Mechanisms of Regulation of the Expression of miRNAs and lncRNAs by Metformin in Ovarian Cancer
Abstract
:1. Introduction
1.1. Ovarian Cancer
1.2. Current and New Approaches for OC
1.3. Metformin in OC
2. Methodological Approach: Search Strategy and Articles Inclusion Criteria
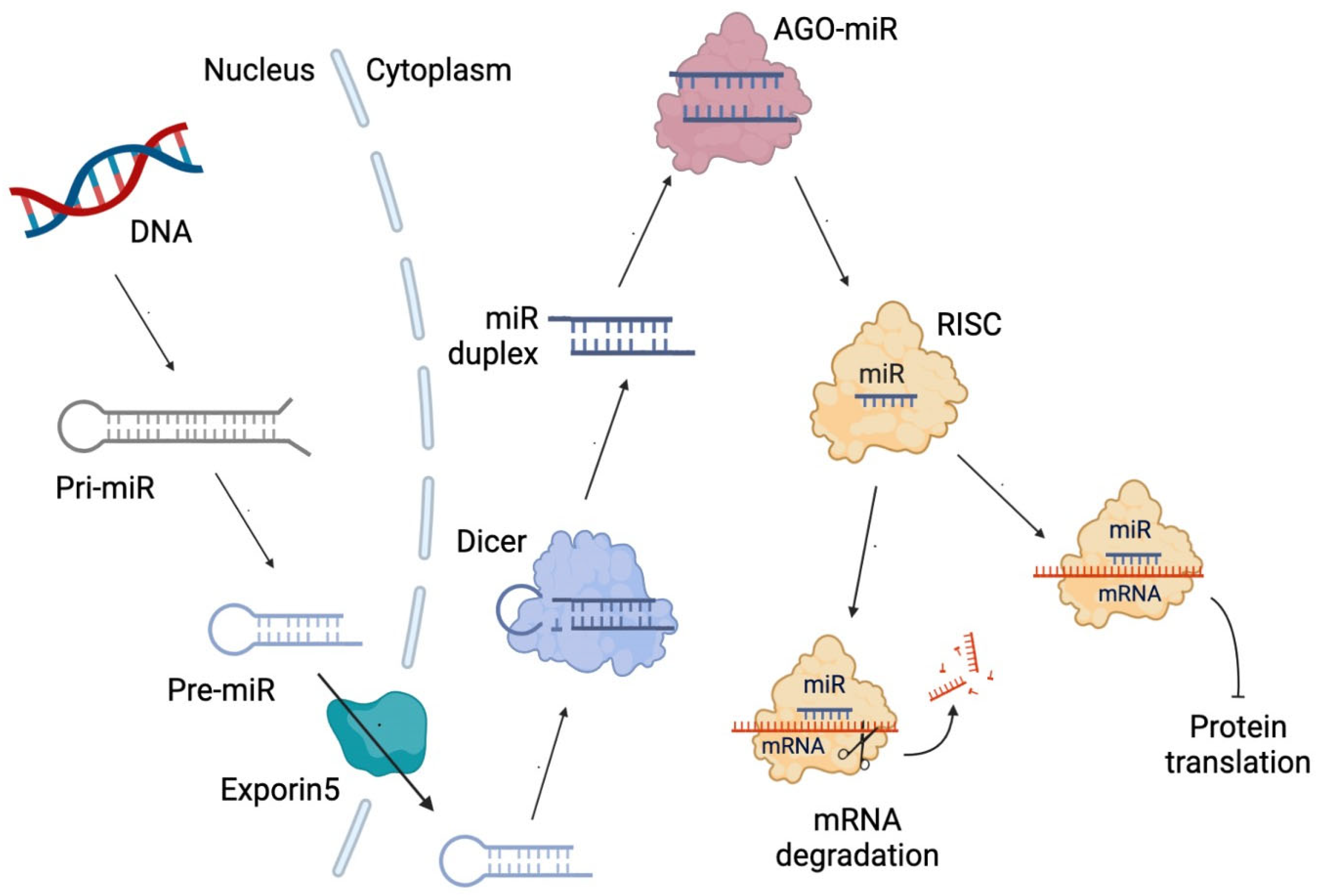
3. Non-Coding RNAs
4. Mechanisms of ncRNA Biosynthesis Regulated by Metformin
4.1. Role of Transcriptional Factors in the Metformin-Mediated Modulation of miRNA Expression
p53-Dependent Metformin Effects
4.2. Role of Metformin on Epigenetic Modification of ncRNAs
4.2.1. DNA Methylation of ncRNAs
4.2.2. Methylation of miRNAs
4.3. Role of Metformin in miRNA Maturation
5. ncRNA-Related Therapeutic Effects of Metformin in OC
5.1. miR-23b and miR-145 in OC
5.2. miR-21 in OC and Other Cancers
5.3. miR-27 in OC and Other Cancers
5.4. lncRNA H19 in EOC and Its Regulation by Metformin
5.5. Metformin Regulation of lncRNA SNHG7 in OC
6. Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cabasag, C.J.; Fagan, P.J.; Ferlay, J.; Vignat, J.; Laversanne, M.; Liu, L.; van der Aa, M.A.; Bray, F.; Soerjomataram, I. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. Int. J. Cancer 2022, 151, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Yancik, R. Ovarian cancer. Age contrasts in incidence, histology, disease stage at diagnosis, and mortality. Cancer 1993, 71, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Arora, T.; Mullangi, S.; Lekkala, M.R. Ovarian Cancer. In StatPearls; StatPearls Publishing LLC: Treasure Island, FL, USA, 2023. [Google Scholar]
- Savant, S.S.; Sriramkumar, S.; O’Hagan, H.M. The Role of Inflammation and Inflammatory Mediators in the Development, Progression, Metastasis, and Chemoresistance of Epithelial Ovarian Cancer. Cancers 2018, 10, 251. [Google Scholar] [CrossRef] [PubMed]
- Gavalas, N.G.; Liontos, M.; Trachana, S.P.; Bagratuni, T.; Arapinis, C.; Liacos, C.; Dimopoulos, M.A.; Bamias, A. Angiogenesis-related pathways in the pathogenesis of ovarian cancer. Int. J. Mol. Sci. 2013, 14, 15885–15909. [Google Scholar] [CrossRef]
- Guo, Y.; Nie, Q.; MacLean, A.L.; Li, Y.; Lei, J.; Li, S. Multiscale Modeling of Inflammation-Induced Tumorigenesis Reveals Competing Oncogenic and Oncoprotective Roles for Inflammation. Cancer Res. 2017, 77, 6429–6441. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Beeghly-Fadiel, A.; Wilson, A.J.; Keene, S.; El Ramahi, M.; Xu, S.; Marnett, L.J.; Fadare, O.; Crispens, M.A.; Khabele, D. Differential cyclooxygenase expression levels and survival associations in type I and type II ovarian tumors. J. Ovarian Res. 2018, 11, 17. [Google Scholar] [CrossRef]
- Daikoku, T.; Wang, D.; Tranguch, S.; Morrow, J.D.; Orsulic, S.; DuBois, R.N.; Dey, S.K. Cyclooxygenase-1 is a potential target for prevention and treatment of ovarian epithelial cancer. Cancer Res. 2005, 65, 3735–3744. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, X.; Jeschke, U.; von Schonfeldt, V. COX-2-PGE(2)-EPs in gynecological cancers. Arch. Gynecol. Obstet. 2020, 301, 1365–1375. [Google Scholar] [CrossRef]
- Pua, K.H.; Chew, C.L.; Lane, D.P.; Tergaonkar, V. Inflammation-associated genomic instability in cancer. Genome Instab. Dis. 2020, 1, 1–9. [Google Scholar] [CrossRef]
- Mei, C.; Gong, W.; Wang, X.; Lv, Y.; Zhang, Y.; Wu, S.; Zhu, C. Anti-angiogenic therapy in ovarian cancer: Current understandings and prospects of precision medicine. Front. Pharmacol. 2023, 14, 1147717. [Google Scholar] [CrossRef]
- Garrido, M.P.; Torres, I.; Vega, M.; Romero, C. Angiogenesis in Gynecological Cancers: Role of Neurotrophins. Front. Oncol. 2019, 9, 913. [Google Scholar] [CrossRef]
- Garrido, M.P.; Fredes, A.N.; Lobos-Gonzalez, L.; Valenzuela-Valderrama, M.; Vera, D.B.; Romero, C. Current Treatments and New Possible Complementary Therapies for Epithelial Ovarian Cancer. Biomedicines 2021, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Ueda, Y.; Naka, T.; Enomoto, T. Therapeutic strategies in epithelial ovarian cancer. J. Exp. Clin. Cancer Res. 2012, 31, 14. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, L.; Nandy, S.K.; Payal, N.; Yadav, S.; Vargas-De-La-Cruz, C.; Anwer, M.K.; Khan, H.; Behl, T.; Bungau, S.G. Molecular Aspects and Therapeutic Implications of Herbal Compounds Targeting Different Types of Cancer. Molecules 2023, 28, 750. [Google Scholar] [CrossRef] [PubMed]
- Sivasankarapillai, V.S.; Madhu Kumar Nair, R.; Rahdar, A.; Bungau, S.; Zaha, D.C.; Aleya, L.; Tit, D.M. Overview of the anticancer activity of withaferin A, an active constituent of the Indian ginseng Withania somnifera. Environ. Sci. Pollut. Res. Int. 2020, 27, 26025–26035. [Google Scholar] [CrossRef] [PubMed]
- Kakar, S.S.; Ratajczak, M.Z.; Powell, K.S.; Moghadamfalahi, M.; Miller, D.M.; Batra, S.K.; Singh, S.K. Withaferin a alone and in combination with cisplatin suppresses growth and metastasis of ovarian cancer by targeting putative cancer stem cells. PLoS ONE 2014, 9, e107596. [Google Scholar] [CrossRef] [PubMed]
- Therachiyil, L.; Anand, A.; Azmi, A.; Bhat, A.; Korashy, H.M.; Uddin, S. Role of RAS signaling in ovarian cancer. F1000Research 2022, 11, 1253. [Google Scholar] [CrossRef]
