Healthcare Facilities as Potential Reservoirs of Antimicrobial Resistant Klebsiella pneumoniae: An Emerging Concern to Public Health in Bangladesh
Abstract
:1. Introduction
2. Results
2.1. Distribution of ESBL-KP and CRKP among the Hospitals
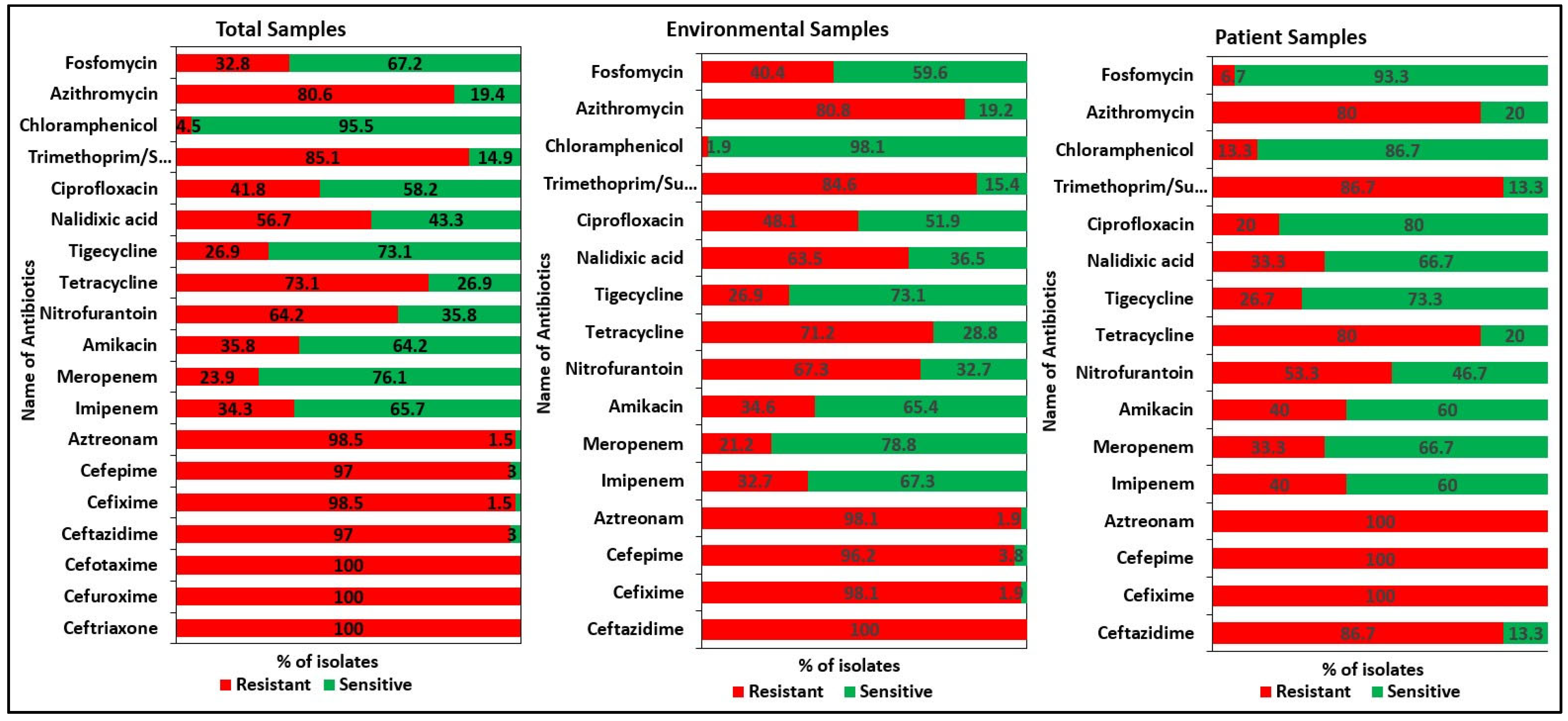
2.2. Antibiotic Resistance Patterns of the Isolates
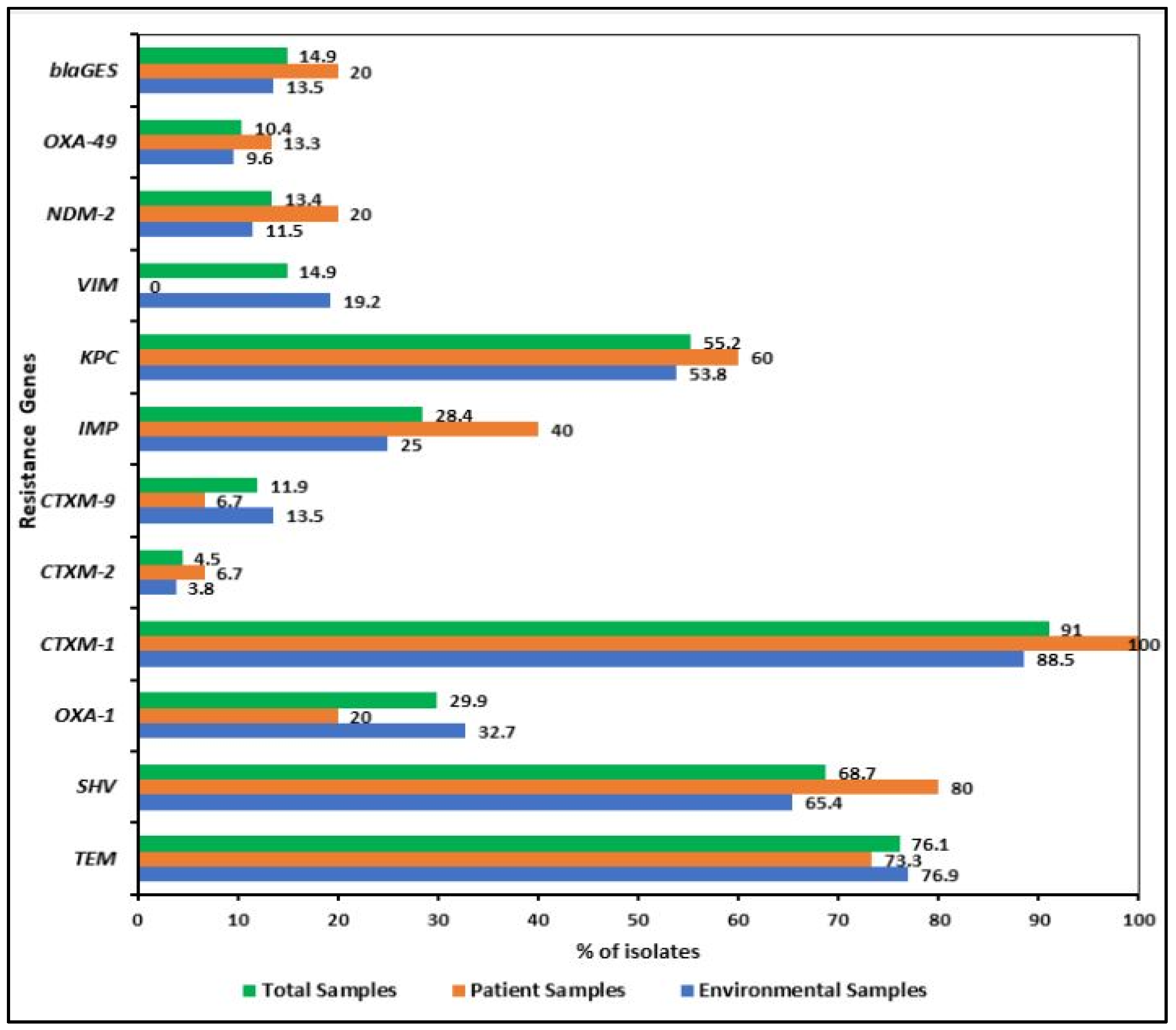
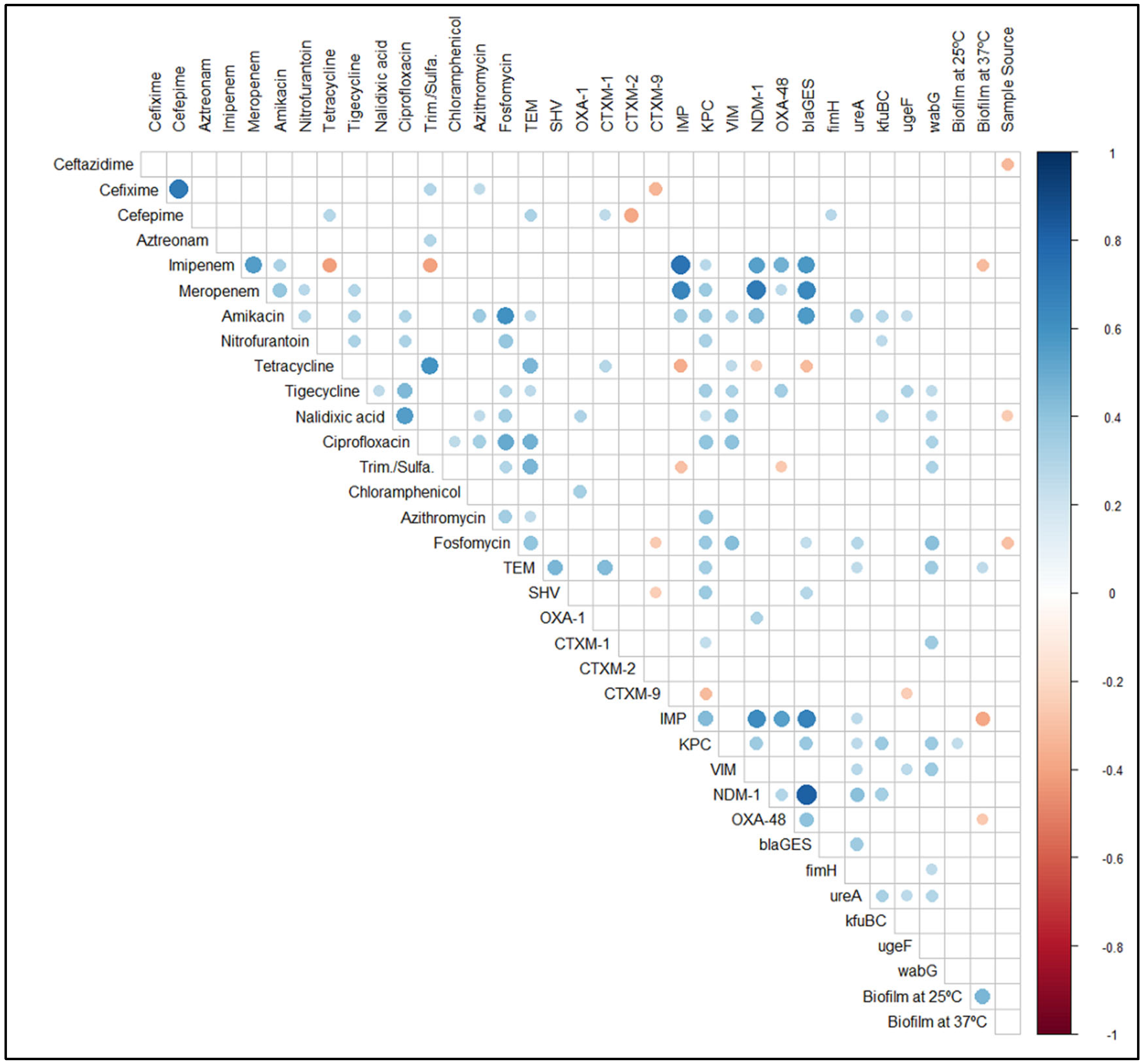
2.3. Detection of Virulence and Resistance Genes
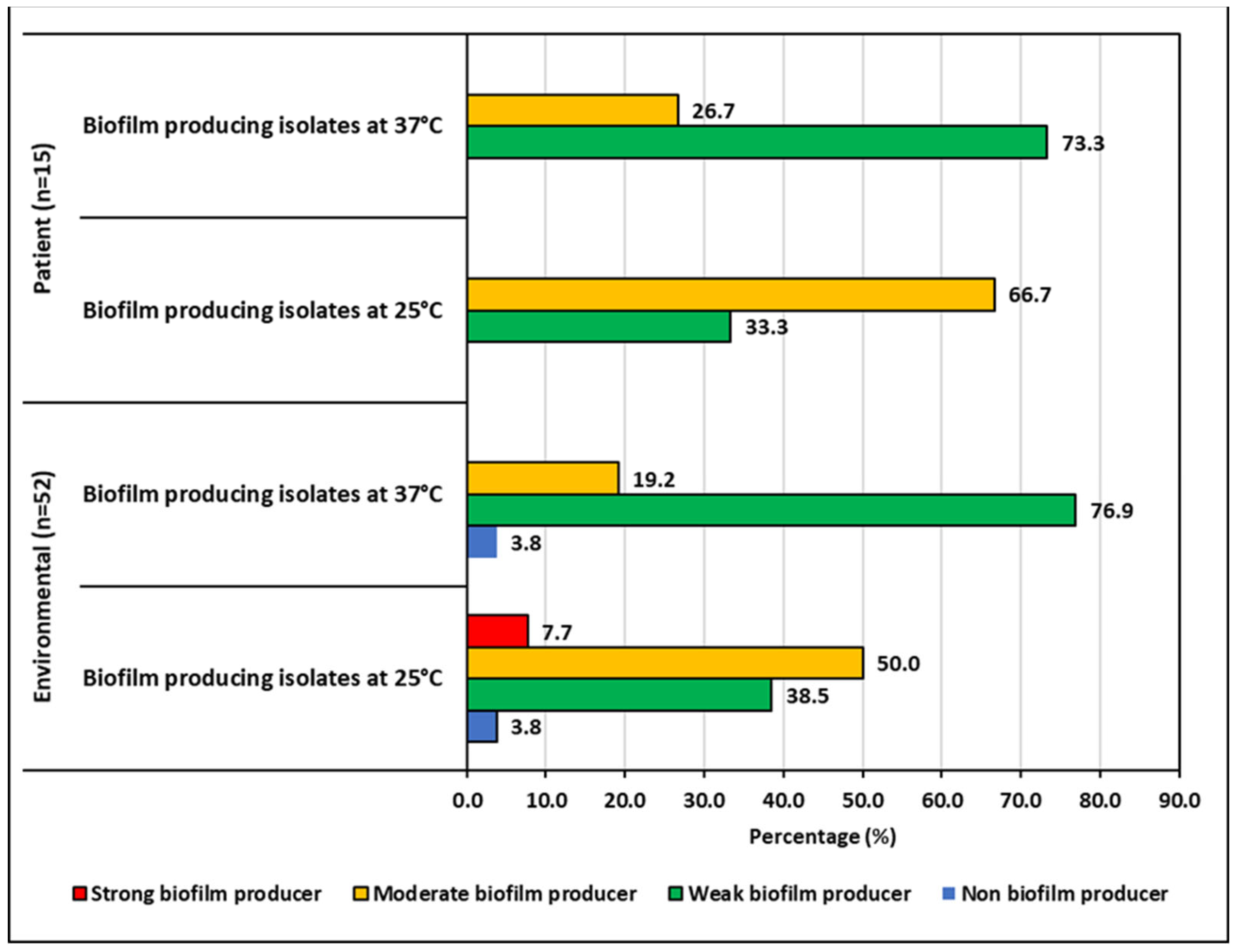
2.4. Biofilm Forming Ability of the ESBL-KP and CRKP Isolates
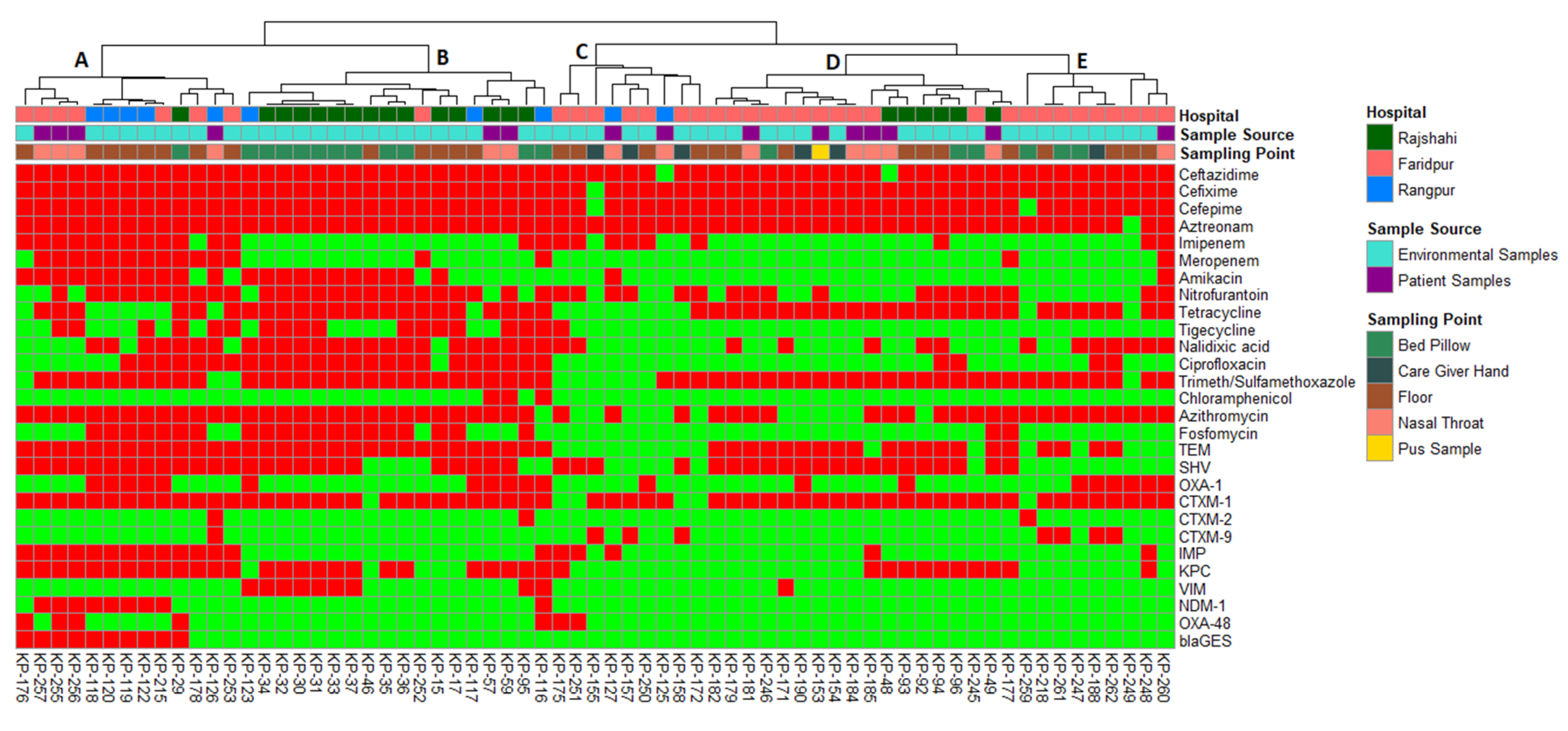
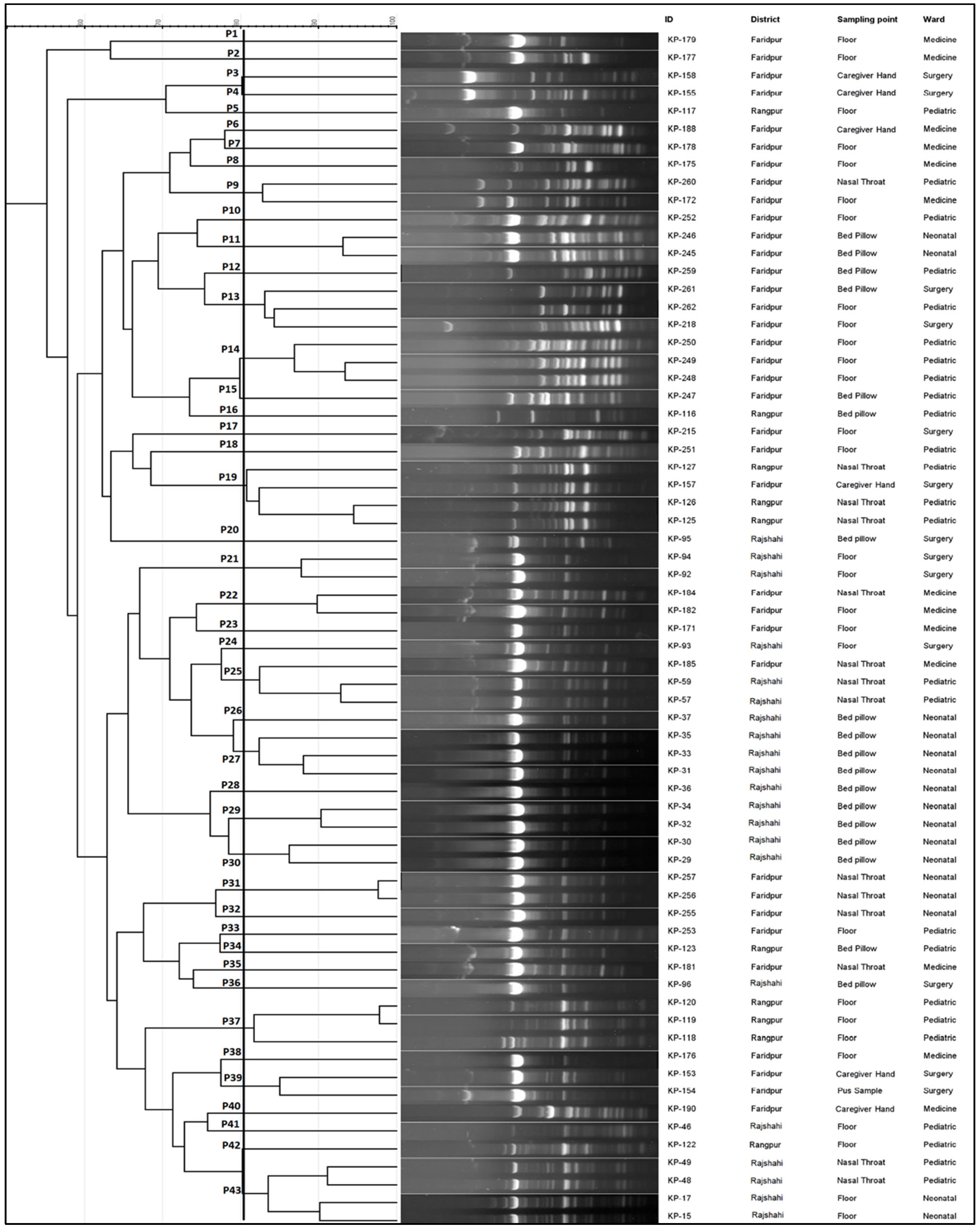
2.5. Molecular Typing of the Isolates
2.6. Associations between Sample Source and Phenotypic and Genotypic Traits
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Isolation of K. pneumoniae
4.2. Detection of Virulence and Resistance Genes
4.3. Molecular Typing of K. pneumoniae
4.4. Biofilm Formation
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-Producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791. [Google Scholar] [CrossRef]
- Temkin, E.; Adler, A.; Lerner, A.; Carmeli, Y. Carbapenem-resistant Enterobacteriaceae: Biology, Epidemiology, and Management. Ann. N. Y. Acad. Sci. 2014, 1323, 22–42. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Sandler, N.; Libby, S.J. Liver Abscess Caused by MagA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 2005, 43, 991–992. [Google Scholar] [CrossRef]
- Fang, C.-T.; Lai, S.-Y.; Yi, W.-C.; Hsueh, P.-R.; Liu, K.-L.; Chang, S.-C. Klebsiella pneumoniae Genotype K1: An Emerging Pathogen That Causes Septic Ocular or Central Nervous System Complications from Pyogenic Liver Abscess. Clin. Infect. Dis. 2007, 45, 284–293. [Google Scholar] [CrossRef]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World Health Organization Releases Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. J. Med. Soc. 2018, 32, 76. [Google Scholar] [CrossRef]
- Podschun, R.; Ullmann, U. Klebsiella Spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef]
- Brinkworth, A.J.; Hammer, C.H.; Olano, L.R.; Kobayashi, S.D.; Chen, L.; Kreiswirth, B.N.; DeLeo, F.R. Identification of Outer Membrane and Exoproteins of Carbapenem-Resistant Multilocus Sequence Type 258 Klebsiella pneumoniae. PLoS ONE 2015, 10, e0123219. [Google Scholar] [CrossRef]
- Yu, W.-L.; Ko, W.-C.; Cheng, K.-C.; Lee, H.-C.; Ke, D.-S.; Lee, C.-C.; Fung, C.-P.; Chuang, Y.-C. Association between RmpA and MagA Genes and Clinical Syndromes Caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 2006, 42, 1351–1358. [Google Scholar] [CrossRef]
- Lawlor, M.S.; O’connor, C.; Miller, V.L. Yersiniabactin Is a Virulence Factor for Klebsiella pneumoniae during Pulmonary Infection. Infect. Immun. 2007, 75, 1463–1472. [Google Scholar] [CrossRef]
