Molecular Insight into Mycobacterium tuberculosis Resistance to Nitrofuranyl Amides Gained through Metagenomics-like Analysis of Spontaneous Mutants
Abstract
:1. Introduction
2. Results
2.1. Chemistry
2.2. Determination of MIC for Compounds 8–13
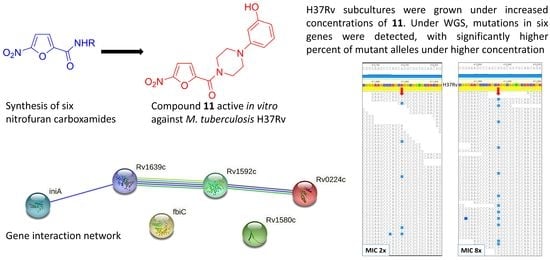
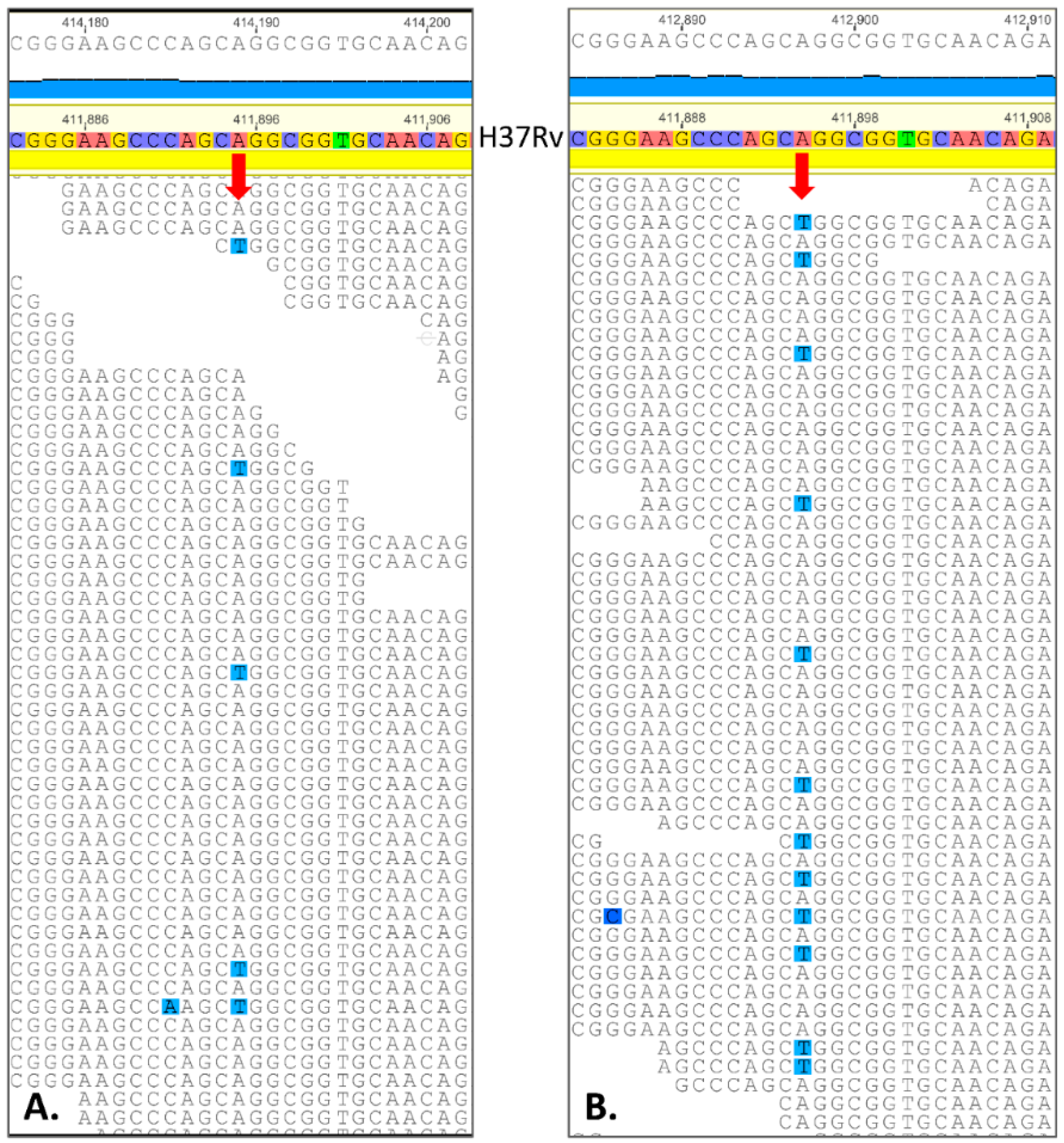
2.3. Spontaneous Mutagenesis, Whole Genome Sequencing, and Bioinformatics
