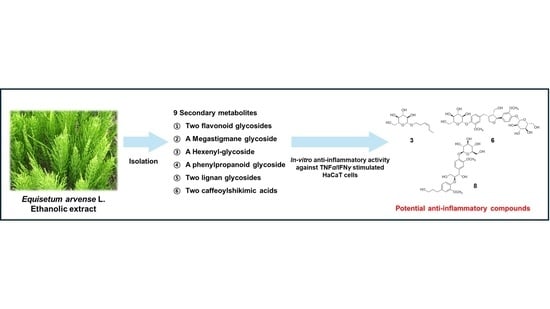
Phytochemical Investigation of Equisetum arvense and Evaluation of Their Anti-Inflammatory Potential in TNFα/INFγ-Stimulated Keratinocytes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Identification of the Compounds
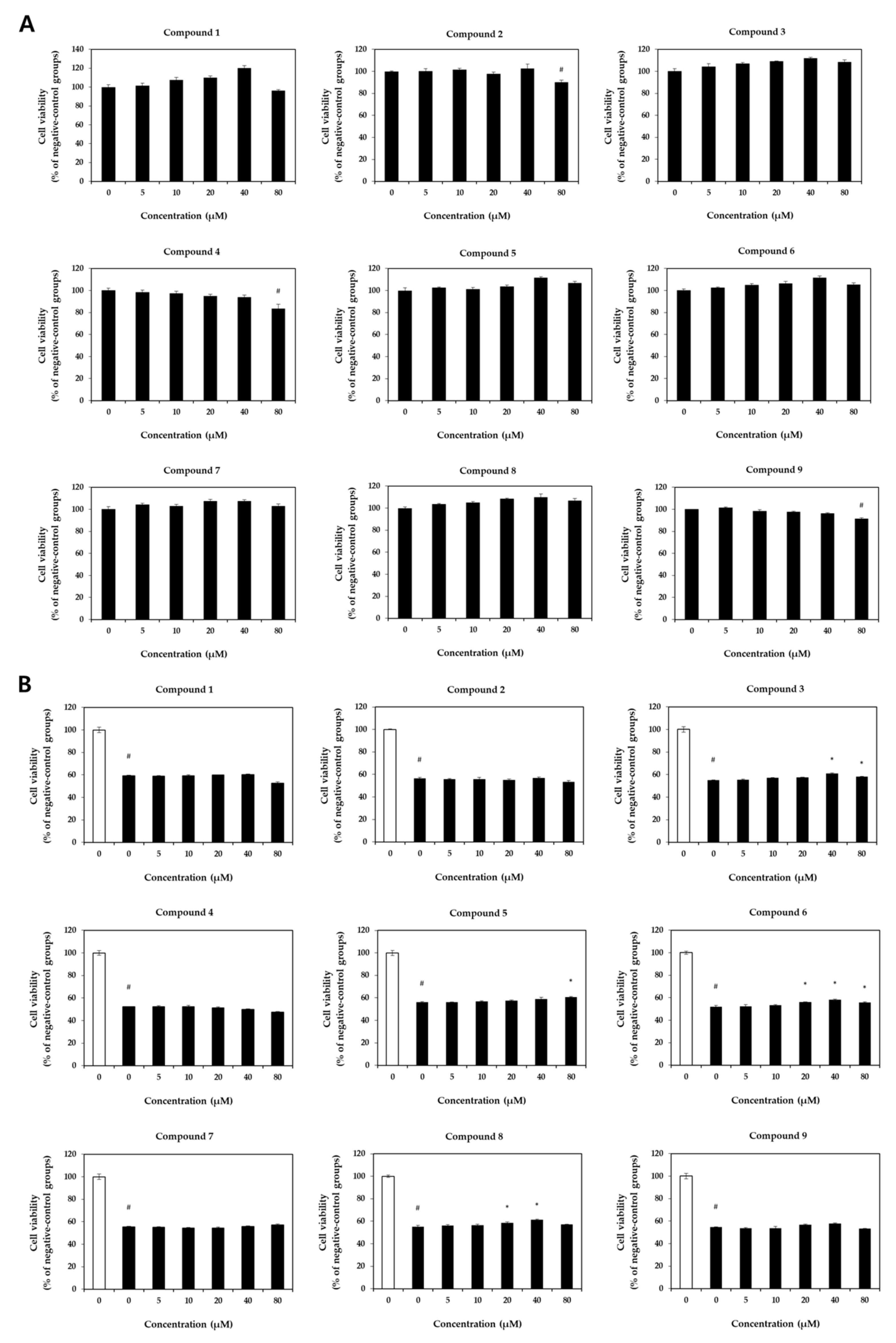
2.2. Cell Viability Assessment
2.3. Expression of Pro-Inflammatory Chemokines and Cytokines
2.4. Activation of Pro-Inflammatory Transcription Factors
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Plant Materials
3.3. Extraction and Isolation
3.4. Chemicals and Reagents
3.5. Cell Culture and Sample Treatment
3.6. Cell Viability
3.7. ELISA and qRT-PCR
3.8. Western Blotting
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, J.K.; Choi, E.; Hong, Y.H.; Kim, H.; Jang, Y.-J.; Lee, J.S.; Choung, E.S.; Woo, B.Y.; Hong, Y.D.; Lee, S. Syk/NF-κB-targeted anti-inflammatory activity of Melicope accedens (Blume) TG Hartley methanol extract. J. Ethnopharmacol. 2021, 271, 113887. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.J.; Amin, A.; Fu, X.-Q.; Chou, J.-Y.; Wu, J.-Y.; Wang, X.-Q.; Chen, Y.-J.; Wu, Y.; Li, J.; Yin, C.-L. The anti-inflammatory effects of an ethanolic extract of the rhizome of Atractylodes lancea, involves Akt/NF-κB signaling pathway inhibition. J. Ethnopharmacol. 2021, 277, 114183. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-S.; Lee, N.-K.; Choi, A.-J.; Choe, J.-S.; Bae, C.H.; Paik, H.-D. Anti-inflammatory potential of probiotic strain Weissella cibaria JW15 isolated from kimchi through regulation of NF-κB and MAPKs pathways in LPS-induced RAW 264.7 cells. J. Microbiol. Biotechnol. 2019, 29, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Hwang, Y.-H.; Gu, M.-J.; Cho, W.-K.; Ma, J.Y. Ethanol extracts of Sanguisorba officinalis L. suppress TNF-α/IFN-γ-induced pro-inflammatory chemokine production in HaCaT cells. Phytomedicine 2015, 22, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Kumar, L.; Mathias, C.; Zurakowski, D.; Oettgen, H.; Gorelik, L.; Geha, R. Toll-like receptor 2 is important for the TH1 response to cutaneous sensitization. J. Allergy Clin. Immunol. 2009, 123, 875–882.e1. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Jin, S.W.; Park, B.H.; Kim, H.G.; Khanal, T.; Han, H.J.; Hwang, Y.P.; Choi, J.M.; Chung, Y.C.; Hwang, S.K. Cultivated ginseng inhibits 2, 4-dinitrochlorobenzene-induced atopic dermatitis-like skin lesions in NC/Nga mice and TNF-α/IFN-γ-induced TARC activation in HaCaT cells. Food Chem. Toxicol. 2013, 56, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Trautmann, A.; Akdis, M.; Kleemann, D.; Altznauer, F.; Simon, H.-U.; Graeve, T.; Noll, M.; Bröcker, E.-B.; Blaser, K.; Akdis, C.A. T cell–mediated Fas-induced keratinocyte apoptosis plays a key pathogenetic role in eczematous dermatitis. J. Clin. Investig. 2000, 106, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lim, J.-Y.; Jo, E.H.; Noh, H.M.; Park, S.; Park, M.C.; Kim, D.-K. Chijabyukpi-Tang Inhibits pro-inflammatory cytokines and chemokines via the Nrf2/HO-1 signaling pathway in TNF-α/IFN-γ-stimulated HaCaT cells and ameliorates 2, 4-dinitrochlorobenzene-induced atopic dermatitis-like skin lesions in mice. Front. Pharmacol. 2020, 11, 1018. [Google Scholar] [CrossRef]