- Ghufran, M.; Khan, H.A.; Ullah, M.; Ghufran, S.; Ayaz, M.; Siddiq, M.; Hassan, S.S.U.; Bungau, S. In Silico Strategies for Designing of Peptide Inhibitors of Oncogenic K-Ras G12V Mutant: Inhibiting Cancer Growth and Proliferation. Cancers 2022, 14, 4884. [Google Scholar] [CrossRef]
- Garrido, M.P.; Vega, M.; Romero, C. Antitumoral Effects of Metformin in Ovarian Cancer. Metformin 2019, 10, 163–180. [Google Scholar] [CrossRef]
- Drzewoski, J.; Hanefeld, M. The Current and Potential Therapeutic Use of Metformin-The Good Old Drug. Pharmaceuticals 2021, 14, 122. [Google Scholar] [CrossRef]
- Lashen, H. Role of metformin in the management of polycystic ovary syndrome. Ther. Adv. Endocrinol. Metab. 2010, 1, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Goswami, S.; Giacomini, K.M.; Altman, R.B.; Klein, T.E. Metformin pathways: Pharmacokinetics and pharmacodynamics. Pharm. Genom. 2012, 22, 820–827. [Google Scholar] [CrossRef]
- Motohashi, H.; Inui, K. Organic cation transporter OCTs (SLC22) and MATEs (SLC47) in the human kidney. AAPS J. 2013, 15, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Ito, K.; Akahira, J.; Sato, N.; Onogawa, T.; Moriya, T.; Unno, M.; Abe, T.; Niikura, H.; Takano, T.; et al. Expression of organic cation transporter SLC22A16 in human epithelial ovarian cancer: A possible role of the adriamycin importer. Int. J. Gynecol. Pathol. 2007, 26, 334–340. [Google Scholar] [CrossRef]
- Andreev, E.; Brosseau, N.; Carmona, E.; Mes-Masson, A.M.; Ramotar, D. The human organic cation transporter OCT1 mediates high affinity uptake of the anticancer drug daunorubicin. Sci. Rep. 2016, 6, 20508. [Google Scholar] [CrossRef] [PubMed]
- Checkley, L.A.; Rudolph, M.C.; Wellberg, E.A.; Giles, E.D.; Wahdan-Alaswad, R.S.; Houck, J.A.; Edgerton, S.M.; Thor, A.D.; Schedin, P.; Anderson, S.M.; et al. Metformin Accumulation Correlates with Organic Cation Transporter 2 Protein Expression and Predicts Mammary Tumor Regression In Vivo. Cancer Prev. Res. 2017, 10, 198–207. [Google Scholar] [CrossRef]
- Bridgeman, S.C.; Ellison, G.C.; Melton, P.E.; Newsholme, P.; Mamotte, C.D.S. Epigenetic effects of metformin: From molecular mechanisms to clinical implications. Diabetes Obes. Metab. 2018, 20, 1553–1562. [Google Scholar] [CrossRef]
- Lengyel, E.; Litchfield, L.M.; Mitra, A.K.; Nieman, K.M.; Mukherjee, A.; Zhang, Y.; Johnson, A.; Bradaric, M.; Lee, W.; Romero, I.L. Metformin inhibits ovarian cancer growth and increases sensitivity to paclitaxel in mouse models. Am. J. Obstet. Gynecol. 2015, 212, 479.e1–479.e10. [Google Scholar] [CrossRef]
- Garrido, M.P.; Vera, C.; Vega, M.; Quest, A.F.G.; Romero, C. Metformin prevents nerve growth factor-dependent proliferative and proangiogenic effects in epithelial ovarian cancer cells and endothelial cells. Ther. Adv. Med. Oncol. 2018, 10, 1758835918770984. [Google Scholar] [CrossRef]
- Rattan, R.; Graham, R.P.; Maguire, J.L.; Giri, S.; Shridhar, V. Metformin suppresses ovarian cancer growth and metastasis with enhancement of cisplatin cytotoxicity in vivo. Neoplasia 2011, 13, 483–491. [Google Scholar] [CrossRef]
- Wu, B.; Li, S.; Sheng, L.; Zhu, J.; Gu, L.; Shen, H.; La, D.; Hambly, B.D.; Bao, S.; Di, W. Metformin inhibits the development and metastasis of ovarian cancer. Oncol. Rep. 2012, 28, 903–908. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.G.; Plas, D.R.; Kubek, S.; Buzzai, M.; Mu, J.; Xu, Y.; Birnbaum, M.J.; Thompson, C.B. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 2005, 18, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Wei, J.; Wan, J.; Wang, W.; Wang, L.; Yuan, Y.; Yang, Z.; Liu, X.; Ming, L. Low glucose and metformin-induced apoptosis of human ovarian cancer cells is connected to ASK1 via mitochondrial and endoplasmic reticulum stress-associated pathways. J. Exp. Clin. Cancer Res. 2019, 38, 77. [Google Scholar] [CrossRef] [PubMed]
- Soraya, H.; Esfahanian, N.; Shakiba, Y.; Ghazi-Khansari, M.; Nikbin, B.; Hafezzadeh, H.; Maleki Dizaji, N.; Garjani, A. Anti-angiogenic Effects of Metformin, an AMPK Activator, on Human Umbilical Vein Endothelial Cells and on Granulation Tissue in Rat. Iran. J. Basic Med. Sci. 2012, 15, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Sun, X.; Ma, Q.; Fu, G.F.; Cong, L.L.; Zhang, H.; Fan, D.F.; Feng, J.; Lu, S.Y.; Liu, J.L.; et al. Metformin’s antitumour and anti-angiogenic activities are mediated by skewing macrophage polarization. J. Cell. Mol. Med. 2018, 22, 3825–3836. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, P.; Wang, H.; Hou, D.; Li, W.; Xiao, G.; Li, C. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44(+)CD117(+) ovarian cancer stem cells. Stem Cell Res. Ther. 2015, 6, 262. [Google Scholar] [CrossRef]
- Grassi, M.L.; Palma, C.S.; Thome, C.H.; Lanfredi, G.P.; Poersch, A.; Faca, V.M. Proteomic analysis of ovarian cancer cells during epithelial-mesenchymal transition (EMT) induced by epidermal growth factor (EGF) reveals mechanisms of cell cycle control. J. Proteom. 2017, 151, 2–11. [Google Scholar] [CrossRef]
- Chowanadisai, W.; Messerli, S.M.; Miller, D.H.; Medina, J.E.; Hamilton, J.W.; Messerli, M.A.; Brodsky, A.S. Cisplatin Resistant Spheroids Model Clinically Relevant Survival Mechanisms in Ovarian Tumors. PLoS ONE 2016, 11, e0151089. [Google Scholar] [CrossRef]
