Biologically Potent Benzimidazole-Based-Substituted Benzaldehyde Derivatives as Potent Inhibitors for Alzheimer’s Disease along with Molecular Docking Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Chemistry
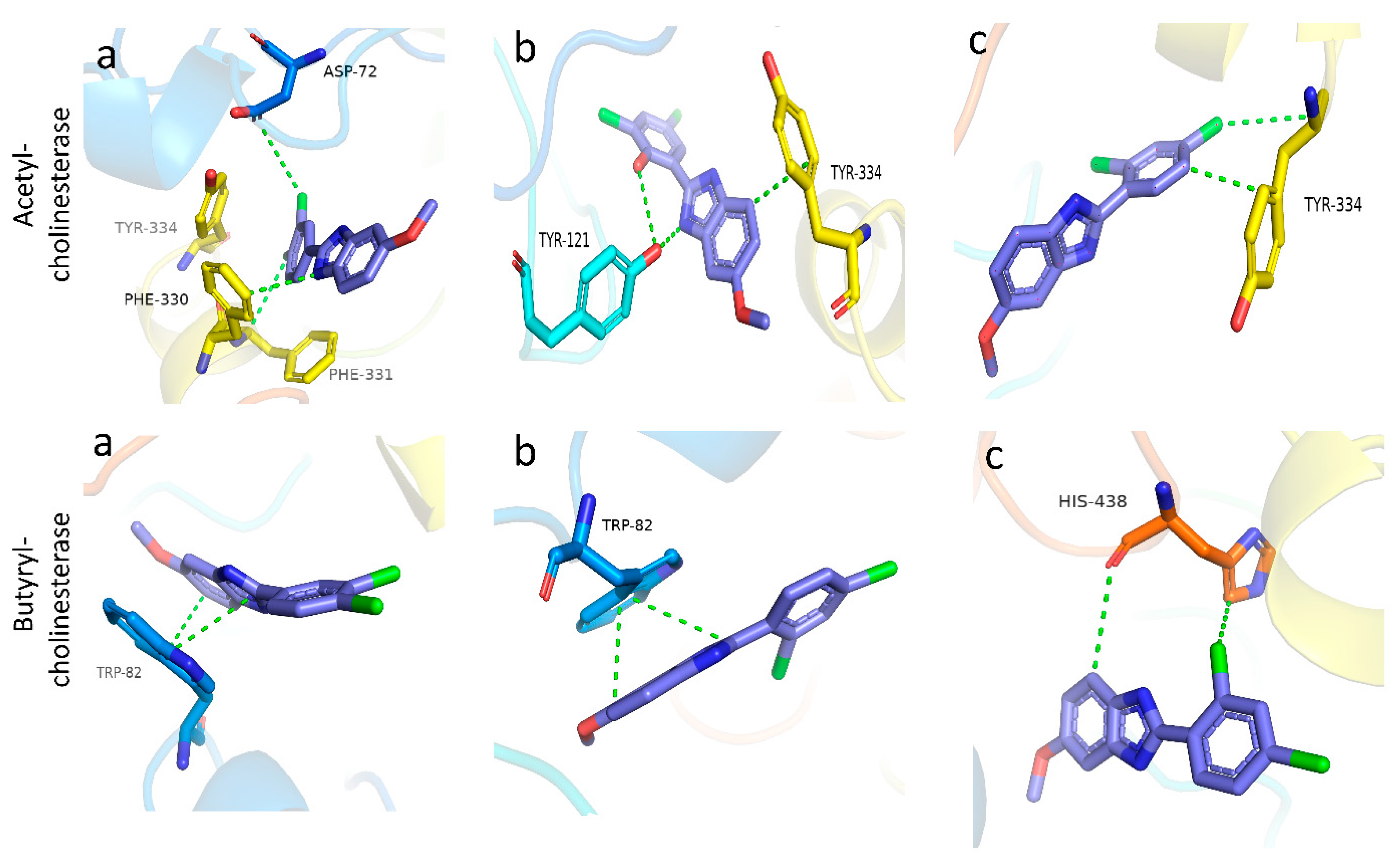
2.2. Molecular Docking Studies
2.3. Post Simulation Analysis
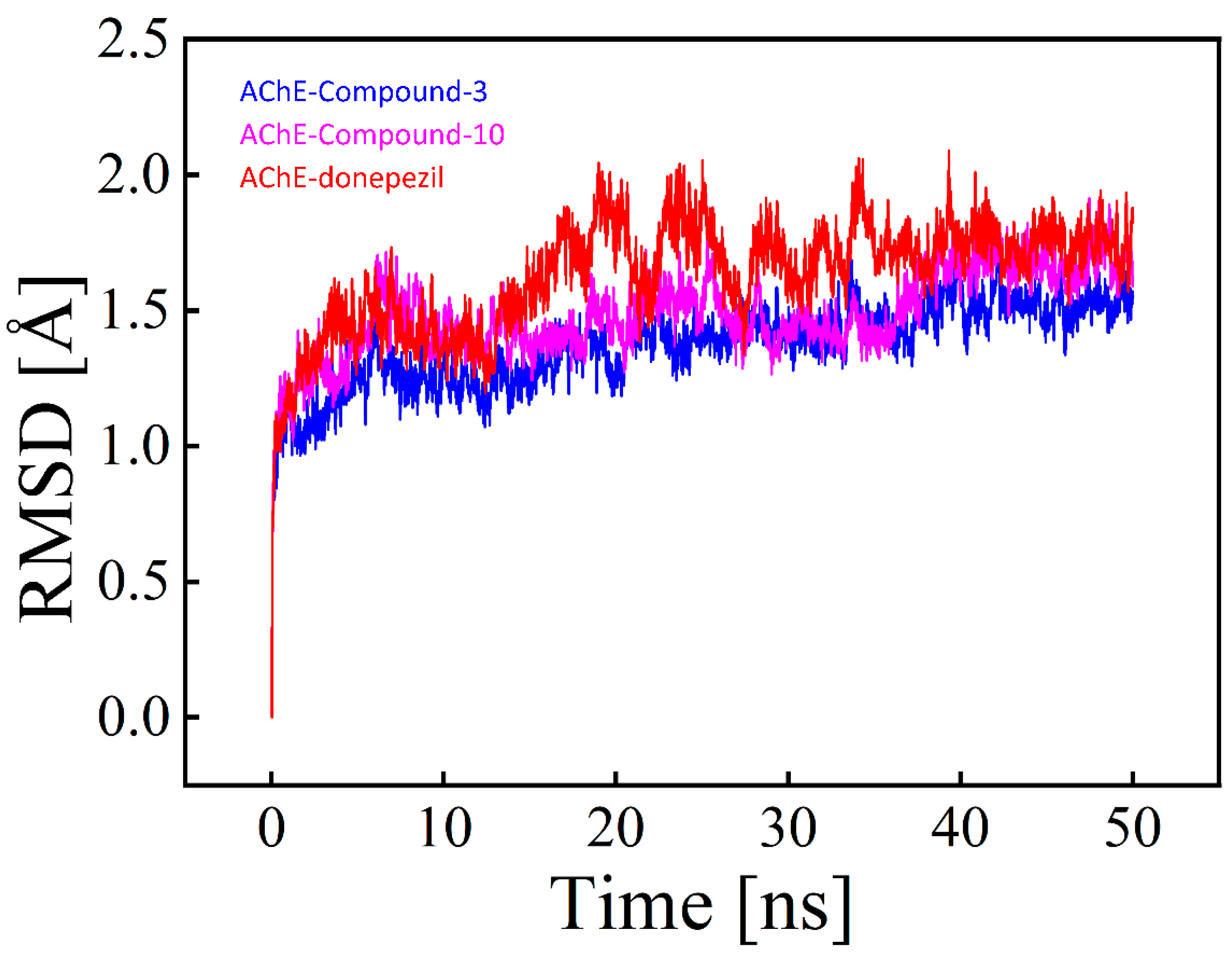
2.3.1. RMSD (Root Mean Square Deviation) Analysis
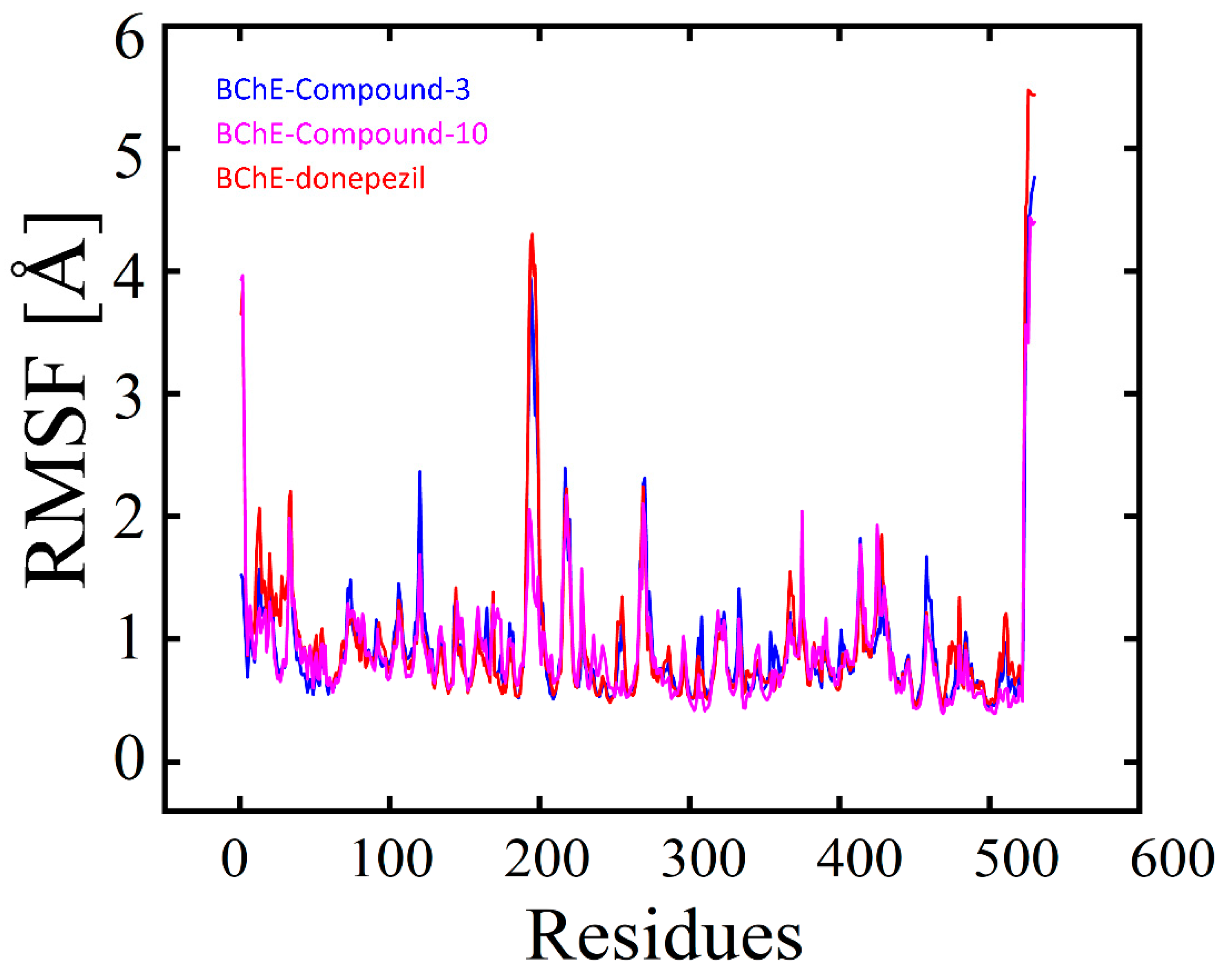
2.3.2. RMSF (Root Mean Square Fluctuation) Analysis
2.4. Binding Energy Calculation
2.5. Pharmacokinetics (ADMET) Properties of Finally Selected Compounds
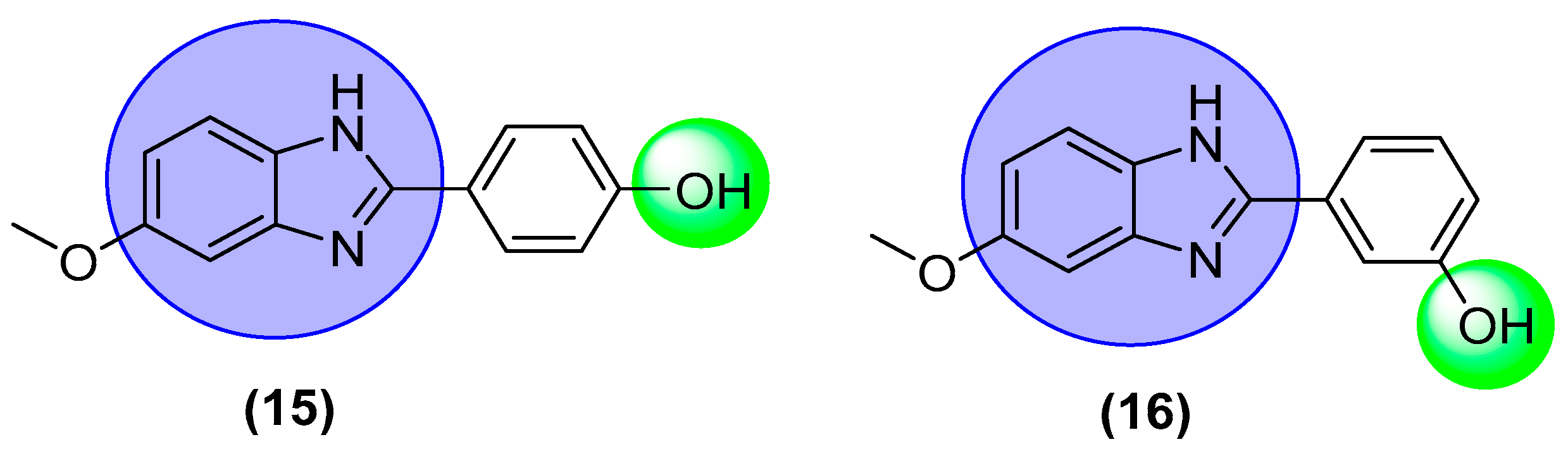
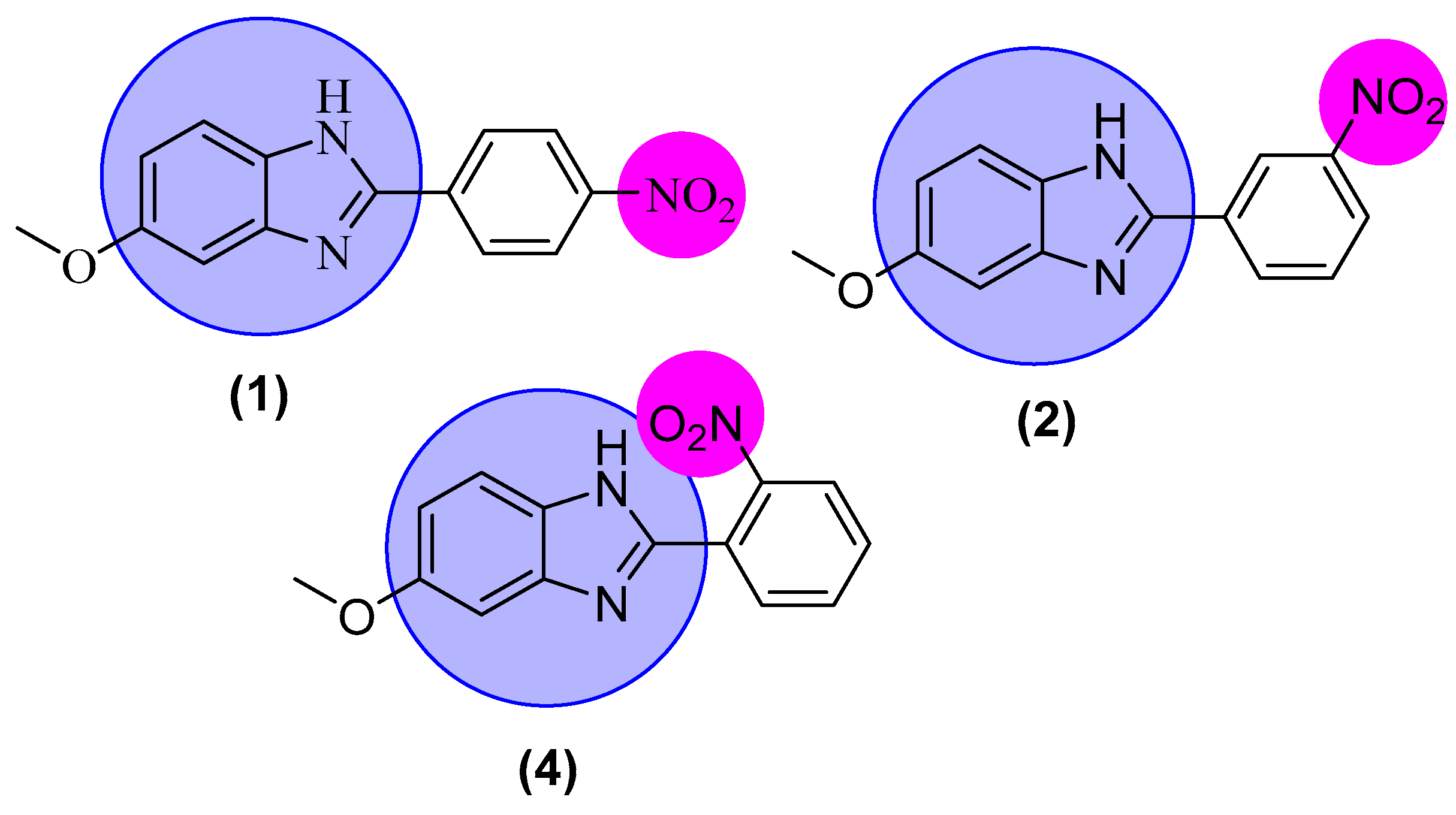
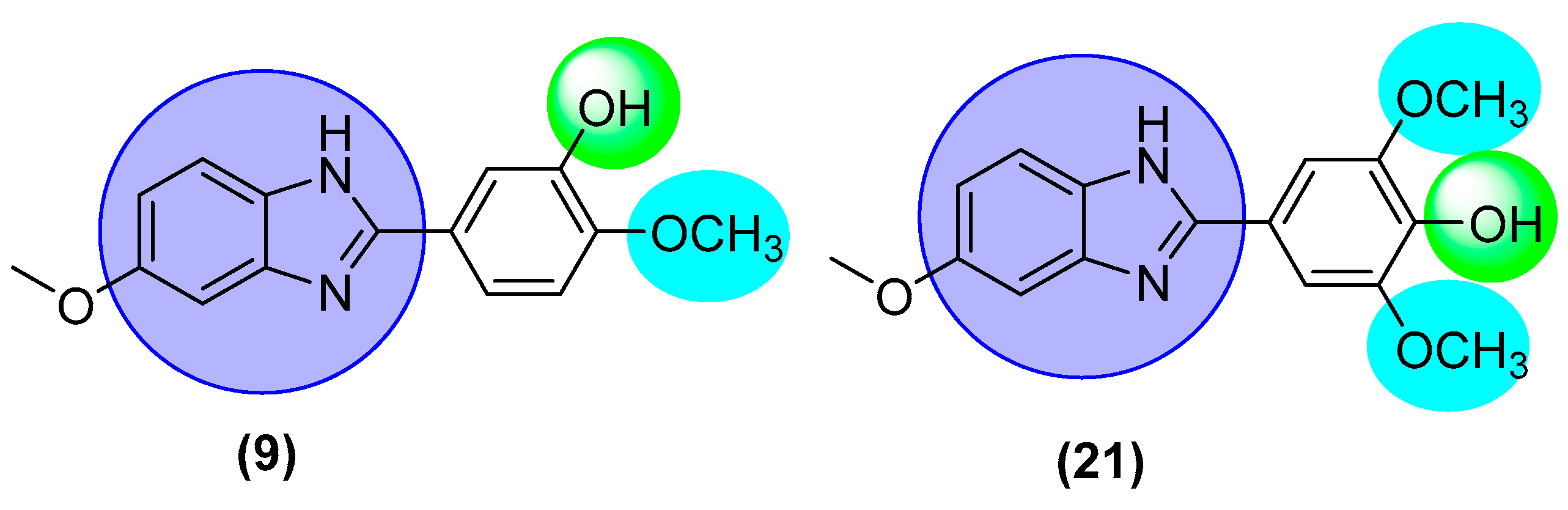
2.6. Structure-Activity Relationship (SAR)
