Chitosan Based MicroRNA Nanocarriers
Abstract
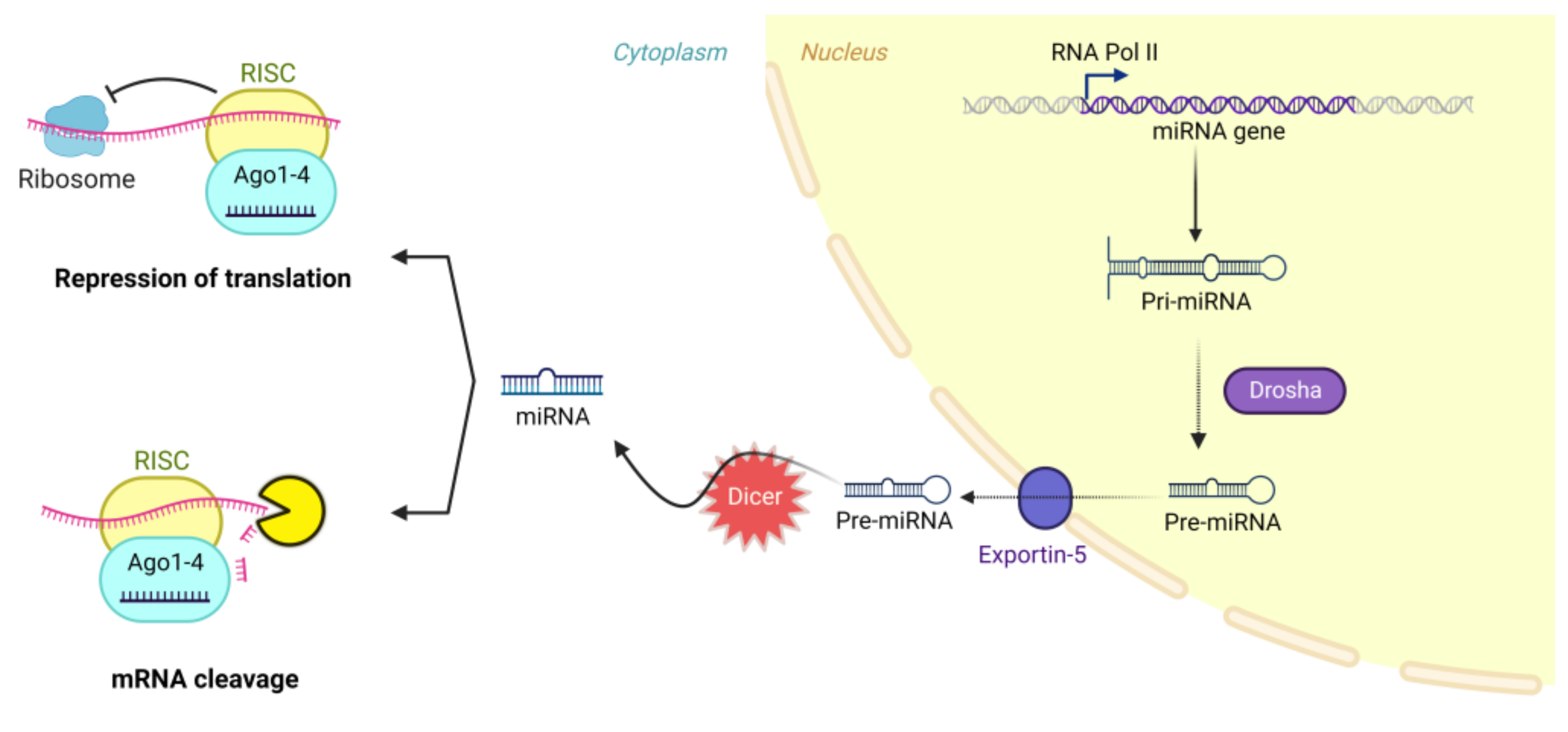
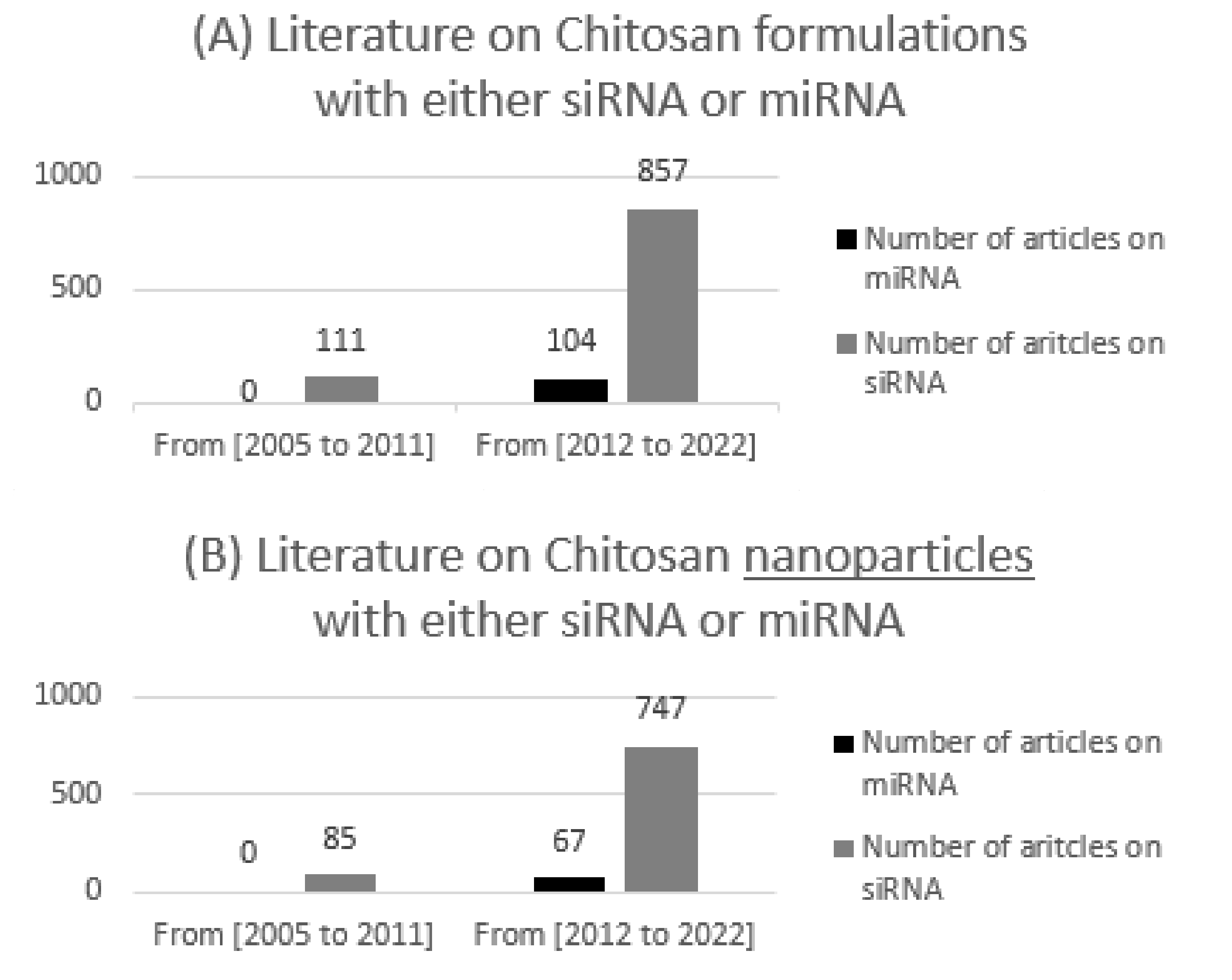
:1. Introduction
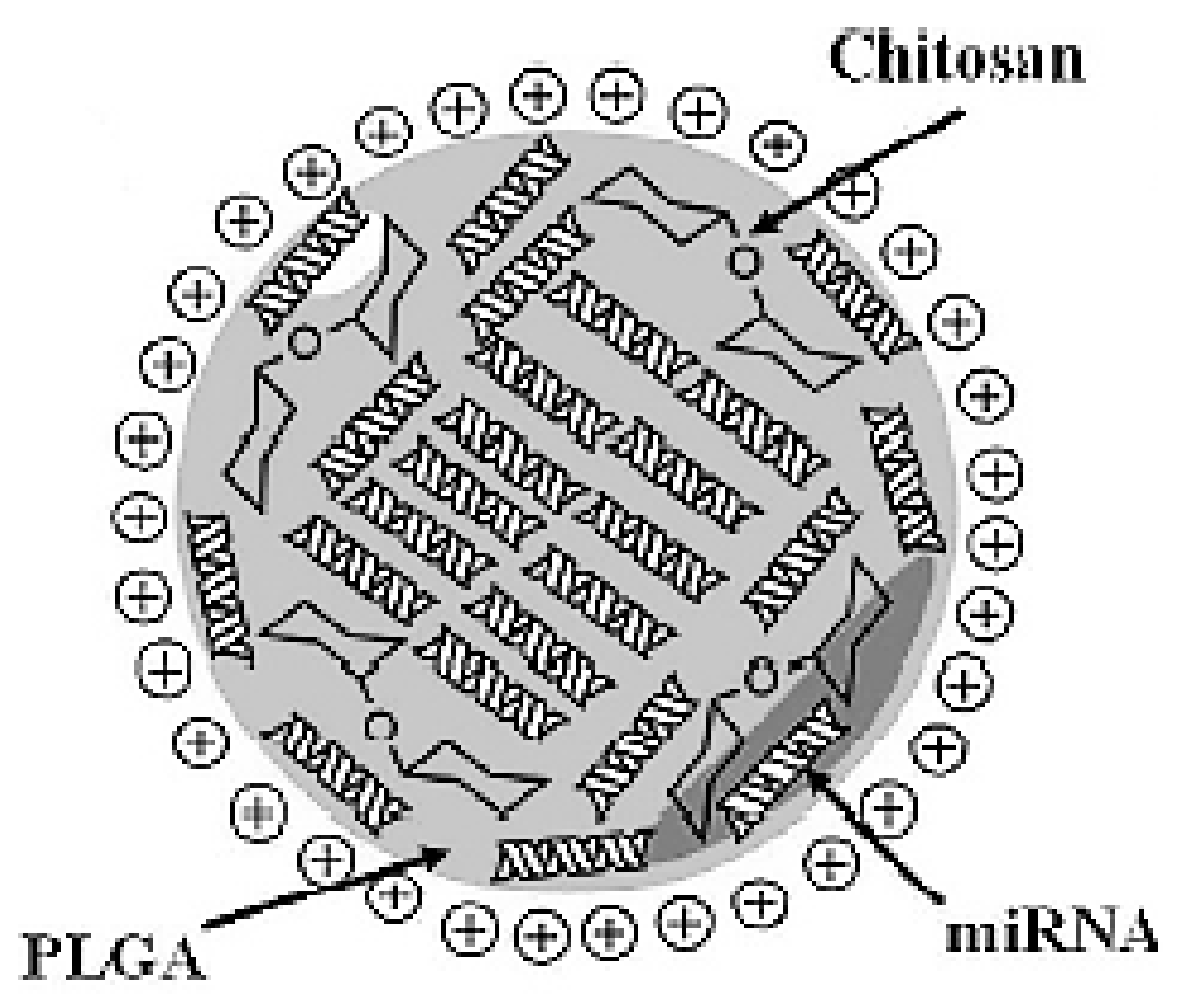
- Nanoparticles formulated by direct complexation between chitosan and miRNA (referred to as nano polyplexes = nanoplexes). Polyplexes are complexes made by electrostatic interaction between a cationic polymer (such as chitosan) and negatively charged genetic materials (such as miRNAs) [17].