- Avena-Woods, C. Overview of atopic dermatitis. Am. J. Manag. Care 2017, 23, S115–S123. [Google Scholar]
- Guttman-Yassky, E.; Nograles, K.E.; Krueger, J.G. Contrasting pathogenesis of atopic dermatitis and psoriasis—Part I: Clinical and pathologic concepts. J. Allergy Clin. Immunol. 2011, 127, 1110–1118. [Google Scholar] [CrossRef]
- Nedoszytko, B.; Sokołowska-Wojdyło, M.; Ruckemann-Dziurdzińska, K.; Roszkiewicz, J.; Nowicki, R. Chemokines and cytokines network in the pathogenesis of the inflammatory skin diseases: Atopic dermatitis, psoriasis and skin mastocytosis. Postepy Dermatol Alergol 2014, 31, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Choi, H.K.; N’deh, K.P.U.; Choi, Y.-J.; Fan, M.; Kim, E.-k.; Chung, K.-H.; An, J.H. Inhibitory effect of Centella asiatica extract on DNCB-induced atopic dermatitis in HaCaT cells and BALB/c mice. Nutrients 2020, 12, 411. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Novak, N. Pathogenesis of atopic dermatitis. Clin. Exp. Allergy 2015, 45, 566–574. [Google Scholar] [CrossRef]
- Wollenberg, A.; Oranje, A.; Deleuran, M.; Simon, D.; Szalai, Z.; Kunz, B.; Svensson, A.; Barbarot, S.; Von Kobyletzki, L.; Taieb, A. ETFAD/EADV Eczema task force 2015 position paper on diagnosis and treatment of atopic dermatitis in adult and paediatric patients. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 729–747. [Google Scholar] [CrossRef]
- Dawid-Pac, R. Medicinal plants used in treatment of inflammatory skin diseases. Postepy Dermatol Alergol 2013, 30, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Al-Snafi, A.E. The pharmacology of Equisetum arvense-A review. IOSR J. Pharm. 2017, 7, 31–42. [Google Scholar] [CrossRef]
- Sandhu, N.S.; Kaur, S.; Chopra, D. Equisetum arvense: Pharmacology and phytochemistry-a review. Asian. J. Pharm. Clin. Res. 2010, 3, 146–150. [Google Scholar]
- Asgarpanah, J.; Roohi, E. Phytochemistry and pharmacological properties of Equisetum arvense L. J. Med. Plant Res. 2012, 6, 3689–3693. [Google Scholar] [CrossRef]
- Gründemann, C.; Lengen, K.; Sauer, B.; Garcia-Käufer, M.; Zehl, M.; Huber, R. Equisetum arvense (common horsetail) modulates the function of inflammatory immunocompetent cells. BMC Complement. Altern. Med. 2014, 14, 283. [Google Scholar] [CrossRef]
- Shiba, F.; Miyauchi, M.; Chea, C.; Furusho, H.; Iwasaki, S.; Shimizu, R.; Ohta, K.; Nishihara, T.; Takata, T. Anti-inflammatory effect of glycyrrhizin with Equisetum arvense extract. Odontology 2021, 109, 464–473. [Google Scholar] [CrossRef]