- Wu, C.-C.; Wang, C.-K.; Chen, Y.-C.; Lin, T.-H.; Jinn, T.-R.; Lin, C.-T. IscR Regulation of Capsular Polysaccharide Biosynthesis and Iron-Acquisition Systems in Klebsiella pneumoniae CG43. PLoS ONE 2014, 9, e107812. [Google Scholar]
- Mizuta, K.; Ohta, M.; Mori, M.; Hasegawa, T.; Nakashima, I.; Kato, N. Virulence for Mice of Klebsiella Strains Belonging to the O1 Group: Relationship to Their Capsular (K) Types. Infect. Immun. 1983, 40, 56–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ofek, I.; Kabha, K.; Athamna, A.; Frankel, G.; Wozniak, D.J.; Hasty, D.L.; Ohman, D.E. Genetic Exchange of Determinants for Capsular Polysaccharide Biosynthesis between Klebsiella pneumoniae Strains Expressing Serotypes K2 and K21a. Infect. Immun. 1993, 61, 4208–4216. [Google Scholar] [CrossRef] [PubMed]
- Ørskov, I.D.A.; FIFE-ASBURY, M.A. New Klebsiella Capsular Antigen, K82, and the Deletion of Five of Those Previously Assigned. Int. J. Syst. Evol. Microbiol. 1977, 27, 386–387. [Google Scholar] [CrossRef]
- Trautmann, M.; Ruhnke, M.; Rukavina, T.; Held, T.K.; Cross, A.S.; Marre, R.; Whitfield, C. O-Antigen Seroepidemiology of Klebsiella Clinical Isolates and Implications for Immunoprophylaxis of Klebsiella Infections. Clin. Diagn. Lab. Immunol. 1997, 4, 550–555. [Google Scholar] [CrossRef]
- Edwards, P.R.; Fife, M.A. Capsule Types of Klebsiella. J. Infect. Dis. 1952, 92–104. [Google Scholar] [CrossRef]
- Edmunds, P.N. Further Klebsiella Capsule Types. J. Infect. Dis. 1954, 94, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Perry, C.; Elgohari, S.; Hampton, C. V PCR Characterization and Typing of Klebsiella pneumoniae Using Capsular Type-Specific, Variable Number Tandem Repeat and Virulence Gene Targets. J. Med. Microbiol. 2010, 59, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-C.; Koh, T.H.; Lee, N.; Fung, C.-P.; Chang, F.-Y.; Tsai, Y.-K.; Ip, M.; Siu, L.K. Genotypes and Virulence in Serotype K2 Klebsiella pneumoniae from Liver Abscess and Non-Infectious Carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014, 6, 1–7. [Google Scholar] [CrossRef]
- Sokurenko, E.V.; Chesnokova, V.; Doyle, R.J.; Hasty, D.L. Diversity of the Escherichia Coli Type 1 Fimbrial Lectin: Differential Binding to Mannosides and Uroepithelial Cells. J. Biol. Chem. 1997, 272, 17880–17886. [Google Scholar] [CrossRef]
- Maroncle, N.; Rich, C.; Forestier, C. The Role of Klebsiella pneumoniae Urease in Intestinal Colonization and Resistance to Gastrointestinal Stress. Res. Microbiol. 2006, 157, 184–193. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial Extracellular Polysaccharides Involved in Biofilm Formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef]