- Nanoparticles based on the interaction between polycationic chitosan with a counter polyanion:
- Nanoparticles made by ionic gelation of polycationic chitosan with a counter polyanionic small molecule such as tripolyphosphate (TPP), adsorbing or encapsulating miRNA (referred to as CS/TPP NPs) [21].
- More complex engineered systems.
2. Parameters Affecting CS/MiRNA NPs
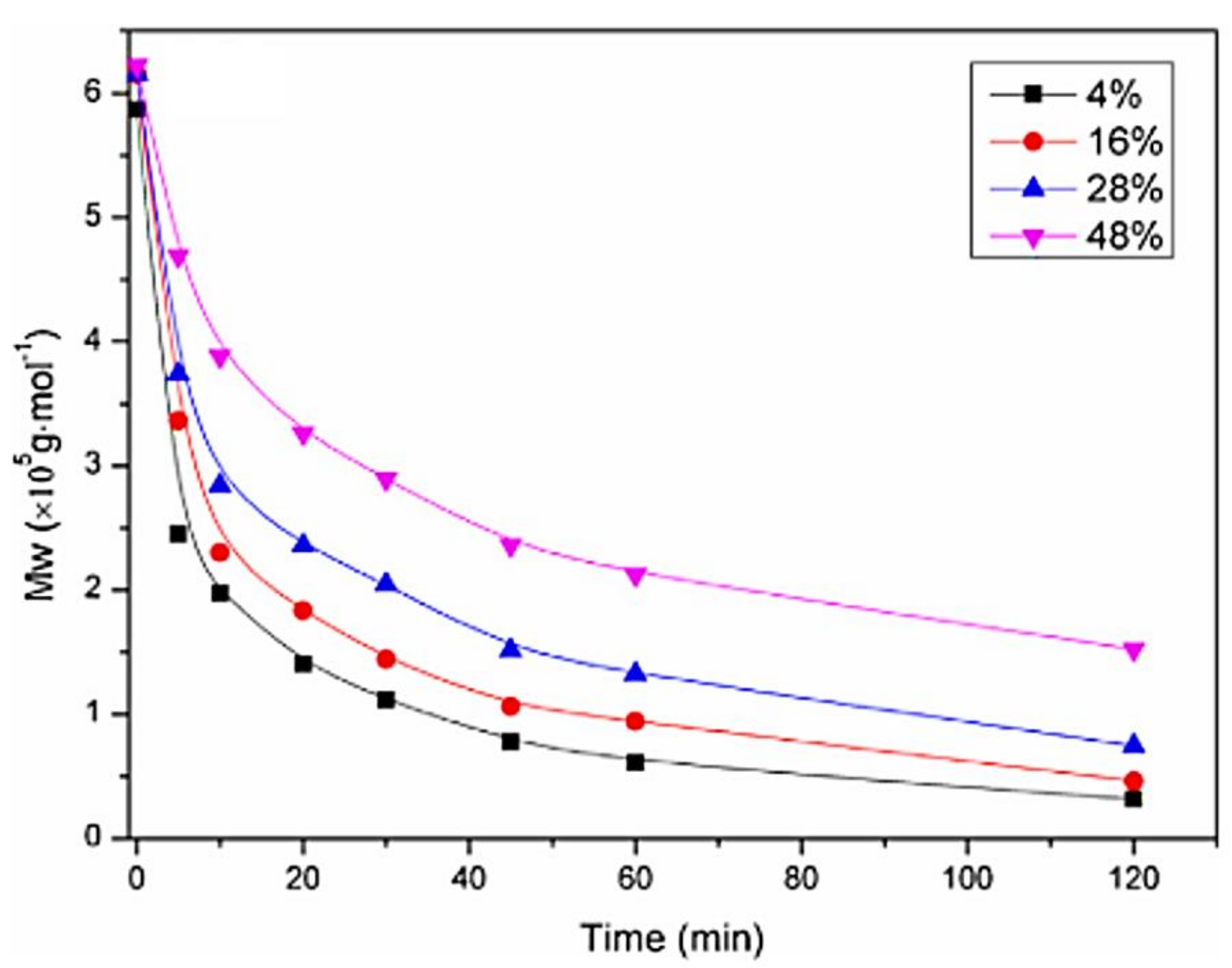
2.1. Chitosan Molar Mass
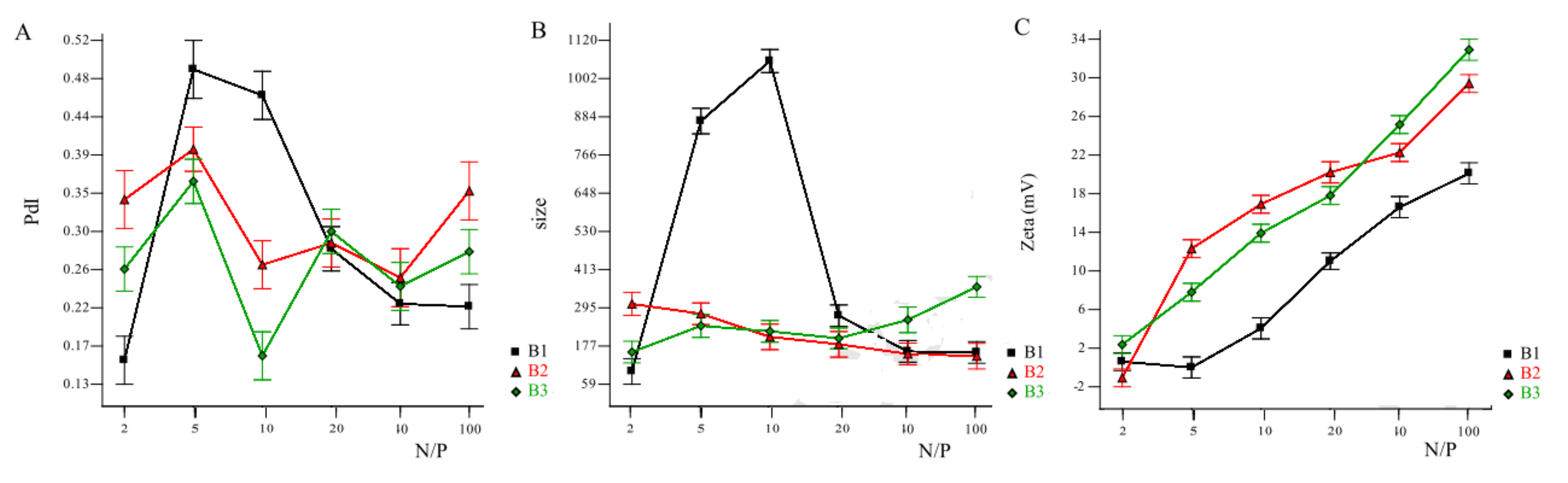
2.1.1. Effect of Chitosan Molar Mass on (CS/Polyanion/MiRNA Nanoparticles)
2.1.2. Effect of Chitosan Molar Mass on Nanoplexes (Chitosan MiRNA Direct Polyplexes)
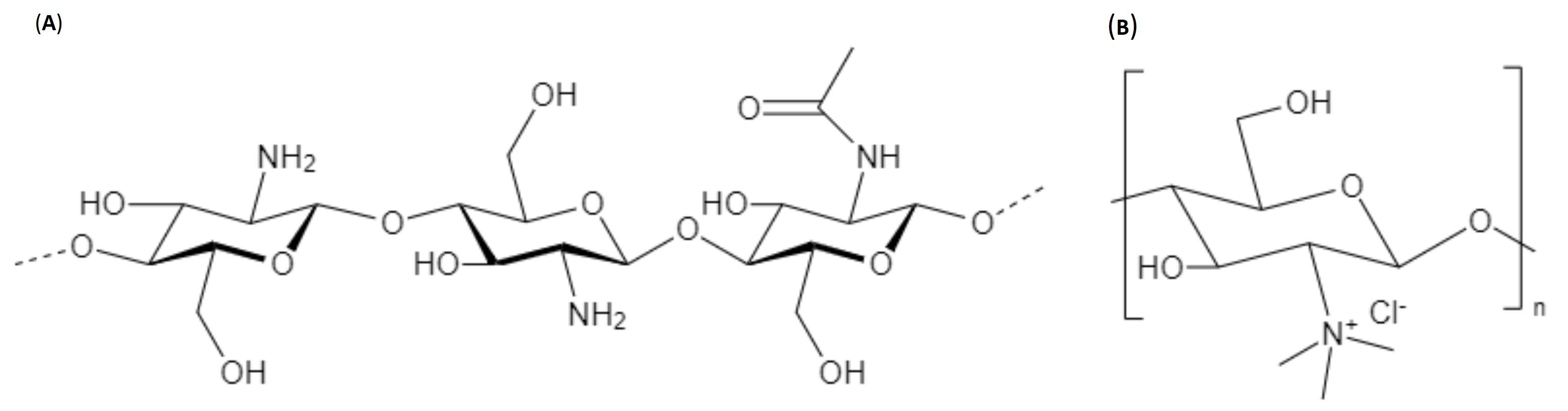
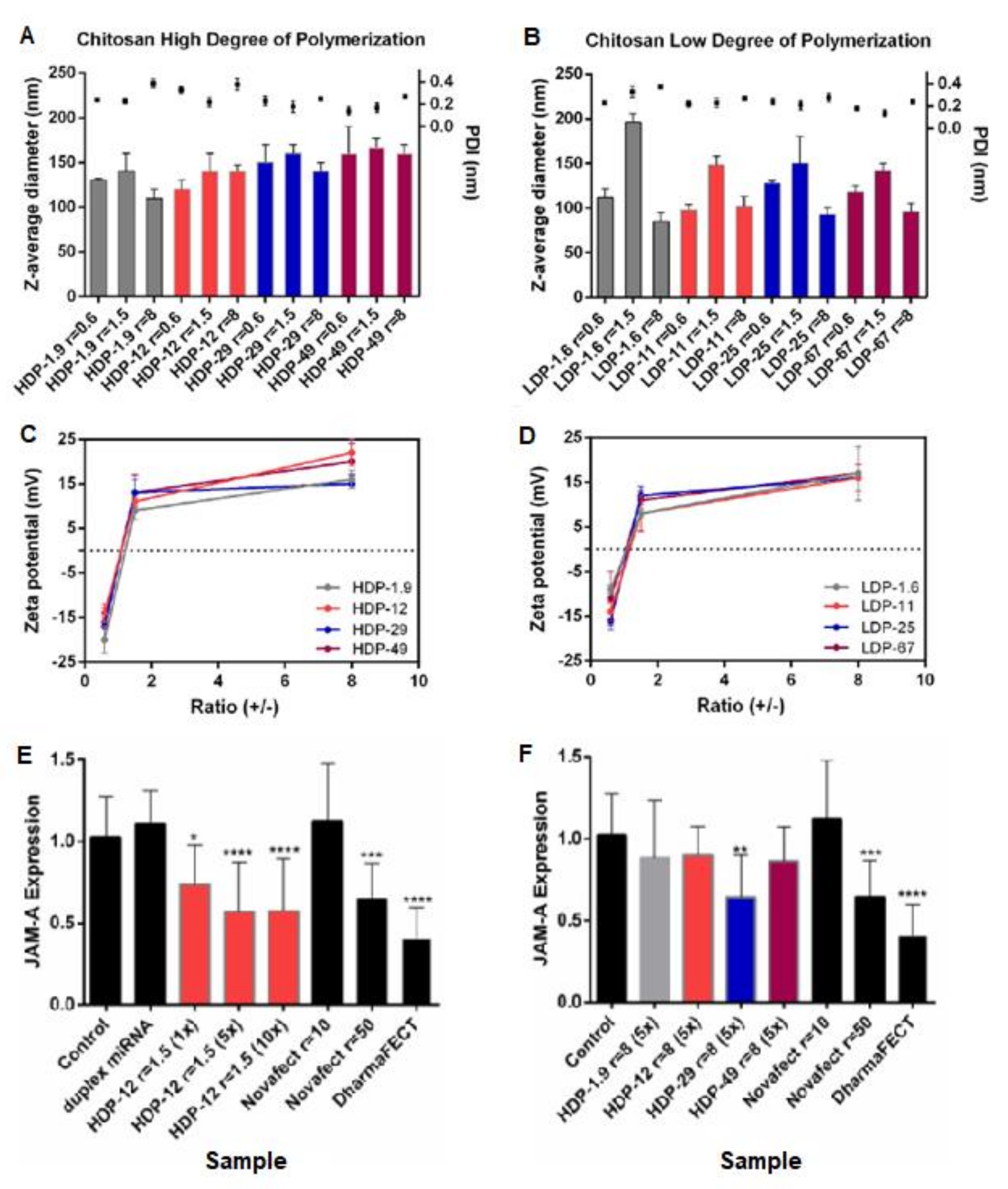
2.2. Degree of Acetylation (DA) of Chitosan/Deacetylation Degree (DD)
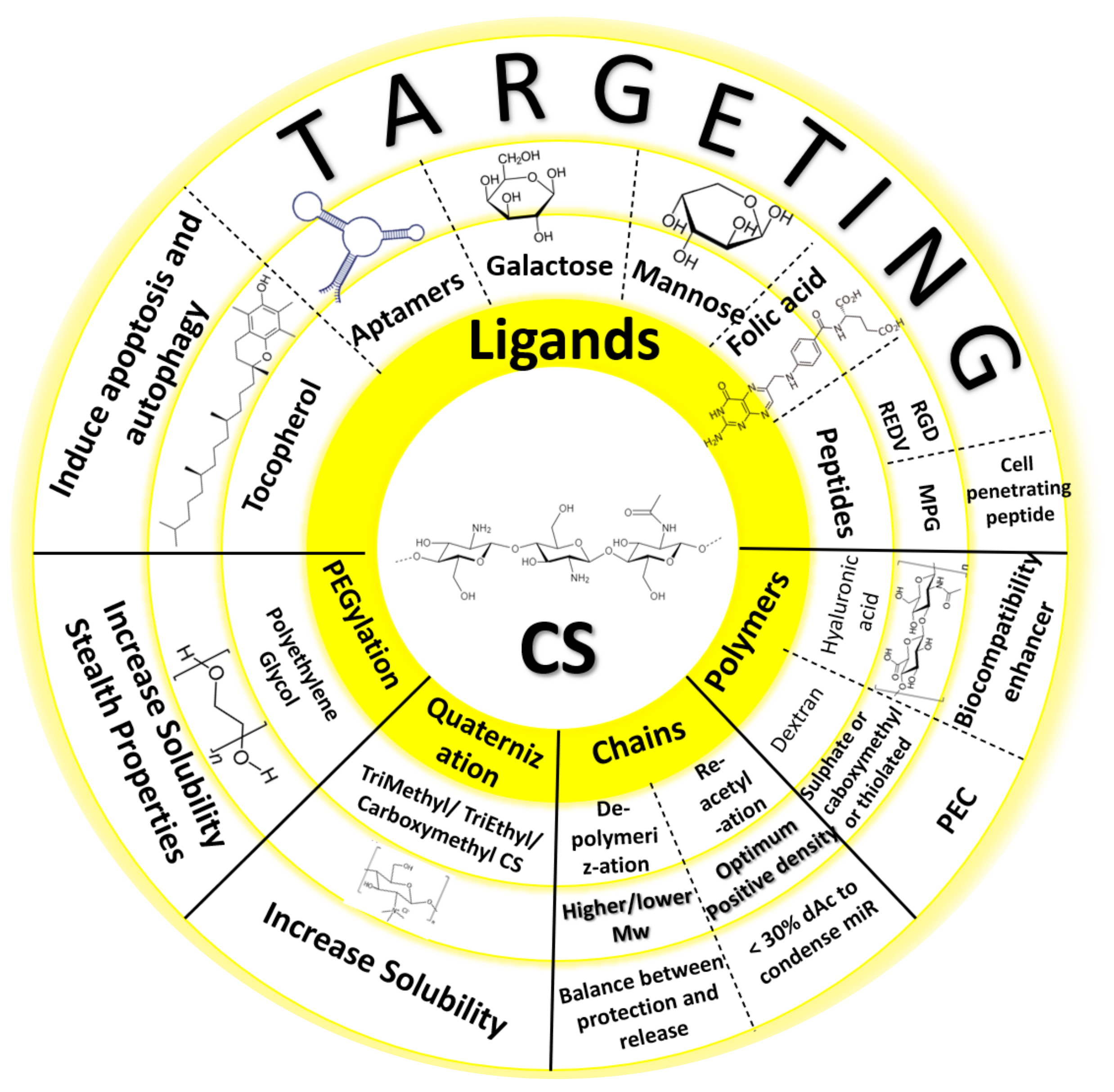
3. Chemical Modifications to Chitosan Reported in MiRNA Formulations
4. Formulation Processes
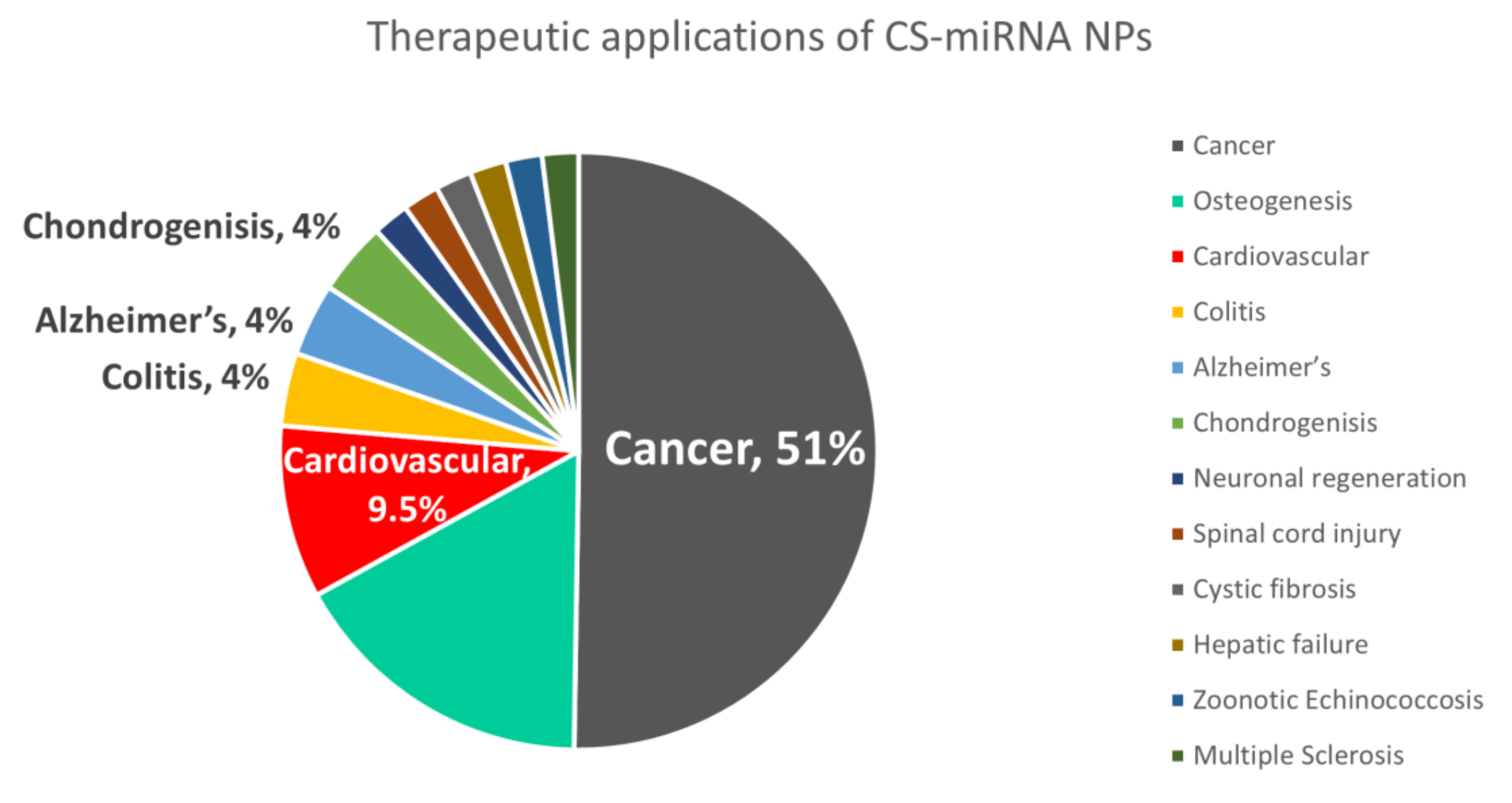
5. Therapeutic Applications of Chitosan MiRNA Delivery Systems
5.1. Cancer
5.2. Regenerative Medicine
5.3. Other Fields of Therapy
6. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, Y.K.; Kim, S.W. Recent Advances in the Development of Gene Delivery Systems. Biomater. Res. 2019, 23, 8. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fuentes, M.; Alonso, M.J. Chitosan-Based Drug Nanocarriers: Where Do We Stand? J. Control. Release 2012, 161, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Peniche, C.; Argüelles-Monal, W.; Goycoolea, F.M. Chitin and Chitosan: Major Sources, Properties and Applications. In Monomers, Polymers and Composites from Renewable Resources; Elsevier Ltd.: Amsterdam, The Netherlands, 2008; pp. 517–542. ISBN 9780080453163. [Google Scholar]
- El Gueddari, N.E.; Moerschbacher, B.M.; Boucher, I.; Jamieson, K. Advances in Chitin Science. Eur. Chitin Soc. Montr. 2004, VII, 56–59. [Google Scholar]
- Rupaimoole, R.; Han, H.D.; Lopez-Berestein, G.; Sood, A.K. MicroRNA Therapeutics: Principles, Expectations, and Challenges. Chin. J. Cancer 2011, 30, 368–370. [Google Scholar] [CrossRef] [PubMed]
- Klausner, E.A.; Zhang, Z.; Chapman, R.L.; Multack, R.F.; Volin, M.V. Ultrapure Chitosan Oligomers as Carriers for Corneal Gene Transfer. Biomaterials 2010, 31, 1814–1820. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and Their Integrated Networks. J. Integr. Bioinform. 2019, 16, 3. [Google Scholar] [CrossRef]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. SiRNA versus MiRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [Green Version]
- Denizli, M.; Aslan, B.; Mangala, L.S.; Jiang, D.; Lopez-berestein, G.; Sood, A.K. Chitosan Nanoparticles for MiRNA Delivery. Methods Mol. Biol. 2017, 1632, 219–230. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent Advances in MiRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, D.Y.; Huang, L. In Vivo Delivery of MiRNAs for Cancer Therapy: Challenges and Strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-Viral Vectors for Gene-Based Therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Petros, R.A.; Desimone, J.M. Strategies in the Design of Nanoparticles for Therapeutic Applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Ragelle, H.; Vandermeulen, G.; Préat, V. Chitosan-Based SiRNA Delivery Systems. J. Control. Release 2013, 172, 207–218. [Google Scholar] [CrossRef]
- Guzman-Villanueva, D.; El-Sherbiny, I.M.; Herrera-Ruiz, D.; Vlassov, A.V.; Smyth, H.D.C. Formulation Approaches to Short Interfering RNA and MicroRNA: Challenges and Implications. J. Pharm. Sci. 2012, 101, 2271–2280. [Google Scholar] [CrossRef]
- Ragelle, H.; Riva, R.; Vandermeulen, G.; Naeye, B.; Pourcelle, V.; Le Duff, C.S.; D’Haese, C.; Nysten, B.; Braeckmans, K.; De Smedt, S.C.; et al. Chitosan Nanoparticles for SiRNA Delivery: Optimizing Formulation to Increase Stability and Efficiency. J. Control. Release 2014, 176, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Vetter, V.C.; Wagner, E. Targeting Nucleic Acid-Based Therapeutics to Tumors: Challenges and Strategies for Polyplexes. J. Control. Release 2022, 346, 110–135. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhu, L.; Li, Y.; Zhang, X.; Xu, S.; Yang, G.; Delair, T. Chitosan-Based Colloidal Polyelectrolyte Complexes for Drug Delivery: A Review. Carbohydr. Polym. 2020, 238, 116126. [Google Scholar] [CrossRef]
- Delair, T. Colloidal Polyelectrolyte Complexes of Chitosan and Dextran Sulfate towards Versatile Nanocarriers of Bioactive Molecules. Eur. J. Pharm. Biopharm. 2011, 78, 10–18. [Google Scholar] [CrossRef]
- Carvalho, S.G.; dos Santos, A.M.; Silvestre, A.L.P.; Meneguin, A.B.; Ferreira, L.M.B.; Chorilli, M.; Gremião, M.P.D. New Insights into Physicochemical Aspects Involved in the Formation of Polyelectrolyte Complexes Based on Chitosan and Dextran Sulfate. Carbohydr. Polym. 2021, 271, 118436. [Google Scholar] [CrossRef]
- Bugnicourt, L.; Ladavière, C. Interests of Chitosan Nanoparticles Ionically Cross-Linked with Tripolyphosphate for Biomedical Applications. Prog. Polym. Sci. 2016, 60, 1–17. [Google Scholar] [CrossRef]
- Buschmann, M.D.; Merzouki, A.; Lavertu, M.; Thibault, M.; Jean, M.; Darras, V. Chitosans for Delivery of Nucleic Acids. Adv. Drug Deliv. Rev. J. 2013, 65, 1234–1270. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Howard, K.A.; Dong, M.; Andersen, M.; Rahbek, U.L.; Johnsen, M.G.; Hansen, O.C.; Besenbacher, F.; Kjems, J. The Influence of Polymeric Properties on Chitosan/SiRNA Nanoparticle Formulation and Gene Silencing. Biomaterials 2007, 28, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Ning, Q.; Liu, Y.F.; Ye, P.J.; Gao, P.; Li, Z.P.; Tang, S.Y.; He, D.X.; Tang, S.S.; Wei, H.; Yu, C.Y. Delivery of Liver-Specific MiRNA-122 Using a Targeted Macromolecular Prodrug toward Synergistic Therapy for Hepatocellular Carcinoma. ACS Appl. Mater. Interfaces 2019, 11, 10578–10588. [Google Scholar] [CrossRef] [PubMed]
- Tekie, F.S.M.; Atyabi, F.; Soleimani, M.; Arefian, E.; Atashi, A.; Kiani, M.; Khoshayand, M.R.; Amini, M.; Dinarvand, R. Chitosan Polyplex Nanoparticle Vector for MiR-145 Expression in MCF-7: Optimization by Design of Experiment. Int. J. Biol. Macromol. 2015, 81, 828–837. [Google Scholar] [CrossRef]
- Tekie, F.S.M.; Kiani, M.; Zakerian, A.; Pilevarian, F.; Assali, A.; Soleimani, M.; Dinarvand, R.; Arefian, E.; Atashi, A.; Amini, M.; et al. Nano Polyelectrolyte Complexes of Carboxymethyl Dextran and Chitosan to Improve Chitosan-Mediated Delivery of MiR-145. Carbohydr. Polym. 2017, 159, 66–75. [Google Scholar] [CrossRef]