- Shiba, F.; Furusho, H.; Takata, T.; Shimizu, R.; Miyauchi, M. Equisetum arvense inhibits alveolar bone destruction in a rat model with lipopolysaccharide (LPS)-induced periodontitis. Int. J. Dent. 2022, 2022, 7398924. [Google Scholar] [CrossRef] [PubMed]
- Mimica-Dukic, N.; Simin, N.; Cvejic, J.; Jovin, E.; Orcic, D.; Bozin, B. Phenolic compounds in field horsetail (Equisetum arvense L.) as natural antioxidants. Molecules 2008, 13, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.; Ramos, A.J.; Sanchis, V.; Marín, S. Equisetum arvense hydro-alcoholic extract: Phenolic composition and antifungal and antimycotoxigenic effect against Aspergillus flavus and Fusarium verticillioides in stored maize. J. Sci. Food Agric. 2013, 93, 2248–2253. [Google Scholar] [CrossRef] [PubMed]
- Kukrić, Z.; Topalić-Trivunović, L.; Pavičić, S.; Žabić, M.; Matoš, S.; Davidović, A. Total phenolic content, antioxidant and antimicrobial activity of Equisetum arvense L. Chem. Ind. Chem. Eng. Q. 2013, 19, 37–43. [Google Scholar] [CrossRef]
- Steinborn, C.; Potterat, O.; Meyer, U.; Trittler, R.; Stadlbauer, S.; Huber, R.; Gründemann, C. In vitro anti-inflammatory effects of Equisetum arvense are not solely mediated by silica. Planta Med. 2018, 50, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.S.; So, H.M.; Kim, S.; Kim, J.K.; Kim, J.-C.; Kang, D.-M.; Ahn, M.-J.; Ko, Y.-J.; Kim, K.H. Comparative evaluation of bioactive phytochemicals in Spinacia oleracea cultivated under greenhouse and open field conditions. Arch. Pharm. Res. 2022, 45, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, K.H.; Han, S.H.; Kim, H.-J.; Cho, I.-H.; Lee, S. Structure determination of heishuixiecaoline A from Valeriana fauriei and its content from different cultivated regions by HPLC/PDA Analysis. Nat. Prod. Sci. 2022, 28, 181–186. [Google Scholar] [CrossRef]
- Yu, J.S.; Jeong, S.Y.; Li, C.; Oh, T.; Kwon, M.; Ahn, J.S.; Ko, S.-K.; Ko, Y.-J.; Cao, S.; Kim, K.H. New phenalenone derivatives from the Hawaiian volcanic soil-associated fungus Penicillium herquei FT729 and their inhibitory effects on indoleamine 2, 3-dioxygenase 1 (IDO1). Arch. Pharm. Res. 2022, 45, 105–113. [Google Scholar] [CrossRef]
- Lee, S.R.; Lee, B.S.; Yu, J.S.; Kang, H.; Yoo, M.J.; Yi, S.A.; Han, J.-W.; Kim, S.; Kim, J.K.; Kim, J.-C. Identification of anti-adipogenic withanolides from the roots of Indian ginseng (Withania somnifera). J. Ginseng Res. 2022, 46, 357–366. [Google Scholar] [CrossRef]
- Lee, K.H.; Kim, J.K.; Yu, J.S.; Jeong, S.Y.; Choi, J.H.; Kim, J.-C.; Ko, Y.-J.; Kim, S.-H.; Kim, K.H. Ginkwanghols A and B, osteogenic coumaric acid-aliphatic alcohol hybrids from the leaves of Ginkgo biloba. Arch. Pharm. Res. 2021, 44, 514–524. [Google Scholar] [CrossRef]