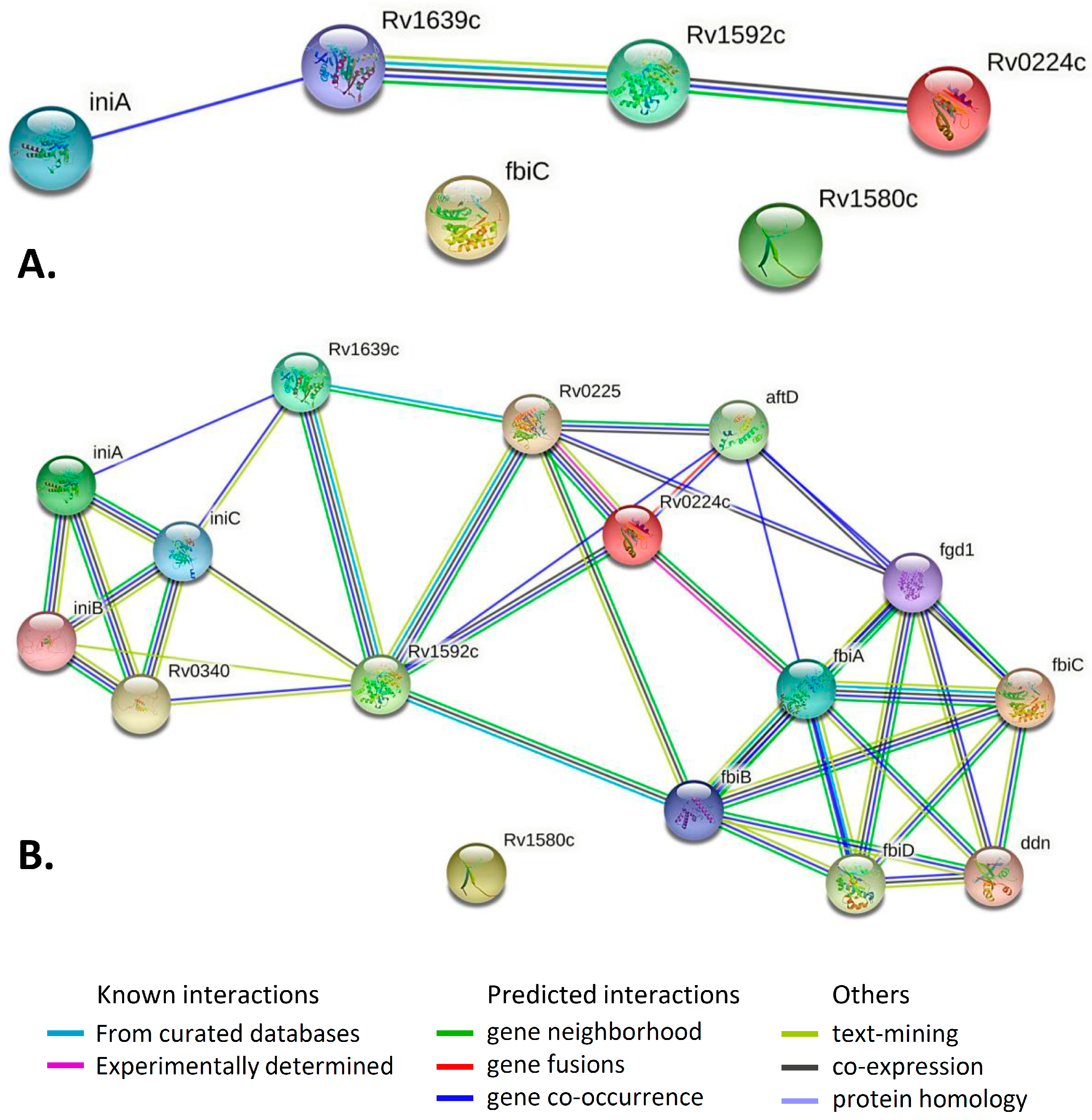
2.4. Gene/Protein Enrichment and Network Analysis
3. Discussion
4. Materials and Methods
4.1. Chemistry Experimental Procedures
4.2. Determination of Minimal Inhibitory Concentration (MIC)
4.3. Spontaneous Mutant Selection, WGS, and Bioinformatics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Tuberculosis Report 2021. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 11 September 2022).