- Davies, D. Understanding Biofilm Resistance to Antibacterial Agents. Nat. Rev. Drug Discov. 2003, 2, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Suleman, L.; Vuotto, C.; Donelli, G. Healthcare-Associated Infections, Medical Devices and Biofilms: Risk, Tolerance and Control. J. Med. Microbiol. 2015, 64, 323–334. [Google Scholar] [CrossRef]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic Resistance of Bacteria in Biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Barus, T.; Hanjaya, I.; Sadeli, J.; LAY, B.W.; Suwanto, A.; Yulandi, A. Genetic Diversity of Klebsiella Spp. Isolated from Tempe Based on Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction (ERIC-PCR). HAYATI J. Biosci. 2013, 20, 171–176. [Google Scholar] [CrossRef]
- Haryani, Y.; Noorzaleha, A.S.; Fatimah, A.B.; Noorjahan, B.A.; Patrick, G.B.; Shamsinar, A.T.; Laila, R.A.S.; Son, R. Incidence of Klebsiella Pneumonia in Street Foods Sold in Malaysia and Their Characterization by Antibiotic Resistance, Plasmid Profiling, and RAPD-PCR Analysis. Food Control 2007, 18, 847–853. [Google Scholar] [CrossRef]
- Sachse, S.; Bresan, S.; Erhard, M.; Edel, B.; Pfister, W.; Saupe, A.; Rödel, J. Comparison of Multilocus Sequence Typing, RAPD, and MALDI-TOF Mass Spectrometry for Typing of β-Lactam-Resistant Klebsiella pneumoniae Strains. Diagn. Microbiol. Infect. Dis. 2014, 80, 267–271. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, W.; Qin, T.; Liu, C.; Ren, H. Defining and Evaluating a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Klebsiella pneumoniae. Front. Microbiol. 2017, 8, 371. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Yang, G.; Ye, Q.; Wu, Q.; Zhang, J.; Huang, Y. Phenotypic and Genotypic Characterization of Klebsiella pneumoniae Isolated from Retail Foods in China. Front. Microbiol. 2018, 9, 289. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, R.; Elkhatib, W.F.; Ashour, H.M. Molecular Typing and Virulence Analysis of Multidrug Resistant Klebsiella pneumoniae Clinical Isolates Recovered from Egyptian Hospitals. Sci. Rep. 2016, 6, 38929. [Google Scholar]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- Okanda, T.; Haque, A.; Koshikawa, T.; Islam, A.; Huda, Q.; Takemura, H.; Matsumoto, T.; Nakamura, S. Characteristics of Carbapenemase-Producing Klebsiella pneumoniae Isolated in the Intensive Care Unit of the Largest Tertiary Hospital in Bangladesh. Front. Microbiol. 2021, 11, 3357. [Google Scholar] [CrossRef]
- Nkuwi, E.J.; Kabanangi, F.; Rugarabamu, S.; Majigo, M. Methicillin-Resistant Staphylococcus Aureus Contamination and Distribution in Patient’s Care Environment at Muhimbili National Hospital, Dar Es Salaam-Tanzania. BMC Res. Notes 2018, 11, 1–6. [Google Scholar] [CrossRef]
- Cheng, V.C.C.; Wong, S.-C.; Chen, J.H.K.; So, S.Y.C.; Wong, S.C.Y.; Ho, P.-L.; Yuen, K.-Y. Control of Multidrug-Resistant Acinetobacter Baumannii in Hong Kong: Role of Environmental Surveillance in Communal Areas after a Hospital Outbreak. Am. J. Infect. Control 2018, 46, 60–66. [Google Scholar] [CrossRef]