- McKiernan, P.J.; Cunningham, O.; Greene, C.M.; Cryan, S.A. Targeting MiRNA-Based Medicines to Cystic Fibrosis Airway Epithelial Cells Using Nanotechnology. Int. J. Nanomed. 2013, 8, 3907–3915. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Cao, M.; Zhang, J.; Hu, K.; Yin, Z.; Zhou, Z.; Xiao, X.; Yang, Y.; Sheng, W.; Wu, Y.; et al. Hyaluronic Acid-Chitosan Nanoparticles for Co-Delivery of MiR-34a and Doxorubicin in Therapy against Triple Negative Breast Cancer. Biomaterials 2014, 35, 4333–4344. [Google Scholar] [CrossRef]
- Gaur, S.; Wen, Y.; Song, J.H.; Parikh, N.U.; Mangala, L.S.; Blessing, A.M.; Ivan, C.; Wu, S.Y.; Varkaris, A.; Shi, Y.; et al. Chitosan Nanoparticle-Mediated Delivery of MiRNA-34a Decreases Prostate Tumor Growth in the Bone and Its Expression Induces Non-Canonical Autophagy. Oncotarget 2015, 6, 29161–29177. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Feng, C.; Hui, G.; Wang, Z.; Tan, J.; Luo, L.; Xue, P.; Wang, Q.; Chen, X. Improving the Osteogenesis of Rat Mesenchymal Stem Cells by Chitosan-Based-MicroRNA Nanoparticles. Carbohydr. Polym. 2016, 138, 49–58. [Google Scholar] [CrossRef]
- Jiang, F.; Yin, F.; Lin, Y.; Xia, W.; Zhou, L.; Pan, C.; Wang, N.; Shan, H.; Zhou, Z.; Yu, X. The Promotion of Bone Regeneration through CS/GP-CTH/Antagomir-133a/b Sustained Release System. Nanomed. Nanotechnol. Biol. Med. 2020, 24, 102116. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, G.; Wei, M.; Liu, Q.; Zhou, J.; Qin, T.; Feng, X.; Liu, H.; Feng, Z.; Zhao, Y. Improving the Osteogenesis of Human Bone Marrow Mesenchymal Stem Cell Sheets by MicroRNA-21-Loaded Chitosan/Hyaluronic Acid Nanoparticles via Reverse Transfection. Int. J. Nanomed. 2016, 11, 2091–2105. [Google Scholar] [CrossRef] [Green Version]
- Tekie, F.S.M.; Soleimani, M.; Zakerian, A.; Dinarvand, M.; Amini, M.; Dinarvand, R.; Arefian, E.; Atyabi, F. Glutathione Responsive Chitosan-Thiolated Dextran Conjugated MiR-145 Nanoparticles Targeted with AS1411 Aptamer for Cancer Treatment. Carbohydr. Polym. 2018, 201, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Çelik, E.; Bayram, C.; Denkbaş, E.B. Chondrogenesis of Human Mesenchymal Stem Cells by MicroRNA Loaded Triple Polysaccharide Nanoparticle System. Mater. Sci. Eng. C 2019, 102, 756–763. [Google Scholar] [CrossRef]
- Cosco, D.; Cilurzo, F.; Maiuolo, J.; Federico, C.; Di Martino, M.T.; Cristiano, M.C.; Tassone, P.; Fresta, M.; Paolino, D. Delivery of MiR-34a by Chitosan/PLGA Nanoplexes for the Anticancer Treatment of Multiple Myeloma. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Carballal, B.; Aaldering, L.J.; Ritzefeld, M.; Pereira, S.; Sewald, N.; Moerschbacher, B.M.; Götte, M.; Goycoolea, F.M. Physicochemical and Biological Characterization of Chitosan-MicroRNA Nanocomplexes for Gene Delivery to MCF-7 Breast Cancer Cells. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef]
- Pereira, P.A.; Tomás, J.F.; Queiroz, J.A.; Figueiras, A.R.; Sousa, F. Recombinant Pre-MiR-29b for Alzheimer’s Disease Therapeutics. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Kaban, K.; Salva, E.; Akbuga, J. The Effects of Chitosan/MiR-200c Nanoplexes on Different Stages of Cancers in Breast Cancer Cell Lines. Eur. J. Pharm. Sci. 2016, 95, 103–110. [Google Scholar] [CrossRef]
- Zhou, F.; Yuan, X. Targeted Delivery of MicroRNA-126 to Vascular Endothelial Cells: Via REDV Peptide Modified PEG-Trimethyl Chitosan. Biomater. Sci. 2016, 4, 849–856. [Google Scholar] [CrossRef]
- Louw, A.M.; Kolar, M.K.; Novikova, L.N.; Kingham, P.J.; Wiberg, M.; Kjems, J.; Novikov, L.N. Chitosan Polyplex Mediated Delivery of MiRNA-124 Reduces Activation of Microglial Cells in Vitro and in Rat Models of Spinal Cord Injury. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Tu, L.; Wang, M.; Zhao, W.Y.; Zhang, Z.Z.; Tang, D.F.; Zhang, Y.Q.; Cao, H.; Zhang, Z.G. MiRNA-218-Loaded Carboxymethyl Chitosan - Tocopherol Nanoparticle to Suppress the Proliferation of Gastrointestinal Stromal Tumor Growth. Mater. Sci. Eng. C 2017, 72, 177–184. [Google Scholar] [CrossRef]
- Kaban, K.; Salva, E.; Akbuga, J. Modulation of the Dual-Faced Effects of MiR-141 with Chitosan/MiR-141 Nanoplexes in Breast Cancer Cells. J. Gene Med. 2019, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xie, H.; Yu, J.; Chen, X.; Tang, S.; Sun, L.; Chen, X.; Peng, D.; Zhang, X.; Zhou, J. Atargeted Therapy for Melanoma by Graphene Oxide Composite with MicroRNA Carrier. Drug Des. Devel. Ther. 2018, 12, 3095–3106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gary, D.J.; Min, J.; Kim, Y.; Park, K.; Won, Y.Y. The Effect of N/P Ratio on the in Vitro and in Vivo Interaction Properties of Pegylated Poly[2-(Dimethylamino)Ethyl Methacrylate]-Based SiRNA Complexes. Macromol. Biosci. 2013, 13, 1059–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alameh, M.; Lavertu, M.; Tran-Khanh, N.; Chang, C.-Y.Y.; Lesage, F.; Bail, M.; Darras, V.; Chevrier, A.; Buschmann, M.D. SiRNA Delivery with Chitosan: Influence of Chitosan Molecular Weight, Degree of Deacetylation, and Amine to Phosphate Ratio on in Vitro Silencing Efficiency, Hemocompatibility, Biodistribution, and in Vivo Efficacy. Biomacromolecules 2018, 19, 112–131. [Google Scholar] [CrossRef] [PubMed]
- Allan, G.G.; Peyron, M. Molecular Weight Manipulation of Chitosan I: Kinetics of Depolymerization by Nitrous Acid. Carbohydr. Res. 1995, 277, 257–272. [Google Scholar] [CrossRef]
- Allan, G.G.; Peyron, M. Molecular Weight Manipulation of Chitosan II: Prediction and Control of Extent of Depolymerization by Nitrous Acid. Carbohydr. Res. 1995, 277, 273–282. [Google Scholar] [CrossRef]
- Vachoud, L.; Zydowicz, N.; Domard, A. Formation and Characterisation of a Physical Chitin Gel. Carbohydr. Res. 1997, 302, 169–177. [Google Scholar] [CrossRef]
- Roux, R.; Ladavière, C.; Montembault, A.; David, L.; Delair, T. Shear Thinning 3D-Colloidal Assemblies of Chitosan and Poly (Lactic Acid) Nanoparticles. J. Phys. Chem. 2013, 117, 7455–7464. [Google Scholar] [CrossRef]
- Shamaeizadeh, N.; Varshosaz, J.; Mirian, M.; Aliomrani, M. Glutathione Targeted Tragacanthic Acid-Chitosan as a Non-Viral Vector for Brain Delivery of MiRNA-219a-5P: An in Vitro/in Vivo Study. Int. J. Biol. Macromol. 2022, 200, 543–556. [Google Scholar] [CrossRef]
- Younes, I.; Rinaudo, M.; Harding, D.; Sashiwa, H. Chitin and Chitosan Preparation from Marine Sources. Structure, Properties and Applications. Mar. Drugs 2015, 13, 1133–1174. [Google Scholar] [CrossRef] [Green Version]