- Wan, C.; Yu, Y.; Zhou, S.; Tian, S.; Cao, S. Isolation and identification of phenolic compounds from Gynura divaricata leaves. Pharmacogn. Mag. 2011, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Woo, K.W.; Lee, K.R. Phytochemical constituents of Allium victorialis var. platyphyllum. Nat. Prod. Sci 2013, 19, 221–226. [Google Scholar]
- Lee, K.H.; Choi, S.U.; Lee, K.R. Sesquiterpenes from Syneilesis palmata and their cytotoxicity against human cancer cell lines in vitro. Arch. Pharm. Res. 2005, 28, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-J.; Yan, L.-L.; Yin, P.-P.; Shi, L.-L.; Zhang, J.-H.; Liu, Y.-J.; Ma, C. Structural characterisation and antioxidant activity evaluation of phenolic compounds from cold-pressed Perilla frutescens var. arguta seed flour. Food. Chem. 2014, 164, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.-B.; Tezuka, Y.; Kikuchi, T.; Nakano, H.; Tamaoki, T.; Park, J.-H. Constituents of a fern, Davallia mariesii Moore. I. Isolation and structures of davallialactone and a new flavanone glucuronide. Chem. Pharm. Bull. 1990, 38, 3218–3225. [Google Scholar] [CrossRef] [PubMed]
- El Gamal, A.; Takeya, K.; Itokawa, H.; Halim, A.; Amer, M.; Saad, H.-E. Lignan bis-glucosides from Galium sinaicum. Phytochemistry 1997, 45, 597–600. [Google Scholar] [CrossRef]
- Saito, T.; Yamane, H.; Murofushi, N.; Takahashi, N.; Phinney, B.O. 4-O-caffeoylshikimic and 4-O-(p-coumaroyl) shikimic acids from the dwarf tree fern, Dicksonia antarctica. Biosci. Biotechnol. Biochem. 1997, 61, 1397–1398. [Google Scholar] [CrossRef]
- Huo, C.; Liang, H.; Zhao, Y.; Wang, B.; Zhang, Q. Neolignan glycosides from Symplocos caudata. Phytochemistry 2008, 69, 788–795. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Yang, L.; Feng, Y.; Liu, Y.; Zeng, X. Studies of phenolic acid constituents from the whole plant of Sarcandra glabra. Zhongyao Xinyao Yu Linchuang Yaoli 2012, 23, 295–298. [Google Scholar]
- Veit, M.; Geiger, H.; Czygan, F.-C.; Markham, K.R. Malonylated flavone 5-O-glucosides in the barren sprouts of Equisetum arvense. Phytochemistry 1990, 29, 2555–2560. [Google Scholar] [CrossRef]
- Francescato, L.N.; Debenedetti, S.L.; Schwanz, T.G.; Bassani, V.L.; Henriques, A.T. Identification of phenolic compounds in Equisetum giganteum by LC–ESI-MS/MS and a new approach to total flavonoid quantification. Talanta 2013, 105, 192–203. [Google Scholar] [CrossRef]
- Wang, Z.; Tian, Y.; Sugimoto, S.; Yamano, Y.; Kawakami, S.; Otsuka, H.; Matsunami, K. Four new glucosides from the aerial parts of Equisetum sylvaticum. J. Nat. Med. 2022, 76, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Kanchanapoom, T.; Otsuka, H.; Ruchirawat, S. Megastigmane glucosides from Equisetum debile and E. diffusum. Chem. Pharm. Bull. 2007, 55, 1277–1280. [Google Scholar] [CrossRef] [PubMed]