- Rawat, D.S. Antituberculosis Drug Research: A Critical Overview. Med. Res. Rev. 2013, 33, 693–764. [Google Scholar]
- Chetty, S.; Ramesh, M.; Singh-Pillay, A.; Soliman, M.E.S. Recent advancements in the development of anti-tuberculosis drugs. Bioorg. Med. Chem. Lett. 2017, 27, 370–386. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.-Z. Recent Natural Products Based Drug Development: A Pharmaceutical Industry Perspective. J. Nat. Prod. 1998, 61, 1053–1071. [Google Scholar] [CrossRef] [PubMed]
- Balunas, M.J.; Kinghorn, A.D. Drug discovery from medicinal plants. Life Sci. 2005, 78, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Sensi, P. History of the Development of Rifampin. Rev. Infect. Dis. 1983, 5, S402–S406. [Google Scholar] [CrossRef] [PubMed]
- Worley, M.V.; Estrada, S.J. Bedaquiline: A Novel Antitubercular Agent for the Treatment of Multidrug-Resistant Tuberculosis. Pharmacotherapy 2014, 34, 1187–1197. [Google Scholar] [CrossRef]
- Field, S.K. Bedaquiline for the treatment of multidrug-resistant tuberculosis: Great promise or disappointment? Ther. Adv. Chronic Dis. 2015, 6, 170–184. [Google Scholar] [CrossRef]
- Gallardo-Macias, R.; Kumar, P.; Jaskowski, M.; Richmann, T.; Shrestha, R.; Russo, R.; Singleton, E.; Zimmerman, M.D.; Ho, H.P.; Dartois, V.; et al. Optimization of N-benzyl-5-nitrofuran-2-carboxamide as an antitubercular agent. Bioorg. Med. Chem. Lett. 2019, 29, 601–606. [Google Scholar] [CrossRef]
- Elsaman, T.; Mohamed, M.S.; Mohamed, M.A. Current development of 5-nitrofuran-2-yl derivatives as antitubercular agents. Bioorg. Chem. 2019, 88, 102969. [Google Scholar] [CrossRef]
- Barry, E.C.; Boshoff, I.M.H.; Dowd, S.C. Prospects for Clinical Introduction of Nitroimidazole Antibiotics for the Treatment of Tuberculosis. Curr. Pharm. Des. 2004, 10, 3239–3262. [Google Scholar] [CrossRef]
- Smith, M.A.; Edwards, D.I. Redox potential and oxygen concentration as factors in the susceptibility of Helicobacter pylori to nitroheterocyclic drugs. J. Antimicrob. Chemother. 1995, 35, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Le, V.V.H.; Rakonjac, J. Nitrofurans: Revival of an “old” drug class in the fight against antibiotic resistance. PLoS Pathog. 2021, 17, e1009663. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, J.; Basil, B.X.; Stewardson, A.; Coenen, S.; Godycki-Cwirko, M.; Adriaenssens, N.; Kowalczyk, A.; Lammens, C.; Harbarth, S.; Goossens, H.; et al. An In Vitro Deletion in ribE Encoding Lumazine Synthase Contributes to Nitrofurantoin Resistance in Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 7225–7233. [Google Scholar] [CrossRef] [PubMed]
- Dobrikov, G.M.; Valcheva, V.; Nikolova, Y.; Ugrinova, I.; Pasheva, E.; Dimitrov, V. Enantiopure antituberculosis candidates synthesized from (−)-fenchone. Eur. J. Med. Chem. 2014, 77, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Tangallapally, P.R.; Yendapally, R.; Daniels, J.A.; Lee, E.B.R.; Lee, E.R. Nitrofurans as Novel Anti-tuberculosis Agents: Identification, Development and Evaluation. Curr. Top. Med. Chem. 2007, 7, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Slavchev, I.; Dobrikov, G.M.; Valcheva, V.; Ugrinova, I.; Pasheva, E.; Dimitrov, V. Antimycobacterial activity generated by the amide coupling of (−)-fenchone derived aminoalcohol with cinnamic acids and analogues. Bioorg. Med. Chem. Lett. 2014, 24, 5030–5033. [Google Scholar] [CrossRef]
- Manjunatha, U.; Boshoff, H.I.; Barry, C.E. The mechanism of action of PA-824: Novel insights from transcriptional profiling. Commun. Integr. Biol. 2009, 2, 215–218. [Google Scholar] [CrossRef]