- Pandit, A.; Indurkar, A.; Deshpande, C.; Jain, R.; Dandekar, P. A Systematic Review of Physical Techniques for Chitosan Degradation. Carbohydr. Polym. Technol. Appl. 2021, 2, 100033. [Google Scholar] [CrossRef]
- Wu, D.; Delair, T. Stabilization of Chitosan/Hyaluronan Colloidal Polyelectrolyte Complexes in Physiological Conditions. Carbohydr. Polym. 2015, 119, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yuan, X. Nanofiber-Mediated MicroRNA-126 Delivery to Vascular Endothelial Cells for Blood Vessel Regeneration. Acta Biomater. 2016, 43, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Wen, M.; Zhou, P.; Zhao, Y.; Jia, X.; Fan, Y. Materials Science & Engineering C Electrospun Membranes of PELCL/PCL-REDV Loading with MiRNA-126 for Enhancement of Vascular Endothelial Cell Adhesion and Proliferation. Mater. Sci. Eng. C 2018, 85, 37–46. [Google Scholar] [CrossRef]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour Angiogenesis Regulation by the MiR-200 Family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatami, F.; Matin, M.M.; Mohammad, N.; Reza, A.; Abnous, K.; Mohammad, S. Targeted Delivery System Using Silica Nanoparticles Coated with Chitosan and AS1411 for Combination Therapy of Doxorubicin and AntimiR-21. Carbohydr. Polym. 2021, 266, 118111. [Google Scholar] [CrossRef] [PubMed]
- Golafzani, F.N.; Vaziri, A.Z.; Javanmardi, M. Delivery of MiRNA-126 through Folic Acid-Targeted Biocompatible Polymeric Nanoparticles for Effective Lung Cancer Therapy. J. Bioactive Compat. Polym. 2022, 37, 168–188. [Google Scholar] [CrossRef]
- Mokri, N.; Sepehri, Z.; Faninam, F.; Khaleghi, S.; Kazemi, N.M.; Hashemi, M. Chitosan-Coated Zn-Metal-Organic Framework Nanocomposites for Effective Targeted Delivery of LNA-Antisense MiR-224 to Colon Tumor: In Vitro Studies. Gene Ther. 2021, 1–11. [Google Scholar] [CrossRef]
- Huang, Z.; Ma, J.; Chen, M.; Jiang, H.; Fu, Y.; Gan, J.; Dong, L.; Zhang, J.; Chen, J. Dual TNF- α/IL-12p40 Interference as a Strategy to Protect Against Colitis Based on MiR-16 Precursors With Macrophage Targeting Vectors. Mol. Ther. 2015, 23, 1611–1621. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; He, S.; Cui, S.; Shi, Y. A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-Targeting Nanoparticles Delivering MiR-146b to Intestinal Macrophages. J. Crohn’s Colitis 2019, 13, 482–494. [Google Scholar] [CrossRef]
- Pereira, P.; Barreira, M.; Cruz, C.; Tom, J.; Lu, Â.; Pedro, A.Q.; Queiroz, A.; Sousa, F. Brain-Targeted Delivery of Pre-MiR-29b Using Polyethyleneimine Polyplexes. Pharmaceuticals 2020, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Safari, S.; Zarrintan, M.H.; Soleimani, M.; Dorkoosh, F.A.; Akbari, H.; Larijani, B.; Tehrani, M.R. Evaluation and Optimization of Chitosan Derivatives-Based Gene Delivery System via Kidney Epithelial Cells. Adv. Pharm. Bull. 2012, 2, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zhou, F.; Cui, C.; Zhao, Y.; Yuan, X. Performance of TMC-g-PEG-VAPG/MiRNA-145 Complexes in Electrospun Membranes for Target-Regulating Vascular SMCs. Colloids Surf. B Biointerfaces 2019, 182, 110369. [Google Scholar] [CrossRef] [PubMed]
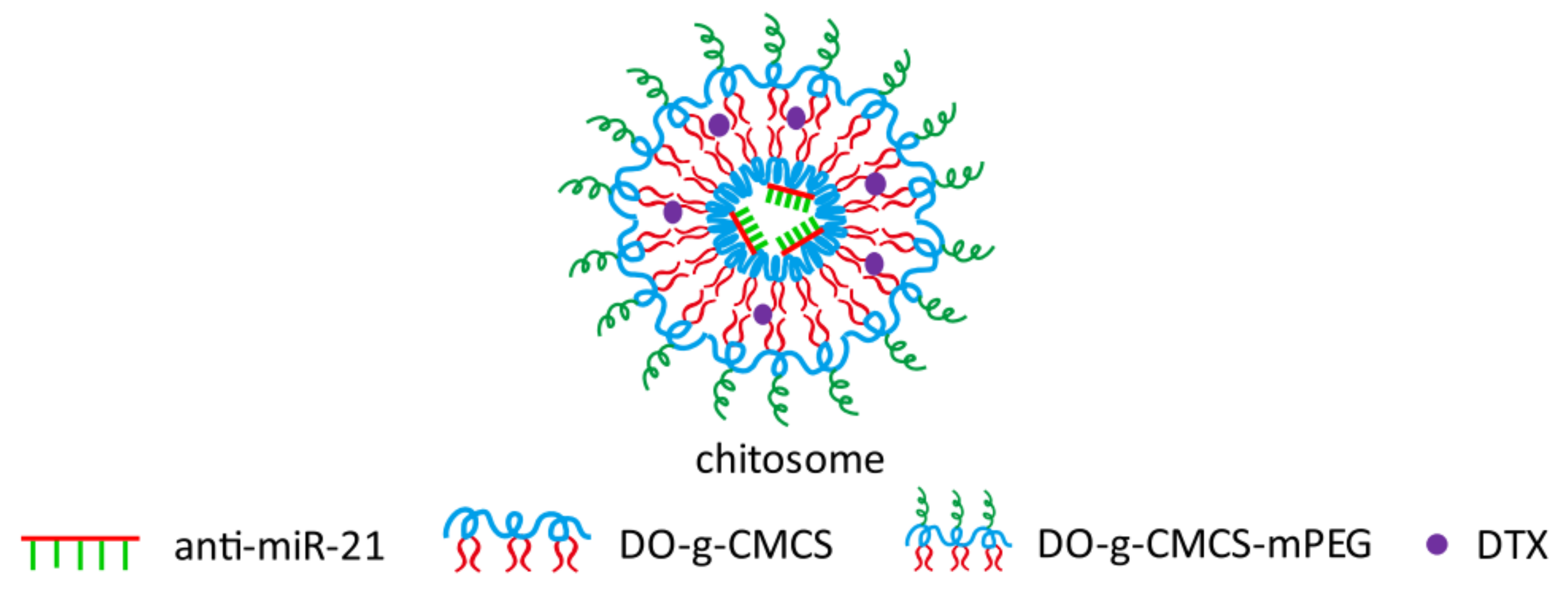
- Sun, X.; Xu, H.; Huang, T.; Zhang, C.; Wu, J.; Luo, S. Simultaneous Delivery of Anti-MiRNA and Docetaxel with Supramolecular Self-Assembled “Chitosome” for Improving Chemosensitivity of Triple Negative Breast Cancer Cells. Drug Deliv. Transl. Res. 2020, 21, 192–204. [Google Scholar] [CrossRef]
- Feczkó, T. Polymeric Nanotherapeutics Acting at Special Regions of Body. J. Drug Deliv. Sci. Technol. 2021, 64, 102597. [Google Scholar] [CrossRef]
- Nguyen, M.A.; Wyatt, H.; Susser, L.; Geoffrion, M.; Rasheed, A.; Duchez, A.C.; Cottee, M.L.; Afolayan, E.; Farah, E.; Kahiel, Z.; et al. Delivery of MicroRNAs by Chitosan Nanoparticles to Functionally Alter Macrophage Cholesterol Efflux In Vitro and In Vivo. ACS Nano 2019, 13, 6491–6505. [Google Scholar] [CrossRef]
- Katas, H.; Alpar, H.O. Development and Characterisation of Chitosan Nanoparticles for SiRNA Delivery. J. Control. Release 2006, 115, 216–225. [Google Scholar] [CrossRef]
- Thipe, V.C.; Raphael, A.; Çakılkaya, P.; Genedy, H.H.; Kaeokhamloed, N.; Roger, E.; Katti, K.V. Green Nanotechnology—An Innovative Pathway towards Biocompatible and Medically Relevant Gold Nanoparticles. J. Drug Deliv. Sci. Technol. 2022, 70, 103256. [Google Scholar] [CrossRef]
- Wu, D.; Ensinas, A.; Verrier, B.; Primard, C.; Cuvillier, A.; Champier, G.; Paul, S.; Delair, T. Zinc-Stabilized Colloidal Polyelectrolyte Complexes of Chitosan/Hyaluronan: A Tool for the Inhibition of HIV-1 Infection. J. Mater. Chem. B 2016, 4, 5455–5463. [Google Scholar] [CrossRef]
- Ban, E.; Kwon, T.H.; Kim, A. Delivery of Therapeutic MiRNA Using Polymer-Based Formulation. Drug Deliv. Transl. Res. 2019, 9, 1043–1056. [Google Scholar] [CrossRef]
- Williams, M.; Cheng, Y.Y.; Blenkiron, C.; Reid, G. Exploring Mechanisms of MicroRNA Downregulation in Cancer. MicroRNA 2017, 6, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-piñeiro, I.; Badiola, I.; Sanchez, A. Nanocarriers for MicroRNA Delivery in Cancer Medicine. Biotechnol. Adv. 2017, 35, 350–360. [Google Scholar] [CrossRef]
- Chen, X.; Gu, S.; Chen, B.; Shen, W.; Yin, Z.; Xu, G.; Hu, J.; Zhu, T.; Li, G.; Wan, C.; et al. Biomaterials Nanoparticle Delivery of Stable MiR-199a-5p Agomir Improves the Osteogenesis of Human Mesenchymal Stem Cells via the HIF1a Pathway. Biomaterials 2015, 53, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Suardi, R.B.; Ysrafil, Y.; Sesotyosari, S.L.; Martien, R.; Wardana, T.; Astuti, I.; Haryana, S.M. The Effects of Combination of Mimic MiR-155-5p and Antagonist MiR-324-5p Encapsulated Chitosan in Ovarian Cancer SKOV3. Asian Pac. J. Cancer Prev. 2020, 21, 2603–2608. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Meng, Z.; Zhao, Z. Silk Fibroin Nanofibrous Scaffolds Incorporated with MicroRNA-222 Loaded Chitosan Nanoparticles for Enhanced Neuronal Differentiation of Neural Stem Cells. Carbohydr. Polym. 2022, 277, 118791. [Google Scholar] [CrossRef]
- Sun, Y.; Kou, Y.; He, X.; Yan, Y.; Guo, X.; Yang, X.; He, N.; Cho, W.C.; Kutyrev, I.; Harandi, M.F.; et al. Efficient Delivery of Echinococcus Multilocularis MiRNAs Using Chitosan Nanoparticles. Biomed. Pharmacother. 2022, 150, 112945. [Google Scholar] [CrossRef]
- Li, K.; Qiu, Y.; Liu, X.; Huang, F. Biomimetic Nanosystems for the Synergistic Delivery of MiR-144/451a for Oral Squamous Cell Carcinoma. Balk. Med. J. 2022, 39, 178–186. [Google Scholar] [CrossRef]