- Cao, X.; Xu, X.; Zhang, Z.; Shen, H.; Chen, J.; Zhang, K. Molecular Characterization of Clinical Multidrug-Resistant Klebsiella pneumoniae Isolates. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 1–5. [Google Scholar] [CrossRef]
- Chouduri, A.U.; Biswas, M.; Haque, M.U.; Arman, M.S.; Uddin, N.; Kona, N.; Akter, R.; Haque, A. Cephalosporin-3G, Highly Prescribed Antibiotic to Outpatients in Rajshahi, Bangladesh: Prescription Errors, Carelessness, Irrational Uses Are the Triggering Causes of Antibiotic Resistance. J. Appl. Pharm. Sci. 2018, 8, 105–112. [Google Scholar]
- Pereira, P.S.; de Araujo, C.F.M.; Seki, L.M.; Zahner, V.; Carvalho-Assef, A.P.D.; Asensi, M.D. Update of the Molecular Epidemiology of KPC-2-Producing Klebsiella pneumoniae in Brazil: Spread of Clonal Complex 11 (ST11, ST437 and ST340). J. Antimicrob. Chemother. 2013, 68, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S. Understanding the Contribution of Environmental Factors in the Spread of Antimicrobial Resistance. Environ. Health Prev. Med. 2015, 20, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Vila, J. Multidrug-Resistant Bacteria without Borders: Role of International Trips in the Spread of Multidrug-Resistant Bacteria. J. Travel Med. 2015, 22, 289–291. [Google Scholar] [CrossRef] [Green Version]
- Begum, N.; Shamsuzzaman, S.M. Emergence of Carbapenemase-Producing Urinary Isolates at a Tertiary Care Hospital in Dhaka, Bangladesh. Tzu Chi Med. J. 2016, 28, 94–98. [Google Scholar] [CrossRef]
- Khan, E.R.; Aung, M.S.; Paul, S.K.; Ahmed, S.; Haque, N.; Ahamed, F.; Sarkar, S.R.; Roy, S.; Rahman, M.M.; Mahmud, M.C. Prevalence and Molecular Epidemiology of Clinical Isolates of Escherichia Coli and Klebsiella pneumoniae Harboring Extended-Spectrum Beta-Lactamase and Carbapenemase Genes in Bangladesh. Microb. Drug Resist. 2018, 24, 1568–1579. [Google Scholar] [CrossRef]
- Islam, M.A.; Talukdar, P.K.; Hoque, A.; Huq, M.; Nabi, A.; Ahmed, D.; Talukder, K.A.; Pietroni, M.A.C.; Hays, J.P.; Cravioto, A. Emergence of Multidrug-Resistant NDM-1-Producing Gram-Negative Bacteria in Bangladesh. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2593–2600. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.; Santos, A.F.; Asensi, M.D.; Peirano, G.; Gales, A.C. First Report of KPC-2-Producing Klebsiella pneumoniae Strains in Brazil. Antimicrob. Agents Chemother. 2009, 53, 333–334. [Google Scholar] [CrossRef]