- Haslehurst, A.M.; Koti, M.; Dharsee, M.; Nuin, P.; Evans, K.; Geraci, J.; Childs, T.; Chen, J.; Li, J.; Weberpals, J.; et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 2012, 12, 91. [Google Scholar] [CrossRef]
- Kajiyama, H.; Shibata, K.; Terauchi, M.; Yamashita, M.; Ino, K.; Nawa, A.; Kikkawa, F. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int. J. Oncol. 2007, 31, 277–283. [Google Scholar] [CrossRef]
- Gonzalez, V.D.; Samusik, N.; Chen, T.J.; Savig, E.S.; Aghaeepour, N.; Quigley, D.A.; Huang, Y.W.; Giangarra, V.; Borowsky, A.D.; Hubbard, N.E.; et al. Commonly Occurring Cell Subsets in High-Grade Serous Ovarian Tumors Identified by Single-Cell Mass Cytometry. Cell Rep. 2018, 22, 1875–1888. [Google Scholar] [CrossRef]
- Zhu, X.; Shen, H.; Yin, X.; Long, L.; Xie, C.; Liu, Y.; Hui, L.; Lin, X.; Fang, Y.; Cao, Y.; et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene 2016, 35, 323–332. [Google Scholar] [CrossRef]
- Cui, Y.; Qin, L.; Tian, D.; Wang, T.; Fan, L.; Zhang, P.; Wang, Z. ZEB1 Promotes Chemoresistance to Cisplatin in Ovarian Cancer Cells by Suppressing SLC3A2. Chemotherapy 2018, 63, 262–271. [Google Scholar] [CrossRef]
- Brown, J.R.; Chan, D.K.; Shank, J.J.; Griffith, K.A.; Fan, H.; Szulawski, R.; Yang, K.; Reynolds, R.K.; Johnston, C.; McLean, K.; et al. Phase II clinical trial of metformin as a cancer stem cell-targeting agent in ovarian cancer. JCI Insight 2020, 5, e133247. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Hock, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Schulten, H.J. Pleiotropic Effects of Metformin on Cancer. Int. J. Mol. Sci. 2018, 19, 2850. [Google Scholar] [CrossRef]
- Alimoradi, N.; Firouzabadi, N.; Fatehi, R. How metformin affects various malignancies by means of microRNAs: A brief review. Cancer Cell Int. 2021, 21, 207. [Google Scholar] [CrossRef]
- Sargolzaei, J.; Etemadi, T.; Alyasin, A. The P53/microRNA network: A potential tumor suppressor with a role in anticancer therapy. Pharmacol. Res. 2020, 160, 105179. [Google Scholar] [CrossRef]
- Do, M.T.; Kim, H.G.; Choi, J.H.; Jeong, H.G. Metformin induces microRNA-34a to downregulate the Sirt1/Pgc-1alpha/Nrf2 pathway, leading to increased susceptibility of wild-type p53 cancer cells to oxidative stress and therapeutic agents. Free Radic. Biol. Med. 2014, 74, 21–34. [Google Scholar] [CrossRef]
- Sun, R.; Ma, X.; Cai, X.; Pan, X.; Liu, D. The effect and mechanism of action of metformin on in vitro FaDu cell proliferation. J. Int. Med. Res. 2016, 44, 1049–1054. [Google Scholar] [CrossRef]
- Sachdeva, M.; Zhu, S.; Wu, F.; Wu, H.; Walia, V.; Kumar, S.; Elble, R.; Watabe, K.; Mo, Y.Y. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA 2009, 106, 3207–3212. [Google Scholar] [CrossRef]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar] [CrossRef]
- Beaufort, C.M.; Helmijr, J.C.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van IJcken, W.F.J.; Heine, A.A.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef]
- Li, C.; Liu, V.W.; Chan, D.W.; Yao, K.M.; Ngan, H.Y. LY294002 and metformin cooperatively enhance the inhibition of growth and the induction of apoptosis of ovarian cancer cells. Int. J. Gynecol. Cancer 2012, 22, 15–22. [Google Scholar] [CrossRef]
- Dong, R.; Liu, X.; Zhang, Q.; Jiang, Z.; Li, Y.; Wei, Y.; Li, Y.; Yang, Q.; Liu, J.; Wei, J.J.; et al. miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget 2014, 5, 10816–10829. [Google Scholar] [CrossRef]
- Rechsteiner, M.; Zimmermann, A.K.; Wild, P.J.; Caduff, R.; von Teichman, A.; Fink, D.; Moch, H.; Noske, A. TP53 mutations are common in all subtypes of epithelial ovarian cancer and occur concomitantly with KRAS mutations in the mucinous type. Exp. Mol. Pathol. 2013, 95, 235–241. [Google Scholar] [CrossRef]
- Gralewska, P.; Gajek, A.; Marczak, A.; Rogalska, A. Metformin Affects Olaparib Sensitivity through Induction of Apoptosis in Epithelial Ovarian Cancer Cell Lines. Int. J. Mol. Sci. 2021, 22, 557. [Google Scholar] [CrossRef]
- Sun, Y.; Tao, C.; Huang, X.; He, H.; Shi, H.; Zhang, Q.; Wu, H. Metformin induces apoptosis of human hepatocellular carcinoma HepG2 cells by activating an AMPK/p53/miR-23a/FOXA1 pathway. OncoTargets Ther. 2016, 9, 2845–2853. [Google Scholar] [CrossRef]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef]
- Gao, Y.; Li, H.; Ma, X.; Fan, Y.; Ni, D.; Zhang, Y.; Huang, Q.; Liu, K.; Li, X.; Wang, L.; et al. E2F3 upregulation promotes tumor malignancy through the transcriptional activation of HIF-2alpha in clear cell renal cell carcinoma. Oncotarget 2017, 8, 54021–54036. [Google Scholar] [CrossRef]
- Pulito, C.; Mori, F.; Sacconi, A.; Goeman, F.; Ferraiuolo, M.; Pasanisi, P.; Campagnoli, C.; Berrino, F.; Fanciulli, M.; Ford, R.J.; et al. Metformin-induced ablation of microRNA 21-5p releases Sestrin-1 and CAB39L antitumoral activities. Cell Discov. 2017, 3, 17022. [Google Scholar] [CrossRef]
- Blandino, G.; Valerio, M.; Cioce, M.; Mori, F.; Casadei, L.; Pulito, C.; Sacconi, A.; Biagioni, F.; Cortese, G.; Galanti, S.; et al. Metformin elicits anticancer effects through the sequential modulation of DICER and c-MYC. Nat. Commun. 2012, 3, 865. [Google Scholar] [CrossRef]
