GEM—10 µM | 64.63 | 62.50 | 38.65 | 26.27 |
| 4l (0.5 µM) + GEM (0.1 µM) | 62.85 | 65.98 | 47.66 | 36.69 |
| 4l (5 µM) + GEM (1 µM) | 61.70 | 64.01 | 25.40 | 28.16 |
| 4l (50 µM) + GEM (10 µM) | 54.27 | 64.09 | 27.79 | 25.19 |
| Web Tool | Description | Database | Target ranking | URL |
|---|---|---|---|---|
| SwissTargetPrediction [20] | A combination of 2D and 3D similarities with known ligands | ChEMBL23 | Similarity threshold of compounds | http://www.swisstargetprediction.ch |
| PLATO [21,22,23,24] | Multifingerprint Similarity Predictive Approach | ChEMBL30 | Similarity between query molecule and known target ligands using different fingerprints | http://plato.uniba.it/ |
| SEA Search [25] | Similarity searching | ChEMBL27 | E-value indicating the reliability of the prediction | https://sea.bkslab.org/ |
| PPB2 [26] | Similarity searching combined with Machine Learning models | ChEMBL22 | Score calculated by the applied model | http://ppb2.gdb.tools/ |
| SuperPred [27] | Similarity searching by ECFP4 fingerprints | ChEMBL29 | Similarity between query molecule and known target ligands | https://prediction.charite.de/subpages/target_prediction.php |
| ChemMapper [28] | Pharmacophore/shape superposition and statistical background distribution | database of 300M drug-like compounds (ChEMBL, BindingDB, DrugBank, KEGG, PDB) | Similarity between query molecule and known target ligands | http://www.lilab-ecust.cn›chemmapper |
| PharmMapper [29] | Reverse Pharmacophore screening | TargetBank DrugBank, BindingDB and PDTD. | Z-score based on fit score (match feature types and positions) | http://www.lilab-ecust.cn/pharmmapper/ |
| PLATO | SwissTargetPrediction | SEA | PPB2 | SuperPRED | PharmMAPPER | ChemMapper |
|---|---|---|---|---|---|---|
| Peroxisome proliferator-activated receptor alpha | Dual specificity mitogen-activated protein kinase1 | Potassium voltage-gated channel subfamily B member 2 | Arachidonate 5-lipoxygenase | Glutaminase kidney isoform, mitochondrial | Cbp/p300-intE4:E27 | Voltage-dependent T-type calcium channel subunit alpha-1H |
| Cathepsin K | ATP-binding cassette sub-family G member 2 | Neuronal calcium sensor 1 | Peroxisome proliferator-activated receptor alpha | Casein kinase II alpha/beta | Coagulation factor XIII A chain | G-protein coupled receptor 55 |
| Cathepsin L | Voltage-gated potassium channel subunit Kv1.5 | Ubiquitin carboxyl-terminal hydrolase BAP1 | G-protein coupled receptor 55 | Muscarinic acetylcholine receptor M3 | Cold shock domain-containing protein E1 | Cannabinoid receptor 2 |
| Tyrosine-protein kinase LCK | Insulin receptor | Survival motor neuron protein | Cannabinoid CB1 receptor | ADAM10 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | Cannabinoid receptor 1 |
| C-C chemokine receptor type 3 | Cannabinoid receptor 1 | Potassium channel subfamily K member 9 | Cannabinoid CB2 receptor | Aurora kinase B/Inner centromere protein | Homeobox protein Hox-B13 | DNA dC->dU-editing enzyme APOBEC-3G |
| G-protein coupled receptor 55 | ALK tyrosine kinase receptor | Cysteinyl leukotriene receptor 1 | Vascular endothelial growth factor receptor 2 | Caspase-8 | Disheveled-associated activator of morphogenesis 1 | Probable DNA dC->dU-editing enzyme APOBEC-3A |
| Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 | Receptor protein-tyrosine kinase erbB-4 | Glutamate receptor ionotropic, kainate 1 | Coagulation factor X | DNA topoisomerase I | Protection of telomeres protein 1 | E3 ubiquitin-protein ligase Mdm2 |
| 11-beta-hydroxysteroid dehydrogenase 1 | Peroxisome proliferator-activated receptor alpha | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 | Sentrin-specific protease 7 | Galectin-3 | Regulator of G-protein signaling 6 | Eukaryotic translation initiation factor 4H |
| G-protein coupled receptor 35 | MAP kinase p38 alpha | Sentrin-specific protease 8 | Epidermal growth factor receptor erbB1 | Indoleamine 2,3-dioxygenase | Heterogeneous nuclear ribonucleoprotein R | Polyadenylate-binding protein 1 |
| Sentrin-specific protease 7 | c-Jun N-terminal kinase 2 | Sentrin-specific protease 7 | Tyrosine-protein kinase SRC | Sphingosine kinase 1 | Calpain-9 | MCOLN3 protein |
| PI3-kinase p110-alpha subunit | Cyclin-dependent kinase 4 | Probable G-protein coupled receptor 139 | Beta-secretase 1 | Adenosine A3 receptor | Glycogen phosphorylase, liver form | Estrogen receptor |
| Caspase-3 | Serine/threonine-protein kinase AKT | Free fatty acid receptor 2 | Adenosine A3 receptor | Integrin alpha-V/beta-3 | Transcription initiation factor TFIID subunit 13 | Putative hexokinase HKDC1 |
| Cannabinoid CB2 receptor | Vascular endothelial growth factor receptor 2 | Sentrin-specific protease 6 | Dopamine D2 receptor | DNA (cytosine-5)-methyltransferase 1 | Proto-oncogene tyrosine-protein kinase Fes/Fps | Hexokinase-1 |
| C-C chemokine receptor type 1 | Ribosomal protein S6 kinase alpha 3 | Solute carrier family 22 member 6 | Serine/threonine-protein kinase Aurora-A | Muscarinic acetylcholine receptor M5 | Ig gamma-1 chain C region secreted form | Coagulation factor XII |
| Sentrin-specific protease 6 | Phosphodiesterase 10A | Acyl-CoA (8–3)-desaturase | Vanilloid receptor | Protein-tyrosine phosphatase 2C | Cytochrome P450 2E1 | Glyceraldehyde-3-phosphate dehydrogenase |
| Sentrin-specific protease 8 | Pregnane X receptor | Trypsin-3 | Induced myeloid leukemia cell differentiation protein Mcl-1 | Muscarinic acetylcholine receptor M1 | Threonine dehydratase biosynthetic | Induced myeloid leukemia cell differentiation protein Mcl-1 |
| 1-acylglycerol-3-phosphate O-acyltransferase beta | Cytochrome P450 19A1 | 10 kDa heat shock protein, mitochondrial | Adenosine A1 receptor | Glutathione S-transferase Pi | Heme oxygenase 1 | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 |
| MAP kinase-activated protein kinase 2 | 60 kDa heat shock protein, mitochondrial | Serine/threonine-protein kinase Aurora-B | Histone deacetylase 4 | Eukaryotic initiation factor 4A-I | Apoptotic protease-activating factor 1 | |
| Carnitine O-palmitoyltransferase 1 liver isoform | Multidrug resistance-associated protein 4 | Calcium sensing receptor | Neprilysin | 72 kDa type IV collagenase | Tumor necrosis factor |
| ID | MW | accptHB | donorHB | QPlogPo/w | Rule OfFive | PSA | #Rotor | CIQP logS |