- Peirano, G.; Seki, L.M.; Val Passos, V.L.; Pinto, M.C.F.G.; Guerra, L.R.; Asensi, M.D. Carbapenem-Hydrolysing β-Lactamase KPC-2 in Klebsiella pneumoniae Isolated in Rio de Janeiro, Brazil. J. Antimicrob. Chemother. 2009, 63, 265–268. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; McElmeel, M.L.; Fulcher, L.C.; Zimmer, B.L. Detection of CTX-M-Type Extended-Spectrum Beta-Lactamase (ESBLs) by Testing with MicroScan Overnight and ESBL Confirmation Panels. J. Clin. Microbiol. 2010, 48, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Bora, A.; Hazarika, N.K.; Shukla, S.K.; Prasad, K.N.; Sarma, J.B.; Ahmed, G. Prevalence of BlaTEM, BlaSHV and BlaCTX-M Genes in Clinical Isolates of Escherichia Coli and Klebsiella pneumoniae from Northeast India. Indian J. Pathol. Microbiol. 2014, 57, 249. [Google Scholar]
- Pitout, J.D.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae Producing Extended-Spectrum β-Lactamases (ESBLs) in the Community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Jacoby, M.D.; Munoz-Price, L. Mechanisms of Disease: The New Beta-Lactamases. N. Engl. J. Med. 2005, 352, 380–391. [Google Scholar]
- Urmi, U.L.; Nahar, S.; Rana, M.; Sultana, F.; Jahan, N.; Hossain, B.; Alam, M.S.; Mosaddek, A.S.M.; McKimm, J.; Rahman, N.A.A. Genotypic to Phenotypic Resistance Discrepancies Identified Involving β-Lactamase Genes, BlaKPC, BlaIMP, BlaNDM-1, and BlaVIM in Uropathogenic Klebsiella pneumoniae. Infect. Drug Resist. 2020, 13, 2863. [Google Scholar] [CrossRef] [PubMed]
- Calhau, V.; Boaventura, L.; Ribeiro, G.; Mendonça, N.; da Silva, G.J. Molecular Characterization of Klebsiella pneumoniae Isolated from Renal Transplanted Patients: Virulence Markers, Extended-Spectrum β-Lactamases, and Genetic Relatedness. Diagn. Microbiol. Infect. Dis. 2014, 79, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Cao, X.L.; Shen, H.; Zhang, Z.F.; Ning, M.Z.; Zhou, W.Q. Investigations on the Virulence, Serotypes and Genotyping of Klebsiella pneumoniae Producing KPC-2. Chin. J. Clin. Lab. Sci 2015, 33, 591–595. [Google Scholar]
- Shah, R.K.; Ni, Z.H.; Sun, X.Y.; Wang, G.Q.; Li, F. The Determination and Correlation of Various Virulence Genes, ESBL, Serum Bactericidal Effect and Biofilm Formation of Clinical Isolated Classical Klebsiella pneumoniae and Hypervirulent Klebsiella pneumoniae from Respiratory Tract Infected Patients. Pol. J. Microbiol. 2017, 66, 501. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.H.; Kao, C.Y.; Yang, D.C.; Tseng, C.C.; Wu, A.B.; Teng, C.H.; Wang, M.C.; Wu, J.J. Clinical and Microbiological Characteristics of Klebsiella pneumoniae from Community-Acquired Recurrent Urinary Tract Infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1533–1539. [Google Scholar] [CrossRef]
- Jung, S.W.; Chae, H.J.; Park, Y.J.; Yu, J.K.; Kim, S.Y.; Lee, H.K.; Lee, J.H.; Kahng, J.M.; Lee, S.O.; Lee, M.K. Microbiological and Clinical Characteristics of Bacteraemia Caused by the Hypermucoviscosity Phenotype of Klebsiella pneumoniae in Korea. Epidemiol. Infect. 2013, 141, 334–340. [Google Scholar] [CrossRef]