- Reimer, D.; Hubalek, M.; Kiefel, H.; Riedle, S.; Skvortsov, S.; Erdel, M.; Hofstetter, G.; Concin, N.; Fiegl, H.; Muller-Holzner, E.; et al. Regulation of transcription factor E2F3a and its clinical relevance in ovarian cancer. Oncogene 2011, 30, 4038–4049. [Google Scholar] [CrossRef]
- Garrido, M.P.; Torres, I.; Avila, A.; Chnaiderman, J.; Valenzuela-Valderrama, M.; Aramburo, J.; Orostica, L.; Duran-Jara, E.; Lobos-Gonzalez, L.; Romero, C. NGF/TRKA Decrease miR-145-5p Levels in Epithelial Ovarian Cancer Cells. Int. J. Mol. Sci. 2020, 21, 7657. [Google Scholar] [CrossRef]
- Hua, M.; Qin, Y.; Sheng, M.; Cui, X.; Chen, W.; Zhong, J.; Yan, J.; Chen, Y. miR-145 suppresses ovarian cancer progression via modulation of cell growth and invasion by targeting CCND2 and E2F3. Mol. Med. Rep. 2019, 19, 3575–3583. [Google Scholar] [CrossRef]
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2012, 81, 303–311. [Google Scholar] [CrossRef]
- Yan, L.; Zhou, J.; Gao, Y.; Ghazal, S.; Lu, L.; Bellone, S.; Yang, Y.; Liu, N.; Zhao, X.; Santin, A.D.; et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 2015, 34, 3076–3084. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Chirshev, E.; Hojo, N.; Bertucci, A.; Sanderman, L.; Nguyen, A.; Wang, H.; Suzuki, T.; Brito, E.; Martinez, S.R.; Castanon, C.; et al. Epithelial/mesenchymal heterogeneity of high-grade serous ovarian carcinoma samples correlates with miRNA let-7 levels and predicts tumor growth and metastasis. Mol. Oncol. 2020, 14, 2796–2813. [Google Scholar] [CrossRef]
- Wu, P.; Tang, Y.; Fang, X.; Xie, C.; Zeng, J.; Wang, W.; Zhao, S. Metformin Suppresses Hypopharyngeal Cancer Growth by Epigenetically Silencing Long Non-coding RNA SNHG7 in FaDu Cells. Front. Pharmacol. 2019, 10, 143. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Y.; Wang, B.; Zhai, J. Metformin Affects Paclitaxel Sensitivity of Ovarian Cancer Cells Through Autophagy Mediated by Long Noncoding RNASNHG7/miR-3127-5p Axis. Cancer Biother. Radiopharm. 2022, 37, 792–801. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, B.; Gu, G.; Yang, X.; Qian, Z. Metformin Increases the Chemosensitivity of Pancreatic Cancer Cells to Gemcitabine by Reversing EMT Through Regulation DNA Methylation of miR-663. OncoTargets Ther. 2020, 13, 10417–10429. [Google Scholar] [CrossRef]
- Bao, X.; Zhao, L.; Guan, H.; Li, F. Inhibition of LCMR1 and ATG12 by demethylation-activated miR-570-3p is involved in the anti-metastasis effects of metformin on human osteosarcoma. Cell Death Dis. 2018, 9, 611. [Google Scholar] [CrossRef]
- Li, J.; Lu, J.; Ye, Z.; Han, X.; Zheng, X.; Hou, H.; Chen, W.; Li, X.; Zhao, L. 20(S)-Rg3 blocked epithelial-mesenchymal transition through DNMT3A/miR-145/FSCN1 in ovarian cancer. Oncotarget 2017, 8, 53375–53386. [Google Scholar] [CrossRef]
- Wu, H.; Xiao, Z.; Wang, K.; Liu, W.; Hao, Q. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem. Biophys. Res. Commun. 2013, 441, 693–700. [Google Scholar] [CrossRef]
- Garrido, M.P.; Salvatierra, R.; Valenzuela-Valderrama, M.; Vallejos, C.; Bruneau, N.; Hernandez, A.; Vega, M.; Selman, A.; Quest, A.F.G.; Romero, C. Metformin Reduces NGF-Induced Tumour Promoter Effects in Epithelial Ovarian Cancer Cells. Pharmaceuticals 2020, 13, 315. [Google Scholar] [CrossRef]
- Cuyas, E.; Fernandez-Arroyo, S.; Verdura, S.; Garcia, R.A.; Stursa, J.; Werner, L.; Blanco-Gonzalez, E.; Montes-Bayon, M.; Joven, J.; Viollet, B.; et al. Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism. Oncogene 2018, 37, 963–970. [Google Scholar] [CrossRef]
- Zhong, T.; Men, Y.; Lu, L.; Geng, T.; Zhou, J.; Mitsuhashi, A.; Shozu, M.; Maihle, N.J.; Carmichael, G.G.; Taylor, H.S.; et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene 2017, 36, 2345–2354. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.B.; Kim, D.; Kim, Y.; Han, J.; Shim, Y.M.; Kim, D.H. Metformin regulates expression of DNA methyltransferases through the miR-148/-152 family in non-small lung cancer cells. Clin. Epigenetics 2023, 15, 48. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chen, C.; Ji, X.; Liu, J.; Zhou, Q.; Wang, G.; Yuan, W.; Kan, Q.; Sun, Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Gao, S.; Ma, L.; Sun, Y.; Peng, Z.Y.; Wu, J.; Du, N.; Ren, H.; Tang, S.C.; Sun, X. Stimulation of Let-7 Maturation by Metformin Improved the Response to Tyrosine Kinase Inhibitor Therapy in an m6A Dependent Manner. Front. Oncol. 2021, 11, 731561. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Guan, H.; Lin, X.; Li, N.; Geng, F.; Li, J. METTL3 serves an oncogenic role in human ovarian cancer cells partially via the AKT signaling pathway. Oncol. Lett. 2020, 19, 3197–3204. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, X.; Huang, Y.Z.; Zhu, Y.L.; Xu, L.Y.; Li, Z.; Dai, X.Y.; Shi, L.; Zhou, X.J.; Wei, J.F.; et al. Metformin exhibits antiproliferation activity in breast cancer via miR-483-3p/METTL3/m(6)A/p21 pathway. Oncogenesis 2021, 10, 7. [Google Scholar] [CrossRef]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Jiang, F.Z.; He, Y.Y.; Wang, H.H.; Zhang, H.L.; Zhang, J.; Yan, X.F.; Wang, X.J.; Che, Q.; Ke, J.Q.; Chen, Z.; et al. Mutant p53 induces EZH2 expression and promotes epithelial-mesenchymal transition by disrupting p68-Drosha complex assembly and attenuating miR-26a processing. Oncotarget 2015, 6, 44660–44674. [Google Scholar] [CrossRef]