- Alswailem, R.; Alqahtani, F.Y.; Aleanizy, F.S.; Alrfaei, B.M.; Badran, M.; Alqahtani, Q.H.; Abdelhady, H.G.; Alsarra, I. MicroRNA-219 Loaded Chitosan Nanoparticles for Treatment of Glioblastoma. Artif. Cells Nanomed. Biotechnol. 2022, 50, 198–207. [Google Scholar] [CrossRef]
- Kaban, K.; Salva, E.; Akbuga, J. In Vitro Dose Studies on Chitosan Nanoplexes for MicroRNA Delivery in Breast Cancer Cells. Nucleic Acid Ther. 2017, 27, 45–55. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Malichewe, C.; Shi, Z.; Wang, L.; Lu, Z. Chitosan Nanoparticle Mediated Upregulation of MicroRNA34a Expression to Suppress the Proliferation, Migration, Invasion of MDA-MB-231 Cells. J. Drug Deliv. Sci. Technol. 2019, 52, 1061–1069. [Google Scholar] [CrossRef]
- Chien, Y.; Chang, Y.L.; Li, H.Y.; Larsson, M.; Wu, W.W.; Chien, C.S.; Wang, C.Y.; Chu, P.Y.; Chen, K.H.; Lo, W.L.; et al. Synergistic Effects of Carboxymethyl-Hexanoyl Chitosan, Cationic Polyurethane-Short Branch PEI in MiR122 Gene Delivery: Accelerated Differentiation of IPSCs into Mature Hepatocyte-like Cells and Improved Stem Cell Therapy in a Hepatic Failure Model. Acta Biomater. 2015, 13, 228–244. [Google Scholar] [CrossRef] [PubMed]
- Sainitya, R.; Sriram, M.; Kalyanaraman, V.; Dhivya, S.; Saravanan, S.; Vairamani, M.; Sastry, T.P.; Selvamurugan, N. Scaffolds Containing Chitosan/Carboxymethyl Cellulose/Mesoporous Wollastonite for Bone Tissue Engineering. Int. J. Biol. Macromol. 2015, 80, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, G.; Feng, Z.; Bai, S.; Dong, Y.; Wu, G.; Zhao, Y. Microarc-Oxidized Titanium Surfaces Functionalized with MicroRNA-21-Loaded Chitosan/Hyaluronic Acid Nanoparticles Promote the Osteogenic Differentiation of Human Bone Marrow Mesenchymal Stem Cells. Int. J. Nanomed. 2015, 10, 6675–6687. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Liu, C.; Zhao, J.; Li, X.; Li, Z.; Wang, J.; Wang, R.; Liu, Y.; Yuan, X.; Cui, Z.; et al. An Injectable MiRNA-Activated Matrix for Effective Bone Regeneration: In Vivo. J. Mater. Chem. B 2016, 4, 6942–6954. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Agarwal, P.; Zhao, S.; Yu, J.; Lu, X.; He, X. A Near-Infrared Laser-Activated “Nanobomb” for Breaking the Barriers to MicroRNA Delivery. Adv. Mater. 2016, 28, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Balagangadharan, K.; Viji Chandran, S.; Arumugam, B.; Saravanan, S.; Devanand Venkatasubbu, G.; Selvamurugan, N. Chitosan/Nano-Hydroxyapatite/Nano-Zirconium Dioxide Scaffolds with MiR-590-5p for Bone Regeneration. Int. J. Biol. Macromol. 2018, 111, 953–958. [Google Scholar] [CrossRef]
- Wu, K.; Liu, M.; Li, N.; Zhang, L.; Meng, F.; Zhao, L.; Liu, M. Chitosan-MiRNA Functionalized Microporous Titanium Oxide Surfaces via a Layer-by-Layer Approach with a Sustained Release Profile for Enhanced Osteogenic Activity. J. Nanobiotechnol. 2020, 18, 127. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, S.; Jia, L.; Li, Q.; Qiao, J.; Peng, X. Interleukin-1 Receptor Antagonist Protein (IL-1Ra) and MiR-140 Overexpression via PNNS-Conjugated Chitosan-Mediated Gene Transfer Enhances the Repair of Full-Thickness Cartilage Defects in a Rabbit Model. Bone Jt. Res. 2019, 8, 165–178. [Google Scholar] [CrossRef]
- Sukumar, U.K.; Gambhir, S.S.; Massoud, T.F.; Paulmurugan, R. Intranasal Delivery of Targeted Polyfunctional Gold-Iron Oxide Nanoparticles Loaded with Therapeutic MicroRNAs for Combined Theranostic Multimodality Imaging and Presensitization of Glioblastoma to Temozolomide. Biomaterials 2020, 218, 119342. [Google Scholar] [CrossRef]
- Motiei, M.; Aboutalebi, F.; Forouzanfar, M.; Dormiani, K. Materials Science & Engineering C Smart Co-Delivery of MiR-34a and Cytotoxic Peptides (LTX-315 and Melittin) by Chitosan Based Polyelectrolyte Nanocarriers for Specific Cancer Cell Death Induction. Mater. Sci. Eng. C 2021, 128, 112258. [Google Scholar] [CrossRef]
- Ballarín-gonzález, B.; Ebbesen, M.F.; Howard, K.A. Polycation-Based Nanoparticles for RNAi-Mediated Cancer Treatment. Cancer Lett. 2014, 352, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Yagi, K.; Iwakawa, S.; Hirai, M. Chitosan Induces Apoptosis via Caspase-3 Activation in Bladder Tumor Cells. Jpn. J. Cancer Res. 2001, 92, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, H.S.; Yadav, P.N. Anticancer Activity of Chitosan, Chitosan Derivatives, and Their Mechanism of Action. Int. J. Biomater. 2018, 2018, 2952085. [Google Scholar] [CrossRef] [Green Version]
- Abedia, Z.; Moghadamnia, A.A.; Zabihi, E.; Pourbagher, R.; Ghasemi, M.; Nouri, H.R.; Tashakorian, H.; Jenabian, N. Anticancer Properties of Chitosan against Osteosarcoma, Breast Cancer and Cervical Cancer Cell Lines. Casp. J. Intern. Med. 2019, 10, 439. [Google Scholar] [CrossRef]
- García, L.; Urbiola, K.; Düzgüneş, N.; Tros De Ilarduya, C. Lipopolyplexes as Nanomedicines for Therapeutic Gene Delivery. Methods Enzymol. 2012, 509, 327–338. [Google Scholar] [CrossRef]
- Chen, W.; Li, H.; Liu, Z.; Yuan, W. Lipopolyplex for Therapeutic Gene Delivery and Its Application for the Treatment of Parkinson’s Disease. Front. Aging Neurosci. 2016, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Sultankulov, B.; Berillo, D.; Sultankulova, K.; Tokay, T.; Saparov, A. Progress in the Development of Chitosan-Based Biomaterials for Tissue Engineering and Regenerative Medicine. Biomolecules 2019, 9, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campardelli, R.; Baldino, L.; Reverchon, E. Supercritical Fluids Applications in Nanomedicine. J. Supercrit. Fluids 2015, 101, 193–214. [Google Scholar] [CrossRef]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [Green Version]
| siRNA | miRNA | |
|---|---|---|
| Composition | 21–23 nucleotides duplex RNA | 19–25 nucleotides duplex RNA |
| Complementarity to target mRNA | Complete | Partial |
| Target mRNA | Single | Multiple |
| Mechanism | Endo-nucleolytic cleavage of mRNA | - Endo-nucleolytic cleavage of mRNA - Translational repression |
| Clinical use | Therapeutic | - Therapeutic - Drug target - Biomarker |
| CS sample | HDP-1.9 | LDP-1.6 | HDP-12 | LDP-11 | HDP-29 | LDP-25 | HDP-49 | LDP-67 |
|---|---|---|---|---|---|---|---|---|
| DA% | 1.9 | 1.6 | 12 | 11 | 29 | 25 | 49 | 67 |
| Mv (kDa) | 26.1 | 1.3 | 25.5 | 1.2 | 20.2 | 1.14 | 18 | 1.95 |
| Formulation | CS Characteristics | MiRNA | Application | In Vitro | In Vivo | Ref. |
|---|---|---|---|---|---|---|
| (A) CS/Polyanion/MiRNA Nanoparticles | ||||||