- Girase, P.S.; Dhawan, S.; Kumar, V.; Shinde, S.R.; Palkar, M.B.; Karpoormath, R. An appraisal of anti-mycobacterial activity with structure-activity relationship of piperazine and its analogues: A review. Eur. J. Med. Chem. 2021, 210, 112967. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E.; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Mahrt, N.; Tietze, A.; Künzel, S.; Franzenburg, S.; Barbosa, C.; Jansen, G.; Schulenburg, H. Bottleneck size and selection level reproducibly impact evolution of antibiotic resistance. Nat. Ecol. Evol. 2021, 5, 1233–1242. [Google Scholar] [CrossRef]
- Zhang, T.; Rao, G.; Gao, X. Identification of Hub Genes in Tuberculosis via Bioinformatics Analysis. Comput. Math. Methods Med. 2021, 2021, 8159879. [Google Scholar] [CrossRef] [PubMed]
- Udhaya Kumar, S.; Saleem, A.; Kumar, D.T.; Preethi, V.A.; Younes, S.; Zayed, H.; Tayubi, I.A.; Doss, C.G.P. Chapter Eleven—A systemic approach to explore the mechanisms of drug resistance and altered signaling cascades in extensively drug-resistant tuberculosis. Adv. Protein Chem. Struct. Biol. 2021, 127, 343–364. [Google Scholar] [PubMed]
- Sinha, S.; Lynn, A.M.; Desai, D.K. Implementation of homology based and non-homology based computational methods for the identification and annotation of orphan enzymes: Using Mycobacterium tuberculosis H37Rv as a case study. BMC Bioinform. 2020, 21, 466. [Google Scholar] [CrossRef]
- Yempalla, K.R.; Munagala, G.; Singh, S.; Magotra, A.; Kumar, S.; Rajput, V.S.; Bharate, S.S.; Tikoo, M.; Singh, G.D.; Khan, I.A.; et al. Nitrofuranyl Methyl Piperazines as New Anti-TB Agents: Identification, Validation, Medicinal Chemistry, and PK Studies. ACS Med. Chem. Lett. 2015, 6, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.C. Maximizing bactericidal activity with combinations of bioreduced drugs. Future Med. Chem. 2010, 2, 1253–1271. [Google Scholar] [CrossRef] [PubMed]
- Krasavin, M.; Lukin, A.; Vedekhina, T.; Manicheva, O.; Dogonadze, M.; Vinogradova, T.; Zabolotnykh, N.; Rogacheva, E.; Kraeva, L.; Sharoyko, V.; et al. Attachment of a 5-nitrofuroyl moiety to spirocyclic piperidines produces non-toxic nitrofurans that are efficacious in vitro against multidrug-resistant Mycobacterium tuberculosis. Eur. J. Med. Chem. 2019, 166, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Tangallapally, R.; Yendapally, R.; McNeil, M.; Lenaerts, A. Heterocyclic Amides with Anti-Tuberculosis Activity. U.S. Patent 20050222408A1, 6 October 2005. [Google Scholar]
- Magotra, A.; Sharma, A.; Singh, S.; Ojha, P.K.; Kumar, S.; Bokolia, N.; Wazir, P.; Sharma, S.; Khan, I.A.; Singh, P.P.; et al. Physicochemical, pharmacokinetic, efficacy and toxicity profiling of a potential nitrofuranyl methyl piperazine derivative IIIM-MCD-211 for oral tuberculosis therapy via in-silico-in-vitro-in-vivo approach. Pulm. Pharmacol. Ther. 2018, 48, 151–160. [Google Scholar] [CrossRef]
- Jian, Y.; Forbes, H.E.; Hulpia, F.; Risseeuw, M.D.P.; Caljon, G.; Munier-Lehmann, H.; Boshoff, H.I.M.; van Calenbergh, S. 2-((3,5-Dinitrobenzyl)thio)quinazolinones: Potent Antimycobacterial Agents Activated by Deazaflavin (F420)-Dependent Nitroreductase (Ddn). J. Med. Chem. 2021, 64, 440–457. [Google Scholar] [CrossRef]
- Dosanjh, N.S.; Rawat, M.; Chung, J.-H.; Av-Gay, Y. Thiol specific oxidative stress response in Mycobacteria. FEMS Microbiol. Lett. 2005, 249, 87–94. [Google Scholar] [CrossRef]