- Regué, M.; Hita, B.; Piqué, N.; Izquierdo, L.; Merino, S.; Fresno, S.; Benedí, V.J.; Tomás, J.M. A Gene, Uge, Is Essential for Klebsiella pneumoniae Virulence. Infect. Immun. 2004, 72, 54–61. [Google Scholar] [CrossRef]
- Izquierdo, L.; Coderch, N.; Piqué, N.; Bedini, E.; Corsaro, M.M.; Merino, S.; Fresno, S.; Tomás, J.M.; Regué, M. The Klebsiella pneumoniae WabG Gene: Role InBiosynthesis of the Core Lipopolysaccharide AndVirulence. J. Bacteriol. 2003, 185, 7213–7221. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-C.; Fang, C.-T.; Lee, C.-Z.; Shun, C.-T.; Wang, J.-T. Genomic Heterogeneity in Klebsiella pneumoniae Strains Is Associated with Primary Pyogenic Liver Abscess and Metastatic Infection. J. Infect. Dis. 2005, 192, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Anes, J.; Hurley, D.; Martins, M.; Fanning, S. Exploring the Genome and Phenotype of Multi-Drug Resistant Klebsiella pneumoniae of Clinical Origin. Front. Microbiol. 2017, 8, 1913. [Google Scholar] [CrossRef]
- Seifi, K.; Kazemian, H.; Heidari, H.; Rezagholizadeh, F.; Saee, Y.; Shirvani, F.; Houri, H. Evaluation of Biofilm Formation among Klebsiella pneumoniae Isolates and Molecular Characterization by ERIC-PCR. Jundishapur J. Microbiol. 2016, 9, e30682. [Google Scholar] [CrossRef] [PubMed]
- Cherif-Antar, A.; Moussa–Boudjemâa, B.; Didouh, N.; Medjahdi, K.; Mayo, B.; Flórez, A.B. Diversity and Biofilm-Forming Capability of Bacteria Recovered from Stainless Steel Pipes of a Milk-Processing Dairy Plant. Dairy Sci. Technol. 2016, 96, 27–38. [Google Scholar] [CrossRef]
- Sahly, H.; Aucken, H.; Benedi, V.J.; Forestier, C.; Fussing, V.; Hansen, D.S.; Ofek, I.; Podschun, R.; Sirot, D.; Tomás, J.M. Increased Serum Resistance in Klebsiella pneumoniae Strains Producing Extended-Spectrum β-Lactamases. Antimicrob. Agents Chemother. 2004, 48, 3477–3482. [Google Scholar] [CrossRef] [PubMed]
- Sahly, H.; Navon-Venezia, S.; Roesler, L.; Hay, A.; Carmeli, Y.; Podschun, R.; Hennequin, C.; Forestier, C.; Ofek, I. Extended-Spectrum β-Lactamase Production Is Associated with an Increase in Cell Invasion and Expression of Fimbrial Adhesins in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2008, 52, 3029–3034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between Biofilm Formation and Antimicrobial Resistance in Gram-Negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-C.; Yang, S.-L.; Peng, H.-L.; Chang, H.-Y. Identification of Genes Present Specifically in a Virulent Strain of Klebsiella pneumoniae. Infect. Immun. 2000, 68, 7149–7151. [Google Scholar] [CrossRef]
- Large Detailed Map of Bangladesh with Cities. Available online: https://ontheworldmap.com/bangladesh/large-detailed-map-of-bangladesh-with-cities.html (accessed on 27 August 2022).