| CS- (FD)-miRNA Fluorescein isothiocyanate Dextran | DA 3% Low Mw -alkyl modified CS: *N, N- Diethyl N- Methyl (DEMC) and N- Triethyl CS (TEC) | fluorescent labeled miR | Cancer | High extent transfection to human embryonic Kidney epithelial cell line (HEK 293T) | [63] | |
| CS-TPP 115 nm | DA 10–25% CS glutamate (Protasan® UP G-113, Mw 160 kDa, | miR-126 | Cystic fibrosis | In vitro on human cystic fibrosis bronchial epithelial cells (CFBE41o-) Safe (nontoxic to cells) but not effective | [27] | |
| RGD labelled CS/TPP NPs | 50–190 kDa Mw | miR-200 | Cancer (Preventing tumor angiogenesis) | Decrease tumor cells invasion Vascular normalization Successful uptake by HeyA8 ovarian cancer cell line. | [56] | |
| DOX-miRNA-34a co-loaded HA-CS TPP NPs | CS-HCl salt 110 kDa | miR-34a | Cancer synergistic effects | 80% intracellular uptake | [28] | |
| CS/TPP NPs | - | miR-34a | Bone-metastatic Prostate Cancer | PCa cell lines of high metastatic potential | [29] | |
| CS/PLGA NPs 160 nm +45 mV surface charge | DA 15% low Mw < 10 kDa copolymer of β (1 → 4) linked 2-acetamido-2-deoxy-β -D-glucopyranose and 2-amino- 2-deoxy-β -D-glucopyranose. | miR-34a | Multiple myeloma | High transfection efficiency → By FACS analysis of SKMM1 (human multiple myleoma cell line) after 6 h treatment with the NPs | Decreased tumor size | [35] |
| CS/TPP NPs | CS HCl (Cl113) Mw 110 kDa | miR-199a-5p agomir | Bone regeneration | Successful transfection of cultured hMSCs | Improved bone regeneration of in rat tibia defect model. | [74] |
| microRNA-21-loaded CS/HA NPs | DA 10% Mw 100 kDa | miR-21 | Enhancing osteogenesis of (hBMMSC) human bone marrow mesenchymal stem cell sheets | Higher than 90% transfection efficacy after 24 h of treatment and more than 75% on the 14th day. Significant improvement in the in vitro osteogenic differentiation (through increased calcification-related gene expression, improved production of alkaline phosphatase, more secretion of collagen, and mineralized nodule formation. | [32] | |
| CS/TPP/HA NPs | DA 10% Mw 100 kDa | AntimiR-138 | Improving osteogenesis | Rat bone marrow mesenchymal stem cells (MSCs) Cytocompatibility with MSCs. Around 70% transfection efficiency significantly improving MSCs osteogenesis | [30] | |
| CS-TPP NPs | low Mw Cs | miR-34a | Cancer | Prostate tumor PC3MM2 cells | [9] | |
| CS-carboxymethyl dextran PECs | CS 9, 18, and 45 kDa obtained by NaNO2 depolymerization of 400 kDa Mw CS | miR-145 | Cancer | Aim to optimize PECs dissociation rate and stability Distinct proliferation reduction of MCF-7(human breast cancer cell line) | [26] | |
| Glutathione responsive CS-thiolated dextran conjugated miRNA-145 NPs targeted with AS1411 aptamer (40–270 nm) | DA 11% 18 kDa | miR-145 | Cancer | Breast cancer MCF7 cell line A significant increase of miRNA-145 in the NP -treated cells A prominent decrease of MUC1 protein after treatment A decrease in relative cell viability to 80% | [33] | |
| CS-TPP NPs 150−200 nm | DA 15-25% Mw (50–190 kDa) | miR-33 | Prevention and treatment of cardiovascular diseases (CVD) by delivering NPs to macrophages where they can function to regulate ABCA1 expression and cholesterol efflux, - target atherosclerotic lesions. | Transfer exogenous miRNA-33 to naive macrophages and reduce the expression of ABCA1, a potent miRNA-33 target gene, both in vitro and in vivo | Mice treated with miRNA33-chNPs showed decreased reverse cholesterol transport (RCT) to the plasma, liver, and feces. - Improvement of cholesterol efflux into the RCT pathway and ABCA1 expression when treated with CS NPs loaded with efflux-promoting miR. | [67] |
| Triple polysaccharide (chondroitin sulfate/hyaluronic acid/CS) nanoparticles | DA 20% 150 kDa | miR-149-5p | Chondrogenesis of human mesenchymal stem cells (osteoarthritis) | 60.97% gene loading efficiency 82.98% increase in miRNA level compared to control group, inhibiting the target gene by 61.57% No toxicity to human mesenchymal stem cells Increased miRNA levels in cells Target gene inhibition Induce chondrogenic markers synthesis and responsible genes upregulation. | [34] | |
| CS/TPP/HA NPs (100–300 nm) in CS/glycerophosphate (GP) gel | - | antimiR-133a/b | Bone regeneration | No cytotoxicity to murine bone marrow stromal cells (BMSCs) Enhanced osteogenic differentiation. | Promote osteogenesis in vivo in mouse calvarial bone defect model. | [31] |
| CS/TPP NPs | Medium Mw (non-specified) | miR-155-5p & AntagomiR-324-5p | Ovarian Cancer | In vitro in cell line SKOV3 Targets’ (HIF1α & GLI)1 expression was downregulated after treatment | [75] | |
| CS/TPP NPs (350 nm) incorporated with silk fibroin (SF) nanofibrous scaffolds | CS Mw of 300 kDa DA 15% | miR-222 | Neural stem cells (NSCs) transplantation therapy for neural tissue regeneration | Good cytocompatibility Improve NSCs neuronal differentiation. | [76] | |
| CS/TPP NPs 150 nm | CS (Macklin) | E. multilocularis miR-4989 | Treatment of Alveolar echinococcosis “Zoonotic disease” | Low cytotoxicity Strong miRNA protection No significant effects on cell proliferation nor apoptosis. | Remarkable liver tropism. UBE2N expression significantly decreased in the liver on both mRNA and protein levels. | [77] |
| CS/TPP NPs camouflaged with macrophage-derived exosomes [MEXO] 143.2 ± 14 nm /−10.3 ± 1.6 mV | CS HCl | miR-144/451a | Oral squamous cell carcinoma (OSCC) | MiR protection from degradation. Cytotoxic to OSCC (in UM-SCC083A and UPCI-SCC029B cell lines). Inhibition of the expression of calcium-binding protein 39 (CAB39) and migration inhibitory factor (MIF). | [78] | |
| Folic acid targeted CS/TPP NPs ~100nm +7.3 ± 2 mV | 50–190 kDa | miR-126 | Lung Cancer | Significantly cytotoxic to A549 (folic acid receptor-positive lung cancer cell line) Cytocompatible with MRC5 (Normal human diploid cell line) | [58] | |
| CS/TPP NPs 112.2 ± 2.27 nm 33.9 ± 0.9mV | 50–190 kDa DA% 15–25% | miR-219 | Glioblastoma | Reduced survival of human GBM cell line (U87MG) No cytotoxic effect on fibroblasts | [79] | |
| (B) CS-miRNA nano polyplexes (Nanoplexes) | ||||||
| CS nanoplexes: preparation via coacervation method | DA 15% Medium Mw CS 9, 18, and 45 kDa | miR-145 | Cancer | Expression in MCF-7 breast cancer cells | [25] | |
| CS–has-miRNA-145 nanoplexes | DA = 1.5%, Mw = 543 kDa modified into: [2–26 kDa] [2–67 % DA%] | miR-145 | Cancer | MCF-7 breast cancer cell line. Nontoxic Determined transfection efficiency by successful downregulation of the expression of target (JAM-A) mRNA in MCF-7 cells. | [36] | |