- Zhu, L.; Zhong, J.; Jia, X.; Liu, G.; Kang, Y.; Dong, M.; Zhang, X.; Li, Q.; Yue, L.; Li, C.; et al. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 2016, 44, 730–743. [Google Scholar] [CrossRef]
- Fujiwara, M.; Kawasaki, M.; Hariguchi, N.; Liu, Y.; Matsumoto, M. Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis 2018, 108, 186–194. [Google Scholar] [CrossRef]
- Booker, S.J.; Grove, T.L. Mechanistic and functional versatility of radical SAM enzymes. F1000 Biol. Rep. 2010, 2, 52. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, P.J.; Perdigao, J.; Gomes, P.; Puyen, Z.M.; Santos-Lazaro, D.; Napier, G.; Hibberd, M.L.; Viveiros, M.; Portugal, I.; Campino, S.; et al. Genetic diversity of candidate loci linked to Mycobacterium tuberculosis resistance to bedaquiline, delamanid and pretomanid. Sci. Rep. 2021, 11, 19431. [Google Scholar] [CrossRef] [PubMed]
- Haver, H.L.; Chua, A.; Ghode, P.; Lakshminarayana, S.B.; Singhal, A.; Mathema, B.; Wintjens, R.; Bifani, P. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 5316–5323. [Google Scholar] [CrossRef] [PubMed]
- Colangeli, R.; Helb, D.; Sridharan, S.; Sun, J.; Varma-Basil, M.; Hazbón, M.H.; Harbacheuski, R.; Megjugorac, N.J.; Jacobs, W.R., Jr.; Holzenburg, A.; et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 2005, 55, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Guo, X.; Yang, X.; Zhang, B.; Ren, J.; Liu, A.; Ran, Y.; Yan, B.; Chen, F.; Guddat, L.W.; et al. Mycobacterial dynamin-like protein IniA mediates membrane fission. Nat. Commun. 2019, 10, 3906. [Google Scholar] [CrossRef]
- Jimenez Ana, J.; Maiuri, P.; Lafaurie-Janvore, J.; Divoux, S.; Piel, M.; Perez, F. ESCRT Machinery Is Required for Plasma Membrane Repair. Science 2014, 343, 1247136. [Google Scholar] [CrossRef]
- Kumar, A.; Anand, K.P.; Chandel, S.; Shrivatava, A.; Kaur, J. Molecular Dynamics Assisted Mechanistic Insight of Val430-Ala Mutation of Rv1592c Protein in Isoniazid Resistant Mycobacterium Tuberculosis. Curr. Comput. Aided Drug Des. 2021, 17, 95–106. [Google Scholar] [CrossRef]
- Becq, J.; Gutierrez, M.C.; Rosas-Magallanes, V.; Rauzier, J.; Gicquel, B.; Neyrolles, O.; Deschavanne, P. Contribution of Horizontally Acquired Genomic Islands to the Evolution of the Tubercle Bacilli. Mol. Biol. Evol. 2007, 24, 1861–1871. [Google Scholar] [CrossRef]
- Kwok, H.F.; Scott, C.J.; Snoddy, P.; Buick, R.J.; Johnston, J.A.; Olwill, S.A. Expression and purification of diagnostically sensitive mycobacterial (Mycobacterium bovis) antigens and profiling of their humoral immune response in a rabbit model. Res. Vet. Sci. 2010, 89, 41–47. [Google Scholar] [CrossRef]
- Kruh, N.A.; Troudt, J.; Izzo, A.; Prenni, J.; Dobos, K.M. Portrait of a Pathogen: The Mycobacterium tuberculosis Proteome IN Vivo. PLoS ONE 2010, 5, e13938. [Google Scholar] [CrossRef] [PubMed]
- Maloney, E.; Lun, S.; Stankowska, D.; Guo, H.; Rajagopalan, M.; Bishai, W.; Madiraju, M. Alterations in Phospholipid Catabolism in Mycobacterium Tuberculosis LysX Mutant. Front. Microbiol. 2011, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Rainczuk, A.K.; Klatt, S.; Yamaryo-Botté, Y.; Brammananth, R.; McConville, M.J.; Coppel, R.L.; Crellin, P.K. MtrP, a putative methyltransferase in Corynebacteria, is required for optimal membrane transport of trehalose mycolates. J. Biol. Chem. 2020, 295, 6108–6119. [Google Scholar] [CrossRef]