3. Materials and Methods
3.1. Procedure for the Synthesis of Benzimidazole Analogs (1–21)
3.1.1. 5-Methoxy-2-(4-Nitrophenyl)-1H-Benzo[d]-Imidazole (1)
3.1.2. 5-Methoxy-2-(3-Nitrophenyl)-1H-Benzo[d]-Imidazole (2)
3.2. Inhibition Assay Protocol of Acetylcholinesterase and Butyrylcholinesterase
3.3. Molecular Docking
3.4. Molecular Dynamics simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adalat, B.; Rahim, F.; Taha, M.; Alshamrani, F.J.; Anouar, E.H.; Uddin, N.; Zakaria, Z.A. Synthesis of Benzimidazole–Based Analogs as Anti Alzheimer’s Disease Compounds and Their Molecular Docking Studies. Molecules 2020, 25, 4828. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, X.; Geng, M. Alzheimer’s disease hypothesis and related theraies. Transl. Neurodegener. 2018, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Bartolini, M.; Bertucci, C.; Cavrini, V.; Andrisano, V. β-Amyloid aggregation induced by human acetylcholinesterase: Inhibition studies. Biochem. Pharmacol. 2003, 65, 407–416. [Google Scholar] [CrossRef]
- Cheong, J.E.; Zaffagni, M.; Chung, I.; Xu, Y.; Wang, Y.; Jernigan, F.E.; Zetter, B.R.; Sun, L. Synthesis and anticancer activity of novel water soluble benzimidazole carbamates. Eur. J. Med. Chem. 2018, 144, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef]
- Da Costa Leite, L.F.C.; Mourão, R.H.V.; de Lima, M.D.C.A.; Galdino, S.L.; Hernandes, M.Z.; Neves, F.D.A.R.; da Rocha, P.I. Synthesis, biological evaluation and molecular modeling