| Galactosylated low Mw CS (G-LMWC) nano complexes | low Mw (non-specified) | miR-16 precursors | Colitis Crohn’s diseases | MiR-16 specific upregulation of in colonic macrophages of colitic mice which significantly reduces TNF-α and IL-12p40 expression, stopping mucosal inflammation and leading to relief of colitis symptoms. | [60] | |
| CS/pre-miRNA-29b nanoplexes~130 nm | Mw = 50–190 kDa | Recombinant pre-miRNA-29b | Alzheimer’s disease (AD) | Efficient delivery of the pre-miR to N2a695 (mouse neuroblasts cell line)78% decrease of hBACE1 protein expression44% decrease of Aβ42 levels | [37] | |
| CS/miRNA-200c nanoplexes (294 nm) | DA 15–25% 75 kDa | miR-200c | Breast cancer | Reduced angiogenesis, invasion, and metastasis. Increase in apoptosis by 3.1, 1.3, and 3-fold in the MCF-7, MDA-MB-231, MDA-MB-435 breast cancer cell lines. | [38] | |
| CS/miRNA-141 CS/miRNA-200c nanoplexes [294–380 nm] | miR-200c- and miRNA-141- | Breast cancer (Dose study) | 100% transfection efficiency in the MCF-7, MDA-MB-231, and MDA-MB-435 breast cancer cell lines. | [80] | ||
| REDV peptide modified PEG trimethyl CS TMC-g-PEG-REDV/miRNA nanoplexes | DA 5% Mw 50 kDa modified into TMC | miR-126 | Vascular endothelial cells for cardiovascular and cancer therapeutics | VECs and VSMCs (primary culture from Sprague-Dawley rat aorta, passage 3–5) MiRNA expression efficiency up to 3.4-fold compared to control group. Cytocompatibility High transfection efficiency, increase in endothelial cellular uptake, and improved VEC proliferation. | [39] | |
| CS nanoplexes 222.0 nm | DA 5% Mw 30 kDa, 150 kDa and 250 kDa | miR-124 | Spinal cord injury | Decreased microglial cells activation No significant decrease in viability - (Ex vivo in Neonatal rat microglia) Decrease of TNF-α and reactive oxygen species (ROS) Decreased MHC-II expression. | Decreased microglial cells activation in rat models (adult female Sprague-Dawley rats). Significant decrease in ED-1 positive macrophages in spinal cord injury | [40] |
| miRNA-218-loaded carboxymethyl CS-Tocopherol NPs ~110 nm | DA 10% O-carboxymethyl CS (OCMC) tocopherol polymer conjugate | miR-218 | GIT stromal tumor | Human GIST cell lines (GIST882) Inhibited proliferation and exhibited superior apoptosis. Inhibited cell invasion Promoted GIST cancer cells apoptosis | [41] | |
| GO-CS-MPG-miRNA33a/miRNA199a nanoplexesGO: Graphene oxide MPG: Cell penetrating peptide | - | miR33a/miR199a | Melanoma | Human melanoma A375 and L929 cells. Well compatible with L929 cells with low cytotoxicity at 80 mg/mL. Transfection: A375 pinocytosis of FITC-GO-CS-MPG microspheres particles could suppress melanoma A375 cells growth. | Subcutaneous tumor implantation in nude mice Significant suppression of the tumor volume | [43] |
| CS/miRNA-141 nanoplexes 296 nm | 75 kDa DA 15–25% | miR-141 | Breast cancer | MDA-MB-231 and MDA-MB-435 cells Relative miRNA expression increased by 7.5-fold compared to control group in MDA-MB-435 cells. Decrease in metastasis, VEGF and invasion. Increased apoptosis levels by 1.5- and 2.4-fold. | [42] | |
| galactosylated-CS-5-fluorouracil (GC-FU) NPs 178.5 nm | DA 4% Mw 500 kDa | Liver-Specific miR-122 | Synergistic Therapy for Hepatocellular Carcinoma | ~95% transfection efficiency Enhanced blood and salt stability, Marked induction of HepG2 cells apoptosis, cell cycle arrest. Decreased HCC cells proliferation, migration, and invasion Downregulation of Bcl-2 and ADAM17 expression in HepG2 cells | By subcutaneous xenografts in the armpit of BALB/c nude mice. Suppressed tumor growth. High biocompatibility. No weight loss nor alteration of liver, heart, and kidney functions. | [24] |
| CS/miR-34a nanoplex ~135 nm +34 mV | Mw 50 kDa; 15–25% DA | miR-34a | Breast Cancer | Increased relative expression level of the miR in MDA-MB-231 cell line. Nontoxic to HUVECs normal cells. Inhibited growth, migration, and invasion of MDA-MB-231. | [81] | |
| mannose-modified trimethyl CS NPs [MTC-miR146b] NPs 213.6 nm +28.3 mV | Trimethyl CS TMC quaternization degree =50% Mw 100 kDa | miR146b | Ulcerative Colitis | Strongly inhibited M1 macrophage activation leading to suppression of pro-inflammatory cytokines induction. Overexpression in bone marrow-derived macrophages [BMDMs]. Improvement of colon epithelial cells proliferation. | After mucosal damage, NPs restored body weight and function of mucosal barrier. Protected miR-146b-deficient mice from dextran sodium sulphate [DSS] injury and consequent cancer | [61] |
| Lactoferrin-Stearic Acid-Modified-CS Polyplexes CS-SA-Lf/pre-miR-29b 325.60 ± 30.99 nm +33.50 ± 2.31 mV | 190–310 kDa; 15–25% DA | Pre-miR-29b | Alzheimer’s disease (AD) treatment | Non-toxic. Enhanced brain-targeting ability Suppression of BACE1 mRNA involved in AD. NPs with two ligands had higher internalization than those with one or no ligands, in neuronal cells. Efficient crossing of BBB | [62] | |
| Glutathione (Glu) targeted tragacanthic acid (TA)- CS Polyplexes | 12 kDa CS (Obtained by NaNO2 depolymerization from 50 kDa) | miRNA-219a-5P | Multiple sclerosis (MS) Brain delivery | In vivo injected into the cuprizone model of MS mice Decreased inflammation and increased brain cell regeneration Overexpression of miR-219, upregulation of crystallin alpha B, and downregulation of apolipoprotein E. | [50] | |
| (C) Scaffolds and other systems | ||||||
| nanostructured amphipathic carboxymethyl–hexanoyl CS (CHC) | miR-122 liver specific | Stem cell therapy for hepatic failure | High transfection efficiency. Facilitated differentiation of human dental. pulp-derived iPSCs (DP-iPSCs) into iPSC-Heps (miRNA122-iPSC-Heps) with functional mature hepatocytes. Hepatoprotection in fulminant hepatic failure experimental model. | [82] | ||
| Scaffolds containing CS/carboxymethyl cellulose/mesoporous wollastonite | Low Mw CS | _ | Bone tissue engineering | Human osteoblastic cells (MG-63) Significant enhancement in protein adsorption and biomineralization properties. Cytocompatibility with human osteoblastic cells. Osteogenic potential proved by calcium deposition and expression of an osteoblast specific miR. | [83] | |
| Microarc-oxidized titanium surfaces decorated with miR-21-loaded CS/HA NPs 160 ± 10 nm, positive ZP | DA 10% Mw 100 kDa | miR-21 | (Dental) promote the osteogenic differentiation of human bone marrow mesenchymal stem cells | Human bone marrow mesenchymal stem cells (hBMMSCs). Transfection efficiency of more than 90%. Nontoxic | [84] | |