- Rickman, L.; Scott, C.; Hunt, D.M.; Hutchinson, T.; Menéndez, M.C.; Whalan, R.; Hinds, J.; Colston, M.J.; Green, J.; Buxton, R.S. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 2005, 56, 1274–1286. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, A.; Kaur, G.; Makkar, P.; Kaur, J. Functional characterization of hypothetical proteins of Mycobacterium tuberculosis with possible esterase/lipase signature: A cumulative in silico and in vitro approach. J. Biomol. Struct. Dyn. 2017, 35, 1226–1243. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.B.; Dubnau, E.; Kolesnikova, I.; Laval, F.; Daffe, M.; Smith, I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 2006, 60, 312–330. [Google Scholar] [CrossRef] [PubMed]
- Loo Sara, L.; Ong, A.; Kyaw, W.; Loïc, M.T.; Lan, R.; Mark, M.T.; Kivisaar, M. Nonsynonymous Polymorphism Counts in Bacterial Genomes: A Comparative Examination. Appl. Environ. Microb. 2020, 87, e02002-20. [Google Scholar]
- Perdigão, J.; Silva, H.; Machado, D.; Macedo, R.; Maltez, F.; Silva, C.; Jordao, L.; Couto, I.; Mallard, K.; Coll, F.; et al. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genom. 2014, 15, 991. [Google Scholar] [CrossRef]
- Schön, T.; Werngren, J.; Machado, D.; Borroni, E.; Wijkander, M.; Lina, G.; Mouton, J.; Matuschek, E.; Kahlmeter, G.; Giske, C.; et al. Multicentre testing of the EUCAST broth microdilution reference method for MIC determination on Mycobacterium tuberculosis. Clin. Microbiol. Infect. 2021, 27, 288.e1–288.e4. [Google Scholar] [CrossRef]
- Palomino, J.-C.; Martin, A.; Camacho, M.; Guerra, H.; Swings, J.; Portaels, F. Resazurin Microtiter Assay Plate: Simple and Inexpensive Method for Detection of Drug Resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2002, 46, 2720–2722. [Google Scholar] [CrossRef]
- Manicheva, O.A.; Dogonadze, M.Z.; Melnikova, N.N.; Vishnevskiy, B.I.; Manichev, S.A. The growth rate phenotypic property of Mycobacterium tuberculosis clinical strains: Dependence on tuberculosis localization, treatment, drug susceptibility. Russ. J. Infect. Immun. 2018, 8, 175–186. (In Russian) [Google Scholar] [CrossRef]
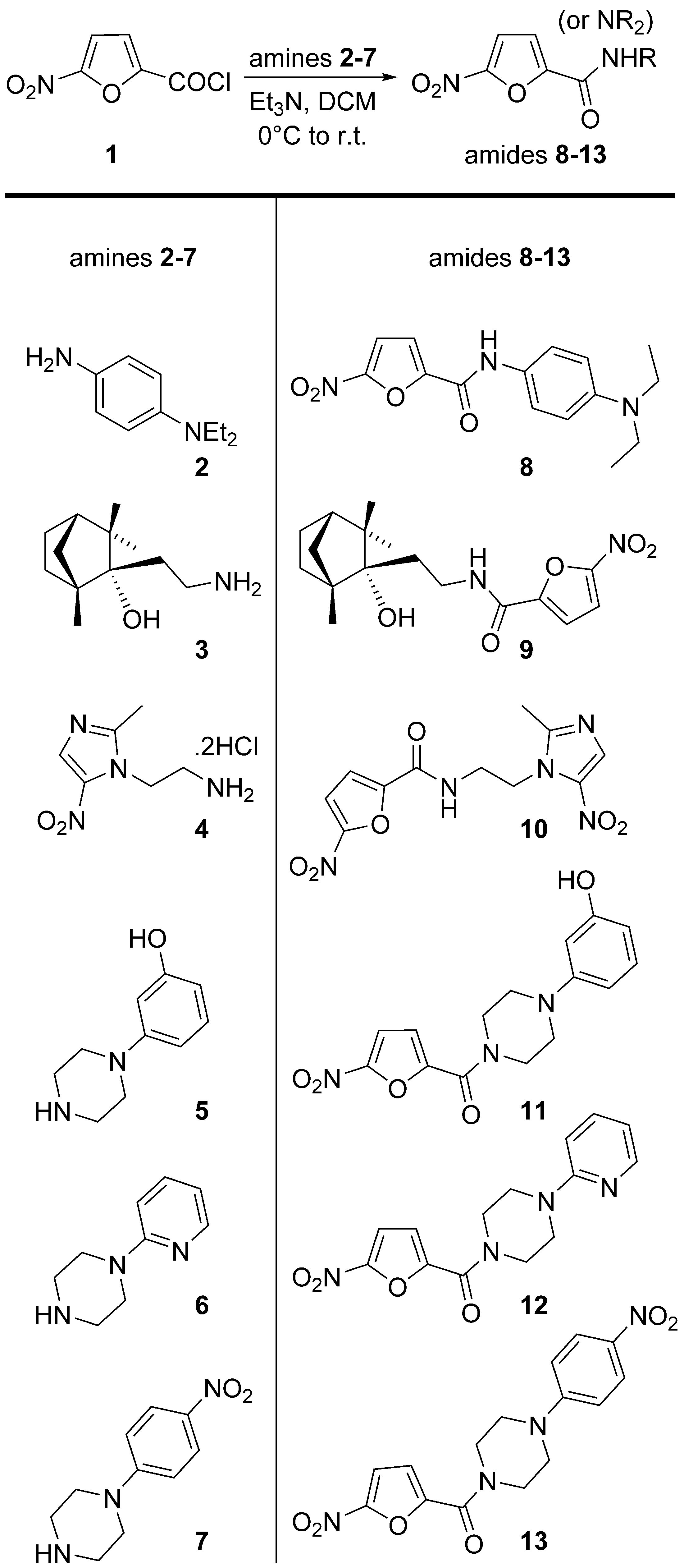
| Compound | MIC (WCMD), μM | MIC (REMA), μM |
|---|---|---|
| 8 | 0.66 | 16.48 |
| 9 | 4.64 | 29.73 |
| 10 | >80 | 162 |
| 11 | 0.19 | 0.50 |
| 12 | 0.026 | 0.20 |
| 13 | <0.012 | 0.36 |
| Compound | MIC (REMA), μM |
|---|---|
| 11 | <1.26; 0.50; 0.50 |
| 12 | <1.32; <0.13; 0.33 |
| 13 | <1.16; <0.12; >0.58; 1.44 |
| Position in Genome | Ref | Mut | Gene | Amino Acid Change | PAM1 * | SIFT p ** | % of Mutant Reads in H37Rv Cultured with 11 (1.00 µM) | % of Mutant Reads in H37Rv Cultured with 11 (2.00 µM) | % of Mutant Reads in H37Rv Cultured with 11 (4.00 µM) | p Value for the Most Contrasting Pairs |
|---|---|---|---|---|---|---|---|---|---|---|
| 268,560 | A | T | Rv0224c | Phe23Tyr | 21 | 0.01 | 9.6 (13/136) | 15.8 (29/183) | 20.0 (25/125) | 0.02 |
| 411,895 | A | T | Rv0342 (iniA) | Gln353Leu | 6 | 0.02 | 12.7 (21/165) | 8.9 (16/180) | 23.8 (35/147) | 0.0004 |
| 1,305,250 | C | G | Rv1173 (fbiC) | Arg774Gly | 1 | 0.00 | 14.0 (18/129) | 12.8 (21/164) | 17.2 (22/128) | 0.3 |
| 1,783,849 | G | C | Rv1580c | Ala15Gly | 21 | 0.00 *** | 22.2 (43/193) | 16.7 (32/192) | 26.0 (44/169) | 0.03 |
| 1,793,445 | T | G | Rv1592c | Glu99Ala | 17 | 0.32 | 6.2 (7/112) | 6.0 (7/117) | 20.0 (23/115) | 0.002 |
| 1,847,247 | G | C | Rv1639c | silent Thr404 | 9871 | - | 13.3 (15/113) | 17.2 (25/145) | 19.8 (23/116) | 0.2 |
| Pair of Genes | Evidence Suggesting a Functional Link | Combined Score |
|---|---|---|
| iniA–Rv1639c | Cooccurrence Across Genomes: Yes, score 0.151 | 0.151 |