- Aurna, S.T. Rapid Identification of Klebsiella pneumoniae Using PCR Based Method Targeting 16S RRNA Gene. Bachelor’s Thesis, BRAC University, Dhaka, Bangladesh, 2017. [Google Scholar]
- Mahmud, Z.H.; Kassu, A.; Mohammad, A.; Yamato, M.; Bhuiyan, N.A.; Nair, G.B.; Ota, F. Isolation and Molecular Characterization of Toxigenic Vibrio Parahaemolyticus from the Kii Channel, Japan. Microbiol. Res. 2006, 161, 25–37. [Google Scholar] [CrossRef]
- Sanders, E.R. Aseptic Laboratory Techniques: Plating Methods. JoVE J. Vis. Exp. 2012, 63, e3064. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Koyano, S.; Nagai, R.; Okamura, N.; Moriya, K.; Koike, K. Evaluation of a Chromogenic Agar Medium for the Detection of Extended-spectrum Β-lactamase-producing Enterobacteriaceae. Lett. Appl. Microbiol. 2010, 51, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100. J. Clin. Microbiol. 2021, 59, e00213–e00221. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Clinical Breakpoints and Dosing of Antibiotics 2020. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 27 August 2022).
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI Suppl. M100 2021, 41, 1–352. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters 2021. pp. 1–116. Available online: https://www.eucast.org/eucast_news/news_singleview/?tx_ttnews%5Btt_news%5D=416&cHash=0af950182d1dea8f854e2be8b11cbbe4 (accessed on 27 August 2022).
- Hossain, M.S.; Sobur Ali, M.H.; Uddin, S.Z.; Moniruzzaman, M.; Islam, M.R.; Shohael, A.M.; Islam, M.S.; Ananya, T.H.; Rahman, M.M.; Rahman, M.A. ESBL Producing Escherichia Coli in Faecal Sludge Treatment Plants: An Invisible Threat to Public Health in Rohingya Camps, Cox’s Bazar, Bangladesh. Front. Public Health 2021, 9, 783019. [Google Scholar] [CrossRef]
- Mahmud, Z.H.; Kabir, M.H.; Ali, S.; Moniruzzaman, M.; Imran, K.M.; Nafiz, T.N.; Islam, M.; Hussain, A.; Hakim, S.A.I.; Worth, M. Extended-Spectrum Beta-Lactamase-Producing Escherichia Coli in Drinking Water Samples from a Forcibly Displaced, Densely Populated Community Setting in Bangladesh. Front. Public Health 2020, 8, 228. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important β-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Brisse, S.; Fevre, C.; Passet, V.; Issenhuth-Jeanjean, S.; Tournebize, R.; Diancourt, L.; Grimont, P. Virulent Clones of Klebsiella pneumoniae: Identification and Evolutionary Scenario Based on Genomic and Phenotypic Characterization. PLoS ONE 2009, 4, e4982. [Google Scholar] [CrossRef] [Green Version]
- Candan, E.D.; Aksöz, N. Klebsiella pneumoniae: Characteristics of Carbapenem Resistance and Virulence Factors. Acta Biochim. Pol. 2015, 62, 4. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, Y.; Ye, L.; Yang, J. Molecular Epidemiology and Virulence Factors of Pyogenic Liver Abscess Causing Klebsiella pneumoniae in China. Clin. Microbiol. Infect. 2014, 20, O818–O824. [Google Scholar] [CrossRef]
- Du, J.; Li, P.; Liu, H.; Lü, D.; Liang, H.; Dou, Y. Phenotypic and Molecular Characterization of Multidrug Resistant Klebsiella pneumoniae Isolated from a University Teaching Hospital, China. PLoS ONE 2014, 9, e95181. [Google Scholar] [CrossRef]
- Siddique, A.B.; Moniruzzaman, M.; Ali, S.; Dewan, M.N.; Islam, M.R.; Islam, M.S.; Amin, M.B.; Mondal, D.; Parvez, A.K.; Mahmud, Z.H. Characterization of Pathogenic Vibrio Parahaemolyticus Isolated from Fish Aquaculture of the Southwest Coastal Area of Bangladesh. Front. Microbiol. 2021, 12, 635539. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.; Usman, J.; Kaleem, F.; Omair, M.; Khalid, A.; Iqbal, M. Evaluation of Different Detection Methods of Biofilm Formation in the Clinical Isolates. Braz. J. Infect. Dis. 2011, 15, 305–311. [Google Scholar] [CrossRef]
- Ali, S.; Hossain, M.; Azad, A.B.; Siddique, A.B.; Moniruzzaman, M.; Ahmed, M.A.; Amin, M.B.; Islam, M.S.; Rahman, M.M.; Mondal, D. Diversity of Vibrio Parahaemolyticus in Marine Fishes of Bangladesh. J. Appl. Microbiol. 2021, 131, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Guo, Y.; Sheng, Q.; Shyr, Y. Heatmap3: An Improved Heatmap Package with More Powerful and Convenient Features. BMC Bioinform. 2014, 15, P16. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef] [Green Version]



| Hospital | Resistance | Sampling Point | ||||
|---|---|---|---|---|---|---|
| Floor | Bed Pillow | Nasal Throat | Pus | Caregivers’ Hand | ||
| Faridpur | ESBL-KP | 47.2% | 13.9% | 19.4% | 2.8% | 16.7% |
| (17/36) | (5/36) | (7/36) | (1/36) | (6/36) | ||
| CRKP | 60.0% | 6.7% | 33.3% | 0.0% | 0.0% | |
| (10/15) | (1/15) | (4/15) | ||||
| Rangpur | ESBL-KP | 50.0% | 20.0% | 30.0% | - | - |
| (5/10) | (2/10) | (3/10) | ||||
| CRKP | 55.6% | 22.2% | 22.2% | - | - | |
| (5/9) | (2/9) | (2/9) | ||||
| Rajshahi | ESBL-KP | 28.6% | 52.4% | 19.0% | - | - |
| (6/21) | (11/21) | (4/21) | ||||
| CRKP | 16.7% | 61.1% | 22.2% | - | - | |
| (3/18) | (11/18) | (4/18) | ||||