| TMC-g-PEG-REDV/miRNA loaded in an electrospun bilayer vascular scaffold | DA 5% Mw ∼50 kDa then modified into TMC | miR-126 | Vascular endothelial cells for blood vessel regeneration | Significant down regulation of SPRED-1 gene expression in VECs on the third day of treatment. The membranes release miR accelerating VEC proliferation within 9 days. | Replacement of rabbit carotid artery for 8 weeks suggested that the PELCL-TPRm scaffold loaded with miRNA could enhance endothelialization in vivo | [54] |
| In situ injectable miR-activated matrix miR-loaded-NPs entrapped into an O-carboxymethyl CS (CMCS) network | Carboxymethylation was 80% (4 kDa) | miR-21 | Bone regeneration/repair. | Human umbilical cord mesenchymal stem cells (hUMSCs). High uptake efficiency (61.6%). Significantly promoted osteogenic differentiation of (hUMSCs) as evidenced by upregulation of osteogenic markers (alkaline phosphatase ALP and runt-related transcription factor (RUNX-2). | 60 male Sprague-Dawley (SD) rats Promoted bone formation (osteogenesis) significantly (2.4-fold) compared to controls | [85] |
| A NIR laser activated Lipopolyplex “Nano-bomb” developed as w/o/w system with miR-34a in the inner hydrophilic phase dispersed in the hydrophobic phase of PLGA, Pluronic F127 & DPPC in an external hydrophilic phase of HA and CS-modified-Pluronic F127. | - | miR-34a | Prostate Cancer | In 2D cultured PC-3 cancer cells and 3D CSCs-enriched prostaspheres. Minimal cytotoxicity | Excellent anticancer safety and efficacy in mice Ability of targeting Significant reduction of tumor volume | [86] |
| CS/nano-hydroxyapatite/nano-zirconium-dioxide scaffolds | - | miR-590-5p | Bone regeneration | Mouse mesenchymal stem cells (C3H10T1/2) Stimulated mMSCs differentiation towards osteoblasts - Biocompatible nature | [87] | |
| miR-126 encapsulated by REDV peptide-modified TMC-g-PEG loading PELCL/PCL-REDV electrospun membranes | TMC-g-PEG-REDV synthesized by a bifunctional PEG linking TMC with a short peptide REDV | miR-126 | Cardiovascular preventing thrombosis | Enhanced VEC adhesion and proliferation Downregulation of SPRED-1 gene expression. | [55] | |
| TMC-g-PEG-VAPG/miRNA-145 NPs in electrospun membranes | DA 10% Mw 50 kDa TMC | miR-145 | target-regulating vascular SMCs - small-diameter blood vessel regeneration. | Low cytotoxicity Enhance cellular uptake in SMCs Inhibit the excessive proliferation and intimal hyperplasia Maintain SMCs in controlled proliferation | [64] | |
| supramolecular self-assembled “chitosome” with anti-miRNA & docetaxel 90 nm | Carboxymethyl CS (CMCS) Mw 10,000 kDa further modified into DO-g-CMCS-mPEG2000 | hydrophilic anti-miR-21 | triple negative breast cancer (TNBC) | Significant improvement in transfection efficiency and stability against enzymatic cleavage. Improved chemosensitivity of TNBC cells (cell line of MDA-MB-231) through synergistic mechanisms. | [65] | |
| polyelectrolyte multilayers (PEMs) of (CS-miRNA) complex & Na-hyaluronate (HA) on microarc-oxidized (MAO) microporous TiO2 surfaces. | Mw 100 kDa 5% DA | antimiR-138 | Bone regeneration “Advanced implants to promote osseointegration” | Significant knockdown of miR-138 Nontoxic Increased osteogenic differentiation of MSCs (increased alkaline phosphatase, collagen production, and ECM mineralization) | Improved osseointegration in rat femur model | [88] |
| A phosphorylatable nuclear localization signal-linked nucleic kinase substrate short peptide (pNNS) conjugated to CS (pNNS-CS) | _ | miRNA-140 | Treating cartilage defects | Transfected into chondrocytes. High miRNA-140 expression levels. Improved proliferation of chondrocytes. Improved production of glycosaminoglycan GAG - Higher levels of TIMP-1, aggrecan and collagen type II alpha 1 chain. Suppression of NO, disintegrin and (MMP)-13 levels. | Target: improve repair of full-thickness cartilage defects in a rabbit model - Injected into the knee joint cavity - High miRNA-140 expression levels - Reduced synovial fluid GAG and NO levels. - Reduced cartilage ADAMTS-5 and MMP-13 levels. - Increased COL2A1, ACAN, and TIMP-1 levels - Decreased cartilage Mankin score in transgenic group. | [89] |
| Au-IONPs coated with β-cyclodextrin-CS (CD-CS) hybrid polymer to co-load both miR-100 and antimiR-21. With PEG-T7 peptide as a surface functionalization. | β-cyclodextrin-CS (CD-CS) hybrid polymer | miR-100 and AntimiR-21 | Cancer (Glioblastoma) Theranostics | Enhanced cellular uptake of miR-100 and AntimiR-21 in GBM. Pre-sensitized GBM Cells to (TMZ) chemotherapy. Synergistic effect activating apoptotic signaling pathway in GBM cells leading to improvement of TMZ therapy. | - Intranasal delivery to U87-MG GBM cell-derived orthotopic xenograft mice models. - Cy5-miRNAs accumulated efficiently in mice - Significant increase in survival of mice co-treated with NPs and systemic TMZ. | [90] |
| polyglutamate PGA grafted CS core, dextran sulfate as a complexing agent, poly-ethylene-imine shell decorated with folic acid. 123 ± 5 nm-36 ± 1 mV ZP | - | miR-34a in the shell and cytotoxic peptides by the core. | Cancer | No cytotoxicity A synergistic effect leading to multiple cell death in chemoresistance human breast adenocarcinoma cell line, MDA-MB-231. - enhanced smart death induction by 54%. | [91] | |
| DOX/Mesoporous Silica NPs coated with CS to electrostatically attach antimir-21 and Aptamer AS1411 | 50 kDa –190 kDa 15-25% DA | antimiR-21 | Cancer | Cytotoxic to nucleolin positive (C26, MCF-7, and 4T1). | Anticancer activity in C26 colorectal cancer bearing mice | [57] |
| FA functionalized CS-coated Zn-MOF nanocomposites ~ 200nm | 50–190 kDa Mw | LNA-antisense miR-224 | Colon Cancer | In vitro on HCT116 (FA receptor-positive colon cancer cell line) and CRL1831 (normal colon cell line) Decreased cell viability Upregulated apoptotic genes expression Upregulated autophagy-related genes expression | [59] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genedy, H.H.; Delair, T.; Montembault, A. Chitosan Based MicroRNA Nanocarriers. Pharmaceuticals 2022, 15, 1036. https://doi.org/10.3390/ph15091036
Genedy HH, Delair T, Montembault A. Chitosan Based MicroRNA Nanocarriers. Pharmaceuticals. 2022; 15(9):1036. https://doi.org/10.3390/ph15091036
Chicago/Turabian StyleGenedy, Hussein H., Thierry Delair, and Alexandra Montembault. 2022. "Chitosan Based MicroRNA Nanocarriers" Pharmaceuticals 15, no. 9: 1036. https://doi.org/10.3390/ph15091036
APA StyleGenedy, H. H., Delair, T., & Montembault, A. (2022). Chitosan Based MicroRNA Nanocarriers. Pharmaceuticals, 15(9), 1036. https://doi.org/10.3390/ph15091036