- Merritt, W.M.; Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Spannuth, W.A.; Schmandt, R.; Urbauer, D.; Pennacchio, L.A.; Cheng, J.F.; Nick, A.M.; et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008, 359, 2641–2650. [Google Scholar] [CrossRef]
- Kuang, Y.; Cai, J.; Li, D.; Han, Q.; Cao, J.; Wang, Z. Repression of Dicer is associated with invasive phenotype and chemoresistance in ovarian cancer. Oncol. Lett. 2013, 5, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Cai, L.; Guo, J.; Chen, N.; Yi, X.; Zhao, Y.; Cai, J.; Wang, Z. Depletion of Dicer promotes epithelial ovarian cancer progression by elevating PDIA3 expression. Tumour Biol. 2016, 37, 14009–14023. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.S.; Ip, C.K.; Mak, A.S.; Wong, A.S. A novel p70 S6 kinase-microRNA biogenesis axis mediates multicellular spheroid formation in ovarian cancer progression. Oncotarget 2016, 7, 38064–38077. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.L.; Zhang, Q.H.; Wang, X.W.; He, H. Antidiabetic drug metformin mitigates ovarian cancer SKOV3 cell growth by triggering G2/M cell cycle arrest and inhibition of m-TOR/PI3K/Akt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1169–1175. [Google Scholar] [PubMed]
- Gwak, H.; Kim, Y.; An, H.; Dhanasekaran, D.N.; Song, Y.S. Metformin induces degradation of cyclin D1 via AMPK/GSK3beta axis in ovarian cancer. Mol. Carcinog. 2017, 56, 349–358. [Google Scholar] [CrossRef]
- Retamales-Ortega, R.; Orostica, L.; Vera, C.; Cuevas, P.; Hernandez, A.; Hurtado, I.; Vega, M.; Romero, C. Role of Nerve Growth Factor (NGF) and miRNAs in Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2017, 18, 507. [Google Scholar] [CrossRef]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Zeinali, T.; Mansoori, B.; Mohammadi, A.; Baradaran, B. Regulatory mechanisms of miR-145 expression and the importance of its function in cancer metastasis. Biomed. Pharmacother. 2019, 109, 195–207. [Google Scholar] [CrossRef]
- Hao, B.; Zhang, J. miRNA-21 inhibition suppresses the human epithelial ovarian cancer by targeting PTEN signal pathway. Saudi J. Biol. Sci. 2019, 26, 2026–2029. [Google Scholar] [CrossRef]
- Liu, H.Y.; Zhang, Y.Y.; Zhu, B.L.; Feng, F.Z.; Yan, H.; Zhang, H.Y.; Zhou, B. miR-21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4149–4155. [Google Scholar] [CrossRef]
- Liu, S.; Fang, Y.; Shen, H.; Xu, W.; Li, H. Berberine sensitizes ovarian cancer cells to cisplatin through miR-21/PDCD4 axis. Acta Biochim. Biophys. Sin. 2013, 45, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Yang, X.; Wang, F.; Cui, Z.; Huang, Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int. J. Mol. Med. 2010, 26, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Huo, W.; Zhao, G.; Yin, J.; Ouyang, X.; Wang, Y.; Yang, C.; Wang, B.; Dong, P.; Wang, Z.; Watari, H.; et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J. Cancer 2017, 8, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Kumar, S. Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: Pivotal role of superoxide dismutase (SOD). Cell. Oncol. 2018, 41, 637–650. [Google Scholar] [CrossRef]
- Nangia-Makker, P.; Yu, Y.; Vasudevan, A.; Farhana, L.; Rajendra, S.G.; Levi, E.; Majumdar, A.P. Metformin: A potential therapeutic agent for recurrent colon cancer. PLoS ONE 2014, 9, e84369. [Google Scholar] [CrossRef]
- Kalogirou, C.; Schafer, D.; Krebs, M.; Kurz, F.; Schneider, A.; Riedmiller, H.; Kneitz, B.; Vergho, D. Metformin-Derived Growth Inhibition in Renal Cell Carcinoma Depends on miR-21-Mediated PTEN Expression. Urol. Int. 2016, 96, 106–115. [Google Scholar] [CrossRef]
- Tossetta, G. Metformin Improves Ovarian Cancer Sensitivity to Paclitaxel and Platinum-Based Drugs: A Review of In Vitro Findings. Int. J. Mol. Sci. 2022, 23, 2893. [Google Scholar] [CrossRef]
- Luo, M.; Tan, X.; Mu, L.; Luo, Y.; Li, R.; Deng, X.; Chen, N.; Ren, M.; Li, Y.; Wang, L.; et al. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci. Rep. 2017, 7, 43427. [Google Scholar] [CrossRef]
- Echevarria-Vargas, I.M.; Valiyeva, F.; Vivas-Mejia, P.E. Upregulation of miR-21 in cisplatin resistant ovarian cancer via JNK-1/c-Jun pathway. PLoS ONE 2014, 9, e97094. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, X.; Liu, J.; Sun, B.; Tang, H.; Zhang, H. miR-27a-mediated antiproliferative effects of metformin on the breast cancer cell line MCF-7. Oncol. Rep. 2016, 36, 3691–3699. [Google Scholar] [CrossRef]
- Nair, V.; Pathi, S.; Jutooru, I.; Sreevalsan, S.; Basha, R.; Abdelrahim, M.; Samudio, I.; Safe, S. Metformin inhibits pancreatic cancer cell and tumor growth and downregulates Sp transcription factors. Carcinogenesis 2013, 34, 2870–2879. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Sun, X.; Jiang, X. UCA1 involved in the metformin-regulated bladder cancer cell proliferation and glycolysis. Tumour Biol. 2017, 39, 1010428317710823. [Google Scholar] [CrossRef] [PubMed]