| Rv1592–Rv1639c | Neighborhood in the Genome: None, but homologous genes are neighbors in other genomes (score 0.064). Cooccurrence Across Genomes: Yes (score 0.207). Co-Expression: none, but putative homologs are coexpressed in other organisms (score 0.072). Association in Curated Databases: none, but putative homologs are reported to interact in other organisms (score 0.095). Co-Mentioned in Pubmed Abstracts: none, but putative homologs are mentioned together in other organisms (score 0.098). | 0.335 |
| Rv1592–Rv0224c | Neighborhood in the Genome: None, but homologous genes are neighbors in other genomes (score 0.060). Cooccurrence Across Genomes: Yes (score 0.271). Co-Expression: none, but putative homologs are coexpressed in other organisms (score 0.067). | 0.304 |
| Gene | Functional Category | Product | Comments, Function, Essentiality |
|---|---|---|---|
| Rv0224c | Intermediary metabolism and respiration | Possible methyl-transferase (methylase) | Causes methylation. Essential gene for in vitro growth. Upregulated in response to thiol specific oxidative stress, maybe functionally related to oxidative stress response [31]. Deletion of NCgl2764 in C. glutamicum (Rv0224c in M. tuberculosis) abolished acetyltrehalose monocorynomycolate synthesis, leading to the accumulation of monohydroxycorynomycolate in the inner membrane and delaying its conversion to trehalose dihydroxycorynomycolate [45]. Methylases in prokaryotes have a primary function of DNA methylation in DNA self-recognition via restriction-modification systems that protect against invading DNA. In contrast, “orphan” methylases may play a role in chromosome stability, mismatch repair, replication. Orphan methylase MamA influences M. tuberculosis gene expression and fitness during hypoxia ([32] and references therein). |
| Rv0342 (iniA) | Cell wall and cell processes | Isoniazid inducible gene protein IniA | May function through a MDR-pump like mechanism, although it does not appear to directly transport isoniazid from the cell Isoniazid inducible gene protein IniA; Participates in the development of tolerance to both isoniazid and ethambutol. iniA gene is also induced by the ethambutol, which inhibits cell wall biosynthesis by a mechanism that is distinct from isoniazid. iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol [37]. IniA mediates TB drug resistance through fission activity to maintain plasma membrane integrity. Fission of the compromised areas could be used for cell membrane repair [38,39]. |
| Rv1173 (fbiC) | Intermediary metabolism and respiration | Probable F420 biosynthesis protein FbiC | Probable f420 biosynthesis protein fbic; Catalyzes the radical-mediated synthesis of 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) from 5-amino-6-(D-ribitylamino)uracil and L-tyrosine. Essential for coenzyme F420 production: participates in a portion of the F420 biosynthetic pathway between pyrimidinedione and FO (biosynthesis intermediate), before the deazaflavin ring is formed. fbiC along with ddn, fbiA, fbiB and fgd1 constitute the F420 biosynthesis pathway. Nitrofuran is activated by Ddn enzyme that is F420-dependent nitroreducatase. |
| Rv1580c | Insertion sequences and phages | PhiRv1 phage protein | The M. tuberculosis prophage-like element φRv1 encodes a site-specific recombination system utilizing an integrase of the serine recombinase family. However, no particles containing φRv1 DNA have been described. In a rabbit model (animals challenged with different mycobacterium), Rv1580c elicited a late humoral response 10 weeks post-challenge [42]. |