| 2b | 410.935 | 4 | 1 | 6.005 | 1 | 41.544 | 5 | −7.236 |
| 4d | 478.934 | 4 | 1 | 6.986 | 1 | 41.011 | 5 | −8.646 |
| 4e | 455.933 | 5 | 1 | 5.283 | 1 | 85.806 | 6 | −7.758 |
| 4f | 467.987 | 6.5 | 2 | 4.932 | 0 | 85.243 | 6 | −7.325 |
| 4h | 445.38 | 4 | 1 | 6.535 | 1 | 38.018 | 5 | −7.949 |
| 4i | 428.926 | 4 | 1 | 6.201 | 1 | 42.618 | 5 | −7.608 |
| 4j | 478.934 | 4 | 1 | 6.949 | 1 | 45.48 | 5 | −8.646 |
| 4k | 455.933 | 5 | 1 | 5.082 | 1 | 87.682 | 6 | −7.758 |
| 4l | 467.987 | 6.5 | 2 | 5.289 | 1 | 86.742 | 6 | −7.325 |
| 4m | 479.825 | 4 | 1 | 6.96 | 1 | 37.855 | 5 | −8.665 |
| ID | Percent Human Oral Absorption | QPPCaco | QPPMDCK | QPlogBB | QPlog HERG | QPlogKhsa | CNS | #metab |
| 2b | 100 | 4797.201 | 10,000 | 0.264 | −7.033 | 0.884 | 1 | 4 |
| 4d | 100 | 5598.932 | 10,000 | 0.616 | −6.735 | 1.136 | 2 | 3 |
| 4e | 95.015 | 629.287 | 1753.816 | −0.773 | −6.819 | 0.818 | −1 | 4 |
| 4f | 100 | 675.963 | 1383.764 | −0.86 | −7.087 | 0.69 | −1 | 3 |
| 4h | 100 | 5564.24 | 10,000 | 0.509 | −7.021 | 0.991 | 2 | 4 |
| 4i | 100 | 4890.181 | 10,000 | 0.387 | −6.754 | 0.916 | 1 | 4 |
| 4j | 100 | 3372.682 | 10,000 | 0.324 | −7.26 | 1.185 | 1 | 5 |
| 4k | 90.944 | 433.678 | 857.42 | −1.064 | −7.137 | 0.81 | −2 | 5 |
| 4l | 100 | 896.897 | 2297.395 | −0.712 | −7.352 | 0.763 | −1 | 5 |
| 4m | 100 | 5649.57 | 10,000 | 0.654 | −6.934 | 1.096 | 2 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amoroso, R.; De Lellis, L.; Florio, R.; Moreno, N.; Agamennone, M.; De Filippis, B.; Giampietro, L.; Maccallini, C.; Fernández, I.; Recio, R.; et al. Benzothiazole Derivatives Endowed with Antiproliferative Activity in Paraganglioma and Pancreatic Cancer Cells: Structure–Activity Relationship Studies and Target Prediction Analysis. Pharmaceuticals 2022, 15, 937. https://doi.org/10.3390/ph15080937
Amoroso R, De Lellis L, Florio R, Moreno N, Agamennone M, De Filippis B, Giampietro L, Maccallini C, Fernández I, Recio R, et al. Benzothiazole Derivatives Endowed with Antiproliferative Activity in Paraganglioma and Pancreatic Cancer Cells: Structure–Activity Relationship Studies and Target Prediction Analysis. Pharmaceuticals. 2022; 15(8):937. https://doi.org/10.3390/ph15080937
Chicago/Turabian StyleAmoroso, Rosa, Laura De Lellis, Rosalba Florio, Nazaret Moreno, Mariangela Agamennone, Barbara De Filippis, Letizia Giampietro, Cristina Maccallini, Inmaculada Fernández, Rocío Recio, and et al. 2022. "Benzothiazole Derivatives Endowed with Antiproliferative Activity in Paraganglioma and Pancreatic Cancer Cells: Structure–Activity Relationship Studies and Target Prediction Analysis" Pharmaceuticals 15, no. 8: 937. https://doi.org/10.3390/ph15080937
APA StyleAmoroso, R., De Lellis, L., Florio, R., Moreno, N., Agamennone, M., De Filippis, B., Giampietro, L., Maccallini, C., Fernández, I., Recio, R., Cama, A., Fantacuzzi, M., & Ammazzalorso, A. (2022). Benzothiazole Derivatives Endowed with Antiproliferative Activity in Paraganglioma and Pancreatic Cancer Cells: Structure–Activity Relationship Studies and Target Prediction Analysis. Pharmaceuticals, 15(8), 937. https://doi.org/10.3390/ph15080937
