| Antibiotics | Number of Resistant Isolates | Biofilm 25 °C | Biofilm 37 °C | |||||
|---|---|---|---|---|---|---|---|---|
| Non-Biofilm Producer | Weak Biofilm Producer | Moderate Biofilm Producer | Strong Biofilm Producer | Non-Biofilm Producer | Weak Biofilm Producer | Moderate Biofilm Producer | ||
| Ceftriaxone | 67 | 2 (3%) | 25 (37.3%) | 36 (53.7%) | 4 (6%) | 2 (3%) | 51 (76.1%) | 14 (20.9%) |
| Cefuroxime | 67 | 2 (3%) | 25 (37.3%) | 36 (53.7%) | 4 (6%) | 2 (3%) | 51 (76.1%) | 14 (20.9%) |
| Ceftazidime | 65 | 2 (3.1%) | 24 (36.9%) | 35 (53.8%) | 4 (6.2%) | 2 (3.1%) | 50 (76.9%) | 13 (20%) |
| Cefotaxime | 67 | 2 (3%) | 25 (37.3%) | 36 (53.7%) | 4 (6%) | 2 (3%) | 51 (76.1%) | 14 (20.9%) |
| Cefixime | 66 | 2 (3%) | 25 (37.9%) | 36 (54.5%) | 3 (4.5%) | 2 (3%) | 51 (77.3%) | 13 (19.7%) |
| Cefepime | 65 | 2 (3.1%) | 25 (38.5%) | 35 (53.8%) | 3 (4.6%) | 2 (3.1%) | 50 (76.9%) | 13 (20%) |
| Aztreonam | 66 | 2 (3%) | 24 (36.4%) | 36 (54.5%) | 4 (6.1%) | 2 (3%) | 50 (75.8%) | 14 (21.2%) |
| Imipenem | 23 | 0 (0%) | 13 (56.5%) | 9 (39.1%) | 1 (4.3%) | 1 (4.3%) | 20 (87%) | 2 (8.7%) |
| Meropenem | 19 | 0 (0%) | 11 (57.9%) | 7 (36.8%) | 1 (5.3%) | 1 (5.3%) | 16 (84.2%) | 2 (10.5%) |
| Amikacin | 24 | 0 (0%) | 8 (33.3%) | 16 (66.7%) | 0 (0%) | 0 (0%) | 23 (95.8%) | 1 (4.2%) |
| Nitrofurantoin | 43 | 2 (4.7%) | 14 (32.6%) | 24 (55.8%) | 3 (7%) | 2 (4.7%) | 32 (74.4%) | 9 (20.9%) |
| Tetracycline | 49 | 2 (4.1%) | 16 (32.7%) | 30 (61.2%) | 1 (2%) | 2 (4.1%) | 37 (75.5%) | 10 (20.4%) |
| Tigecycline | 18 | 2 (11.1%) | 5 (27.8%) | 10 (55.6%) | 1 (5.6%) | 1 (5.6%) | 13 (72.2%) | 4 (22.2%) |
| Nalidixic Acid | 38 | 2 (5.3%) | 14 (36.8%) | 21 (55.3%) | 1 (2.6%) | 2 (5.3%) | 33 (86.8%) | 3 (7.9%) |
| Ciprofloxacin | 28 | 2 (7.1%) | 7 (25%) | 18 (64.3%) | 1 (3.6%) | 1 (3.6%) | 23 (82.1%) | 4 (14.3%) |
| Trimethoprim/Sulfamethoxazole | 57 | 2 (3.5%) | 19 (33.3%) | 34 (59.6%) | 2 (3.5%) | 2 (3.5%) | 44 (77.2%) | 11 (19.3%) |
| Chloramphenicol | 3 | 0 (0%) | 0 (0%) | 2 (66.7%) | 1 (33.3%) | 0 (0%) | 1 (33.3%) | 2 (66.7%) |
| Azithromycin | 54 | 2 (3.7%) | 20 (37%) | 31 (57.4%) | 1 (1.9%) | 2 (3%) | 51 (76.1%) | 14 (20.9%) |
| Fosfomycin | 22 | 1 (4.5%) | 5 (22.7%) | 16 (72.7%) | 0 (0%) | 2 (3%) | 51 (76.1%) | 14 (20.9%) |
| Isolate ID for Patient Sample | Hospital | Ward | Sampling Point | Corresponding Environmental Isolate ID | Corresponding Environmental Sampling Point |
|---|---|---|---|---|---|
| KP-48 | Rajshahi | Pediatric | Nasal-Throat | KP-46 | Floor |
| KP-49 | Rajshahi | Pediatric | Nasal-Throat | KP-46 | Floor |
| KP-57 | Rajshahi | Pediatric | Nasal-Throat | KP-46 | Floor |
| KP-59 | Rajshahi | Pediatric | Nasal-Throat | KP-46 | Floor |
| KP-125 | Rangpur | Pediatric | Nasal-Throat | KP-116, KP-117, KP-118, KP-119, KP-120, KP-122, KP-123 | Bed pillow, Floor |
| KP-126 | Rangpur | Pediatric | Nasal-Throat | KP-116, KP-117, KP-118, KP-119, KP-120, KP-122, KP-123 | Bed pillow, Floor |
| KP-127 | Rangpur | Pediatric | Nasal-Throat | KP-116, KP-117, KP-118, KP-119, KP-120, KP-122, KP-123 | Bed pillow, Floor |
| KP-154 | Faridpur | Surgery | Pus Sample | KP-153, KP-155, KP-157, KP-158 | Caregivers’ Hand |
| KP-181 | Faridpur | Medicine | Nasal-Throat | KP-171, KP-172, KP-175, KP-176, KP-177, 178, KP-179 | Floor |
| KP-184 | Faridpur | Medicine | Nasal-Throat | KP-188, KP-189 | Caregivers’ Hand |
| KP-185 | Faridpur | Medicine | Nasal-Throat | KP-188, KP-189 | Caregivers’ Hand |
| KP-255 | Faridpur | Neonatal | Nasal-Throat | KP-245, KP-246 | Bed pillow |
| KP-256 | Faridpur | Neonatal | Nasal-Throat | KP-245, KP-246 | Bed pillow |
| KP-257 | Faridpur | Neonatal | Nasal-Throat | KP-245, KP-246 | Bed pillow |
| KP-260 | Faridpur | Pediatric | Nasal-Throat | KP-259, KP-262 | Bed pillow, Floor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmud, Z.H.; Uddin, S.Z.; Moniruzzaman, M.; Ali, S.; Hossain, M.; Islam, M.T.; Costa, D.T.D.; Islam, M.R.; Islam, M.S.; Hassan, M.Z.; et al. Healthcare Facilities as Potential Reservoirs of Antimicrobial Resistant Klebsiella pneumoniae: An Emerging Concern to Public Health in Bangladesh. Pharmaceuticals 2022, 15, 1116. https://doi.org/10.3390/ph15091116
Mahmud ZH, Uddin SZ, Moniruzzaman M, Ali S, Hossain M, Islam MT, Costa DTD, Islam MR, Islam MS, Hassan MZ, et al. Healthcare Facilities as Potential Reservoirs of Antimicrobial Resistant Klebsiella pneumoniae: An Emerging Concern to Public Health in Bangladesh. Pharmaceuticals. 2022; 15(9):1116. https://doi.org/10.3390/ph15091116
Chicago/Turabian StyleMahmud, Zahid Hayat, Salman Zahir Uddin, M. Moniruzzaman, Sobur Ali, Monir Hossain, Md. Tamzid Islam, Dorin Teresa D. Costa, Mohammad Rafiqul Islam, Md. Shafiqul Islam, Md. Zakiul Hassan, and et al. 2022. "Healthcare Facilities as Potential Reservoirs of Antimicrobial Resistant Klebsiella pneumoniae: An Emerging Concern to Public Health in Bangladesh" Pharmaceuticals 15, no. 9: 1116. https://doi.org/10.3390/ph15091116