| Rv1592c | Lipid metabolism | Lipase/esterase | AB hydrolase superfamily. Lipase family. Rv1592c mRNA was identified by DNA microarray analysis (gene induced by hypoxia or by isoniazid (INH) or ethionamide treatment). DNA microarrays show a higher level of expression in M. tuberculosis H37Rv than in Rv3676 mutant [46]. It was demonstrated by both in silico and experimental approaches that Rv1592c could be a possible lipase/esterase [47]. Rv1592c is a lipolytic enzyme and its expression was up-regulated during INH treatment [40]. Lipid/ester catabolism is an important requirement for M. tuberculosis infection and persistence in hosts. Some lipolytic enzymes were shown to promote the survival of bacterium under isoniazid treatment and cell wall lipid remodeling could be the possible reason for drug tolerance ([40] and references therein). |
| Rv1639c | Cell wall and cell processes | Conserved membrane protein | Contains PS00904 protein phenyltransferases alpha subunit repeat signature. Transcriptomics: DNA microarrays and qRT-PCR show a higher level of expression in M. tuberculosis H37Rv than in phoP|Rv0757 mutant [48]. The Rv1639c protein was identified by mass spectrometry in M. tuberculosis H37Rv -infected guinea pig lungs at 90 days post infection but not 30 days [43]. A two-domain lysyltransferase and lysyl-tRNA-synthetase protein encoded by lysX gene of M. tuberculosis are necessary for phospholipid phosphatidylglycerol lysinylation, optimal survival in lungs of mice and guinea pigs, resistance to the host cationic antimicrobial peptides and for maintaining optimal membrane potential [44]. Rv1639c is located immediately downstream lysX and the Rv1639c-lysX intergenic region lacks typical mycobacterial promoter features. Two-domain lysyltransferase-lysyl-tRNA synthetase protein appears to negatively regulate Rv1639c expression [44]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mokrousov, I.; Slavchev, I.; Solovieva, N.; Dogonadze, M.; Vyazovaya, A.; Valcheva, V.; Masharsky, A.; Belopolskaya, O.; Dimitrov, S.; Zhuravlev, V.; et al. Molecular Insight into Mycobacterium tuberculosis Resistance to Nitrofuranyl Amides Gained through Metagenomics-like Analysis of Spontaneous Mutants. Pharmaceuticals 2022, 15, 1136. https://doi.org/10.3390/ph15091136
Mokrousov I, Slavchev I, Solovieva N, Dogonadze M, Vyazovaya A, Valcheva V, Masharsky A, Belopolskaya O, Dimitrov S, Zhuravlev V, et al. Molecular Insight into Mycobacterium tuberculosis Resistance to Nitrofuranyl Amides Gained through Metagenomics-like Analysis of Spontaneous Mutants. Pharmaceuticals. 2022; 15(9):1136. https://doi.org/10.3390/ph15091136
Chicago/Turabian StyleMokrousov, Igor, Ivaylo Slavchev, Natalia Solovieva, Marine Dogonadze, Anna Vyazovaya, Violeta Valcheva, Aleksey Masharsky, Olesya Belopolskaya, Simeon Dimitrov, Viacheslav Zhuravlev, and et al. 2022. "Molecular Insight into Mycobacterium tuberculosis Resistance to Nitrofuranyl Amides Gained through Metagenomics-like Analysis of Spontaneous Mutants" Pharmaceuticals 15, no. 9: 1136. https://doi.org/10.3390/ph15091136
APA StyleMokrousov, I., Slavchev, I., Solovieva, N., Dogonadze, M., Vyazovaya, A., Valcheva, V., Masharsky, A., Belopolskaya, O., Dimitrov, S., Zhuravlev, V., Portugal, I., Perdigão, J., & Dobrikov, G. M. (2022). Molecular Insight into Mycobacterium tuberculosis Resistance to Nitrofuranyl Amides Gained through Metagenomics-like Analysis of Spontaneous Mutants. Pharmaceuticals, 15(9), 1136. https://doi.org/10.3390/ph15091136