- Golshan, M.; Khaleghi, S.; Shafiee, S.M.; Valaee, S.; Ghanei, Z.; Jamshidizad, A.; Dashtizad, M.; Shamsara, M. Metformin modulates oncogenic expression of HOTAIR gene via promoter methylation and reverses epithelial-mesenchymal transition in MDA-MB-231 cells. J. Cell. Biochem. 2021, 122, 385–393. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, Z.; Zhang, J.; Hao, Z.; He, Y.; Wu, Z.; Song, Y.; Yuan, K.; Zheng, S.; Zhao, Q.; et al. lncRNA MALAT1 participates in metformin inhibiting the proliferation of breast cancer cell. J. Cell. Mol. Med. 2021, 25, 7135–7145. [Google Scholar] [CrossRef]
- Chen, J.; Qin, C.; Zhou, Y.; Chen, Y.; Mao, M.; Yang, J. Metformin may induce ferroptosis by inhibiting autophagy via lncRNA H19 in breast cancer. FEBS Open Bio 2022, 12, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qian, T.; Li, S.; Xie, Y.; Tao, M. Metformin reverses tamoxifen resistance through the lncRNA GAS5-medicated mTOR pathway in breast cancer. Ann. Transl. Med. 2022, 10, 366. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Y.; Chen, L.; Li, R.; Ning, Y.; Zhu, X. Role of metformin in functional endometrial hyperplasia and polycystic ovary syndrome involves the regulation of MEG3/miR-223/GLUT4 and SNHG20/miR-4486/GLUT4 signaling. Mol. Med. Rep. 2022, 26, 218. [Google Scholar] [CrossRef]
- Guo, J.; Li, Y.; Duan, H.; Yuan, L. Metformin Suppresses the Proliferation and Promotes the Apoptosis of Colon Cancer Cells Through Inhibiting the Expression of Long Noncoding RNA-UCA1. OncoTargets Ther. 2020, 13, 4169–4181. [Google Scholar] [CrossRef]
- Aminimoghaddam, S.; Fooladi, B.; Noori, M.; Nickhah Klashami, Z.; Kakavand Hamidi, A.; Amoli, M.M. The Effect of Metformin on Expression of Long Non-coding RNA H19 in Endometrial Cancer. Med. J. Islam Repub. Iran 2021, 35, 155. [Google Scholar] [CrossRef]
- Li, P.; Tong, L.; Song, Y.; Sun, J.; Shi, J.; Wu, Z.; Diao, Y.; Li, Y.; Wang, Z. Long noncoding RNA H19 participates in metformin-mediated inhibition of gastric cancer cell invasion. J. Cell. Physiol. 2019, 234, 4515–4527. [Google Scholar] [CrossRef]
- Tseng, H.H.; Chen, Y.Z.; Chou, N.H.; Chen, Y.C.; Wu, C.C.; Liu, L.F.; Yang, Y.F.; Yeh, C.Y.; Kung, M.L.; Tu, Y.T.; et al. Metformin inhibits gastric cancer cell proliferation by regulation of a novel Loc100506691-CHAC1 axis. Mol. Ther. Oncolytics 2021, 22, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Gandhy, S.U.; Imanirad, P.; Jin, U.H.; Nair, V.; Hedrick, E.; Cheng, Y.; Corton, J.C.; Kim, K.; Safe, S. Specificity protein (Sp) transcription factors and metformin regulate expression of the long non-coding RNA HULC. Oncotarget 2015, 6, 26359–26372. [Google Scholar] [CrossRef] [PubMed]
- Sabry, D.; Abdelaleem, O.O.; El Amin Ali, A.M.; Mohammed, R.A.; Abdel-Hameed, N.D.; Hassouna, A.; Khalifa, W.A. Anti-proliferative and anti-apoptotic potential effects of epigallocatechin-3-gallate and/or metformin on hepatocellular carcinoma cells: In vitro study. Mol. Biol. Rep. 2019, 46, 2039–2047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Luo, M.; Zhang, J.; Guo, B.; Singh, S.; Lin, X.; Xiong, H.; Ju, S.; Wang, L.; Zhou, Y.; et al. The role of lncRNA H19 in tumorigenesis and drug resistance of human Cancers. Front. Genet 2022, 13, 1005522. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, L.; Yan, W.; Qiu, L.; Zhang, J.; Jia, X. lncRNA GAS5 Sensitizes Breast Cancer Cells to Ionizing Radiation by Inhibiting DNA Repair. BioMed Res. Int. 2022, 2022, 1987519. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, W.J. Long non-coding RNA-H19 promotes ovarian cancer cell proliferation and migration via the microRNA-140/Wnt1 axis. Kaohsiung J. Med. Sci. 2021, 37, 768–775. [Google Scholar] [CrossRef]
- Mizrahi, A.; Czerniak, A.; Levy, T.; Amiur, S.; Gallula, J.; Matouk, I.; Abu-lail, R.; Sorin, V.; Birman, T.; de Groot, N.; et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 2009, 7, 69. [Google Scholar] [CrossRef]
- Ma, H.; Gao, L.; Yu, H.; Song, X. Long non-coding RNA H19 correlates with unfavorable prognosis and promotes cell migration and invasion in ovarian cancer. Ginekol. Pol. 2022, 93, 68293. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, W.; Zheng, A.; Zhang, Y.; Fang, C.; Zhang, P. Ginsenoside Rg3 suppresses ovarian cancer cell proliferation and invasion by inhibiting the expression of lncRNA H19. Acta Biochim. Pol. 2021, 68, 575–582. [Google Scholar] [CrossRef]
- Zhu, Z.; Song, L.; He, J.; Sun, Y.; Liu, X.; Zou, X. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10082–10091. [Google Scholar]
- Xu, J.; Zheng, L.H.; Hong, Y.N.; Xuan, C.; Yan, S.L.; Lv, G.L.; Jiang, Z.G.; Ding, X.F. Long Non-coding RNA UCA1 Regulates SRPK1 Expression Through miR- 99b-3p in Ovarian Cancer. Protein Pept. Lett. 2022, 29, 829–838. [Google Scholar] [CrossRef]
- Zheng, X.; Zhou, Y.; Chen, W.; Chen, L.; Lu, J.; He, F.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells. Cell Physiol. Biochem. 2018, 51, 1340–1353. [Google Scholar] [CrossRef]
- Tian, X.; Zuo, X.; Hou, M.; Li, C.; Teng, Y. LncRNA-H19 regulates chemoresistance to carboplatin in epithelial ovarian cancer through microRNA-29b-3p and STAT3. J. Cancer 2021, 12, 5712–5722. [Google Scholar] [CrossRef]
- Hashemi, M.; Moosavi, M.S.; Abed, H.M.; Dehghani, M.; Aalipour, M.; Heydari, E.A.; Behroozaghdam, M.; Entezari, M.; Salimimoghadam, S.; Gunduz, E.S.; et al. Long non-coding RNA (lncRNA) H19 in human cancer: From proliferation and metastasis to therapy. Pharmacol. Res. 2022, 184, 106418. [Google Scholar] [CrossRef]