- Boeing, T.; Tafarelo Moreno, K.G.; Gasparotto Junior, A.; Mota da Silva, L.; de Souza, P. Phytochemistry and pharmacology of the genus Equisetum (Equisetaceae): A narrative review of the species with therapeutic potential for kidney diseases. Evid. Based Complement. Alternat. Med. 2021, 2021, 6658434. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Yoo, J.-M.; Lee, E.; Lee, B.; Cho, W.-K.; Park, K.-I.; Ma, J.Y. Anti-inflammatory effects of Perillae Herba ethanolic extract against TNF-α/IFN-γ-stimulated human keratinocyte HaCaT cells. J. Ethnopharmacol. 2018, 211, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Brunner, P.M.; Guttman-Yassky, E.; Leung, D.Y. The immunology of atopic dermatitis and its reversibility with broad-spectrum and targeted therapies. J. Allergy Clin. Immunol. 2017, 139, S65–S76. [Google Scholar] [CrossRef] [PubMed]
- Albanesi, C.; Scarponi, C.; Giustizieri, M.L.; Girolomoni, G. Keratinocytes in inflammatory skin diseases. Curr. Drug. Targets. Inflamm. Allergy 2005, 4, 329–334. [Google Scholar] [CrossRef] [PubMed]
- An, E.-J.; Kim, Y.; Lee, S.-H.; Choi, S.-H.; Chung, W.S.; Jang, H.-J. Ophiopogonin D ameliorates DNCB-induced atopic dermatitis-like lesions in BALB/c mice and TNF-α-inflamed HaCaT cell. Biochem. Biophys. Res. Commun. 2020, 522, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Dorjsembe, B.; Nho, C.W.; Choi, Y.; Kim, J.-C. Extract from black soybean cultivar a63 extract ameliorates atopic dermatitis-like skin inflammation in an oxazolone-induced murine model. Molecules 2022, 27, 2751. [Google Scholar] [CrossRef]
- Kwon, D.-J.; Bae, Y.-S.; Ju, S.M.; Goh, A.R.; Youn, G.S.; Choi, S.Y.; Park, J. Casuarinin suppresses TARC/CCL17 and MDC/CCL22 production via blockade of NF-κB and STAT1 activation in HaCaT cells. Biochem. Biophys. Res. Commun. 2012, 417, 1254–1259. [Google Scholar] [CrossRef]
- Nakayama, T.; Hieshima, K.; Nagakubo, D.; Sato, E.; Nakayama, M.; Kawa, K.; Yoshie, O. Selective induction of Th2-attracting chemokines CCL17 and CCL22 in human B cells by latent membrane protein 1 of Epstein-Barr virus. J. Virol. 2004, 78, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Luyen, B.T.T.; Tai, B.H.; Cuong, N.M.; Kim, Y.C.; Van Minh, C.; Kim, Y.H. Chemical constituents of Miliusa balansae leaves and inhibition of nitric oxide production in lipopolysaccharide-induced RAW 264.7 cells. Bioorg. Med. Chem. Lett. 2015, 25, 3859–3863. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xiao, Y.; Yang, B.; Wang, Z.; Wu, L.; Su, X.; Brantner, A.; Kuang, H.; Wang, Q. Isolation and screened neuroprotective active constituents from the roots and rhizomes of Valeriana amurensis. Fitoterapia 2014, 96, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Ngan, N.T.T.; Quang, T.H.; Tai, B.H.; Song, S.B.; Lee, D.; Kim, Y.H. Anti-inflammatory and PPAR transactivational effects of components from the stem bark of Ginkgo biloba. J. Agric. Food Chem. 2012, 60, 2815–2824. [Google Scholar] [CrossRef] [PubMed]