- Zheng, Z.G.; Xu, H.; Suo, S.S.; Xu, X.L.; Ni, M.W.; Gu, L.H.; Chen, W.; Wang, L.Y.; Zhao, Y.; Tian, B.; et al. The Essential Role of H19 Contributing to Cisplatin Resistance by Regulating Glutathione Metabolism in High-Grade Serous Ovarian Cancer. Sci. Rep. 2016, 6, 26093. [Google Scholar] [CrossRef] [PubMed]
- Medrzycki, M.; Zhang, Y.; Zhang, W.; Cao, K.; Pan, C.; Lailler, N.; McDonald, J.F.; Bouhassira, E.E.; Fan, Y. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014, 74, 6463–6473. [Google Scholar] [CrossRef] [PubMed]
- Malakoti, F.; Alemi, F.; Yeganeh, S.J.; Hosseini, F.; Shabestani, N.; Samemaleki, S.; Maleki, M.; Daneshvar, S.F.; Montazer, M.; Yousefi, B. Long noncoding RNA SNHG7-miRNA-mRNA axes crosstalk with oncogenic signaling pathways in human cancers. Chem. Biol. Drug Des. 2023, 101, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Wu, Y.; Bai, S.; Yan, Y.; Kang, H.; Ma, W.; Zhang, J.; Gao, Y.; Hui, B.; Ma, H.; et al. Long non-coding RNA SNGH7 Is activated by SP1 and exerts oncogenic properties by interacting with EZH2 in ovarian cancer. J. Cell. Mol. Med. 2020, 24, 7479–7489. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, R.; Ye, Y. Long non-coding RNA (LncRNA) SNHG7/Eukaryotic translation initiation factor 4 gamma 2 (EIF4G2) involves in the malignant events of ovarian cancer cells with paclitaxel resistant. Bioengineered 2021, 12, 10541–10552. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Y.; He, J.; Sun, H.; Jin, Z. Long non-coding RNA H19 mediates ovarian cancer cell cisplatin-resistance and migration during EMT. Int. J. Clin. Exp. Pathol. 2019, 12, 2506–2515. [Google Scholar]
- Goodwin, P.J.; Chen, B.E.; Gelmon, K.A.; Whelan, T.J.; Ennis, M.; Lemieux, J.; Ligibel, J.A.; Hershman, D.L.; Mayer, I.A.; Hobday, T.J.; et al. Effect of Metformin vs. Placebo on Invasive Disease-Free Survival in Patients with Breast Cancer: The MA.32 Randomized Clinical Trial. JAMA 2022, 327, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Kordes, S.; Pollak, M.N.; Zwinderman, A.H.; Mathot, R.A.; Weterman, M.J.; Beeker, A.; Punt, C.J.; Richel, D.J.; Wilmink, J.W. Metformin in patients with advanced pancreatic cancer: A double-blind, randomised, placebo-controlled phase 2 trial. Lancet Oncol. 2015, 16, 839–847. [Google Scholar] [CrossRef]
- Lord, S.R.; Cheng, W.C.; Liu, D.; Gaude, E.; Haider, S.; Metcalf, T.; Patel, N.; Teoh, E.J.; Gleeson, F.; Bradley, K.; et al. Integrated Pharmacodynamic Analysis Identifies Two Metabolic Adaption Pathways to Metformin in Breast Cancer. Cell Metab. 2018, 28, 679–688.e674. [Google Scholar] [CrossRef] [PubMed]
- Higurashi, T.; Hosono, K.; Takahashi, H.; Komiya, Y.; Umezawa, S.; Sakai, E.; Uchiyama, T.; Taniguchi, L.; Hata, Y.; Uchiyama, S.; et al. Metformin for chemoprevention of metachronous colorectal adenoma or polyps in post-polypectomy patients without diabetes: A multicentre double-blind, placebo-controlled, randomised phase 3 trial. Lancet Oncol. 2016, 17, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Gutkind, J.S.; Molinolo, A.A.; Wu, X.; Wang, Z.; Nachmanson, D.; Harismendy, O.; Alexandrov, L.B.; Wuertz, B.R.; Ondrey, F.G.; Laronde, D.; et al. Inhibition of mTOR signaling and clinical activity of metformin in oral premalignant lesions. JCI Insight 2021, 6, e147096. [Google Scholar] [CrossRef] [PubMed]
- Laskov, I.; Drudi, L.; Beauchamp, M.C.; Yasmeen, A.; Ferenczy, A.; Pollak, M.; Gotlieb, W.H. Anti-diabetic doses of metformin decrease proliferation markers in tumors of patients with endometrial cancer. Gynecol. Oncol. 2014, 134, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Hadad, S.; Iwamoto, T.; Jordan, L.; Purdie, C.; Bray, S.; Baker, L.; Jellema, G.; Deharo, S.; Hardie, D.G.; Pusztai, L.; et al. Evidence for biological effects of metformin in operable breast cancer: A pre-operative, window-of-opportunity, randomized trial. Breast Cancer Res. Treat. 2011, 128, 783–794. [Google Scholar] [CrossRef]
- Joshua, A.M.; Zannella, V.E.; Downes, M.R.; Bowes, B.; Hersey, K.; Koritzinsky, M.; Schwab, M.; Hofmann, U.; Evans, A.; van der Kwast, T.; et al. A pilot ‘window of opportunity’ neoadjuvant study of metformin in localised prostate cancer. Prostate Cancer Prostatic Dis. 2014, 17, 252–258. [Google Scholar] [CrossRef]
- Mitsuhashi, A.; Kiyokawa, T.; Sato, Y.; Shozu, M. Effects of metformin on endometrial cancer cell growth in vivo: A preoperative prospective trial. Cancer 2014, 120, 2986–2995. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Evaluation of Metformin Effect on the Fertility of Women Treated With 131I for Thyroid Cancer (METHYR). Available online: https://classic.clinicaltrials.gov/ct2/show/NCT05468554 (accessed on 23 August 2023).
- ClinicalTrials.gov. Cancer Chemoprevention by Metformin Hydrochloride in Oral Potentially Malignant Lesions. Available online: https://clinicaltrials.gov/study/NCT03685409 (accessed on 23 August 2023).
- ClinicalTrials.gov. Cancer Chemoprevention by Metformin Hydrochloride Compared to Placebo in Oral Potentially Malignant Lesions. Available online: https://clinicaltrials.gov/study/NCT03684707?id=%22NCT03684707%22&rank=1 (accessed on 23 August 2023).
- ClinicalTrials.gov. Longitudinal Follow-Up in Women with Endometrial Hyperplasia without Atypia. Available online: https://clinicaltrials.gov/study/NCT05292573?tab=results (accessed on 23 August 2023).

| Cancer | Experimental Model | Metformin Dose | lncRNA | Regulation | Target | Biological Effects | Ref. |
|---|---|---|---|---|---|---|---|
| Bladder | 5637 cells | 10 and 20 mM | UCA1 | Down | n/a | Inhibits cell proliferation and glycolysis | [113] |
| Breast | MCF-7 cells | 2 mM | H19 | Down | SAHH | Reduces cell viability | [82] |
| MDA-MB-231 cells | 10 and 20 mM | HOTAIR | Down | n/a | Decreases cell viability, migration, and invasion, and suppresses EMT | [114] | |
| MCF-7 cells | 1, 5, and 20 mM | MALAT1 | Up | n/a | Inhibits cell proliferation and migration, induces apoptosis, and induces autophagy and ER stress markers | [115] | |
| 1 and 20 mM | HOTAIR | ||||||
| 20 mM | DICER1-AS1 | ||||||
| 10 and 20 mM | LINCO01121 | ||||||
| 20 mM | TUG1 | ||||||
| MCF-7 cells | 2, 5, and 10 mM | H19 | Down | n/a | Induces ferroptosis by inhibiting autophagy | [116] | |
| MCF-7 cells resistant to tamoxifen | 5 mM | GAS5 | Up | n/a | Increases sensitivity to tamoxifen by inhibiting cell growth and inducing apoptosis | [117] | |
| Cervical | HCC-94 cells | 100 uM | H19 | Down | n/a | Inhibits cell viability | [118] |
| FTX | |||||||
| Colorectal | SW480 and SW620 cells | 20, 40, and 80 mM | UCA1 | Down | n/a | Suppresses cell proliferation and promotes apoptosis | [119] |
| Endometrial | ARK2 cells and endometrial cancer patients | 2 mM in cells, 750 mg/d up to 2250 mg/d in patients | H19 | Down | SAHH | Alters DNA methylation genome widely. Reduces cell viability and tumor cell proliferation | [82] |
| Endometrial cancer patients | 750 mg/d up to 2250 mg/d | H19 | Down | n/a | Reduces H19 expression | [120] | |
| Gastric | AGS cells | 20 mM | H19 | Down | n/a | Inhibits cell proliferation, invasion, and migration, and suppresses metastasis | [121] |
| HR, AZ-521, NCI-N87, and TSGH cells | 10 mM | H19 | Down | n/a | Inhibits cell proliferation and invasion | [122] | |
| 1, 5, and 10 mM | RBMS3-AS3 | n/a | |||||
| Hypopharyngeal | FaDu cells and xenograft mouse model | 2,4,6,8, and 10 mM in cells and 8 mM in mice | SNHG7 | Down | SAHH | Inhibits cell proliferation and tumor growth. Sensitizes to taxol and radiotherapy | [74] |
| Liver cancer | HepG2, SNU-449, and SK-Hep-1 cells | 10 and 20 mM | HULC | Down | n/a | Decreases cell growth | [123] |
| HepG2 cells | 7.57 μg/ml | AF085935 | Down | n/a | Inhibits cell proliferation | [124] |
| Pathology | Findings in the Metformin-Treated Group | Ref. |
|---|---|---|
| Adenoma and polyp recurrence in patients with a high risk of adenoma recurrence | Reduced the prevalence and number of metachronous adenomas or polyps after polypectomy | [145] |
| Oral pre-malignant lesions | Decrease in cell proliferation in the squamous epithelium and inhibition of mTOR signaling | [146] |
| Endometrial cancer | Decrease in Ki-67 and pS6 staining. Decrease in plasma IGF-1 and IGFBP-7 | [147] |
| Newly diagnosed women with breast cancer | Reduced expression of p53, BRCA1, and cell cycle pathways following metformin treatment | [148] |
| Localized prostatic cancer | Metformin reduced the Ki-67 proliferation index and trended toward prostatic-specific antigen reduction | [149] |
| Endometrial cancer | Decreased phosphorylated extracellular signal-regulated kinase (ERK1/2), cyclin D1, Ki-67, and topoisomerase IIα. Increased p27 | [150] |
| Study | Patient | Treatment | Aim of Investigation | miRNA |
|---|---|---|---|---|
| NCT05468554 Thyroid cancer (Phase 3) [151] | Women of reproductive age diagnosed with thyroid carcinoma | Metformin 500 mg, 3 times a day | Evaluate the metformin effect on the fertility of women treated with 131I for thyroid cancer | Difference in the expression of selected miRNAs (uninformed) |
| NCT03685409 Oral cancer (Phase 3) [152] | Both genders (20–70 years), clinically diagnosed and histologically confirmed as having potential oral malignant lesions | Metformin 500 mg, twice a day | Effect of systemic metformin hydrochloride on the millimeter change in the largest diameter of the potential oral malignant lesion | Numerical differences between miR-21 and miR-200 in tissue biopsies versus saliva at baseline and at 3 months |
| NCT03684707 Oral cancer (Phase 4) [153] | Both genders (20–70 years), clinically diagnosed and histologically confirmed as having potential oral malignant lesions | Metformin 500 mg, daily | Evaluate lesion size in millimeters | Measured miRNA31 and 210 in saliva and tissue biopsy |
| NCT05292573 Endometrial cancer (Phase 3) [154] | Women aged ≥20 years with histological diagnosis of simple hyperplasia/complex hyperplasia (SH/CH) | Metformin 500 mg, twice a day | Longitudinal follow-up in women with endometrial hyperplasia without atypia | The area under the receiver operating characteristic curve (ROC curve) (AUC) of the miRNA panel |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alfaro, I.; Vega, M.; Romero, C.; Garrido, M.P. Mechanisms of Regulation of the Expression of miRNAs and lncRNAs by Metformin in Ovarian Cancer. Pharmaceuticals 2023, 16, 1515. https://doi.org/10.3390/ph16111515
Alfaro I, Vega M, Romero C, Garrido MP. Mechanisms of Regulation of the Expression of miRNAs and lncRNAs by Metformin in Ovarian Cancer. Pharmaceuticals. 2023; 16(11):1515. https://doi.org/10.3390/ph16111515
Chicago/Turabian StyleAlfaro, Ignacio, Margarita Vega, Carmen Romero, and Maritza P. Garrido. 2023. "Mechanisms of Regulation of the Expression of miRNAs and lncRNAs by Metformin in Ovarian Cancer" Pharmaceuticals 16, no. 11: 1515. https://doi.org/10.3390/ph16111515
APA StyleAlfaro, I., Vega, M., Romero, C., & Garrido, M. P. (2023). Mechanisms of Regulation of the Expression of miRNAs and lncRNAs by Metformin in Ovarian Cancer. Pharmaceuticals, 16(11), 1515. https://doi.org/10.3390/ph16111515