- Nahar, P.P.; Driscoll, M.V.; Li, L.; Slitt, A.L.; Seeram, N.P. Phenolic mediated anti-inflammatory properties of a maple syrup extract in RAW 264.7 murine macrophages. J. Funct. Foods 2014, 6, 126–136. [Google Scholar] [CrossRef]
- Cho, J.Y.; Kim, A.R.; Park, M.H. Lignans from the rhizomes of Coptis japonica differentially act as anti-inflammatory principles. Planta Med. 2001, 67, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yang, M.; Zuo, Z. Overview of the anti-inflammatory effects, pharmacokinetic properties and clinical efficacies of arctigenin and arctiin from Arctium lappa L. Acta Pharmacol. Sin. 2018, 39, 787–801. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-S.; Kim, W.-J.; Bae, W.-Y.; Lee, N.-K.; Paik, H.-D. Inula britannica inhibits adipogenesis of 3T3-L1 preadipocytes via modulation of mitotic clonal expansion involving ERK 1/2 and Akt signaling pathways. Nutrients 2020, 12, 3037. [Google Scholar] [CrossRef]





| Gene | Primer Sequence |
|---|---|
| CCL17 | Forward: 5′-CAGCTCGAGGGACCAATGTG-3′ |
| Reverse: 5′-CCTGCCCTGCACAGTTACAA-3′ | |
| CCL5 | Forward: 5′-CAGTCGTCTTTGTCACCCGA-3′ |
| Reverse: 5′-TCTTCTCTGGGTTGGCACAC-3′ | |
| CCL22 | Forward: 5′-ACTCCTGGTTGTCCTCGTC-3′ |
| Reverse: 5′-GACGTAATCACGGCAGCAGA-3′ | |
| CCL2 | Forward: 5′-AATCAATGCCCCAGTCACCT-3′ |
| Reverse: 5′-CTTCTTTGGGACACTTGCTGC-3′ | |
| IL1B | Forward: 5′-CAGCTACGAATCTCCGACCAC-3′ |
| Reverse: 5′-GGCAGGGAACCAGCATCTTC-3′ | |
| IL6 | Forward: 5′-TTCGGTCCAGTTGCCTTCTC-3′ |
| Reverse: 5′-TCTTCTCCTGGGGGTACTGG-3′ | |
| CXCL8 | Forward: 5′-TGTCTGGACCCCAAGGAAAAC-3′ |
| Reverse: 5′-TGGCATCTTCACTGATTCTTGG-3′ | |
| G3PD | Forward: 5′-GAAGGTGAAGGTCGGAGTC-3′ |
| Reverse: 5′-GAAGATGGTGATGGGATTTC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.Y.; Yu, H.-S.; Ra, M.-J.; Jung, S.-M.; Yu, J.-N.; Kim, J.-C.; Kim, K.H. Phytochemical Investigation of Equisetum arvense and Evaluation of Their Anti-Inflammatory Potential in TNFα/INFγ-Stimulated Keratinocytes. Pharmaceuticals 2023, 16, 1478. https://doi.org/10.3390/ph16101478
Jeong SY, Yu H-S, Ra M-J, Jung S-M, Yu J-N, Kim J-C, Kim KH. Phytochemical Investigation of Equisetum arvense and Evaluation of Their Anti-Inflammatory Potential in TNFα/INFγ-Stimulated Keratinocytes. Pharmaceuticals. 2023; 16(10):1478. https://doi.org/10.3390/ph16101478
Chicago/Turabian StyleJeong, Se Yun, Hyung-Seok Yu, Moon-Jin Ra, Sang-Mi Jung, Jeong-Nam Yu, Jin-Chul Kim, and Ki Hyun Kim. 2023. "Phytochemical Investigation of Equisetum arvense and Evaluation of Their Anti-Inflammatory Potential in TNFα/INFγ-Stimulated Keratinocytes" Pharmaceuticals 16, no. 10: 1478. https://doi.org/10.3390/ph16101478
APA StyleJeong, S. Y., Yu, H.-S., Ra, M.-J., Jung, S.-M., Yu, J.-N., Kim, J.-C., & Kim, K. H. (2023). Phytochemical Investigation of Equisetum arvense and Evaluation of Their Anti-Inflammatory Potential in TNFα/INFγ-Stimulated Keratinocytes. Pharmaceuticals, 16(10), 1478. https://doi.org/10.3390/ph16101478