studies of arylidene-thiazolidinediones with potential hypoglycemic and hypolipidemic activities. Eur. J. Med. Chem. 2007, 42, 1263–1271. [Google Scholar] [CrossRef]
- Fang, Y.; Zhou, H.; Gu, Q.; Xu, J. Synthesis and evaluation of tetrahydroisoquinoline-benzimidazole hybrids as multifunctional agents for the treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2019, 167, 133–145. [Google Scholar] [CrossRef]
- Ganie, A.M.; Dar, A.M.; Khan, F.A.; Dar, B.A. Benzimidazole derivatives as potential antimicrobial and antiulcer agents: A mini review. Mini-Rev. Med. Chem. 2019, 19, 1292–1297. [Google Scholar] [CrossRef]
- Gurjar, A.S.; Solanki, V.S.; Meshram, A.R.; Vishwakarma, S.S. Exploring beta amyloid cleavage enzyme-1 inhibition and neuroprotective role of benzimidazole analogues as anti-alzheimer agents. J. Chin. Chem. Soc. 2020, 67, 864–873. [Google Scholar] [CrossRef]
- Karaburun, A.Ç.; Kaya Çavuşoğlu, B.; Acar, Ç.U.; Osmaniye, D.; Sağlık, B.N.; Levent, S.; Kaplancıklı, Z.A. Synthesis and antifungal potential of some novel benzimidazole-1, 3, 4-oxadiazole compounds. Molecules 2019, 24, 191. [Google Scholar] [CrossRef]
- Lane, R.M.; Potkin, S.G.; Enz, A. Targeting acetylcholinesterase and butyrylcholinesterase in dementia. Int. J. Neuropsychopharmacol. 2006, 9, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Marucci, G.; Buccioni, M.; Dal, B.D.; Lambertucci, C.; Volpini, R.; Amenta, F. Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. Neuropharmacology 2021, 190, 108352. [Google Scholar] [CrossRef] [PubMed]
- McGleenon, B.M.; Dynan, K.B.; Passmore, A.P. Acetylcholinesterase inhibitors in Alzheimer’s disease. Br. J. Clin. Pharmacol. 1999, 48, 471. [Google Scholar] [CrossRef] [PubMed]
- Debus, H. U ber die einwirkung des ammoniaks auf glyoxal. Justus Liebigs. Ann. Chem. 1858, 107, 199–208. [Google Scholar] [CrossRef]
- Grimmett, M.R. Imidazole and Benzimidazole Synthesis; Academic Press: Cambridge, MA, USA, 1997. [Google Scholar]
- Maghraby, M.T.E.; Abou-Ghadir, O.M.; Abdel-Moty, S.G.; Ali, A.Y.; Salem, O.I. Novel class of benzimidazole-thiazole hybrids: The privileged scaffolds of potent anti-inflammatory activity with dual inhibition of cyclooxygenase and 15-lipoxygenase enzymes. Bioorg. Med. Chem. 2020, 28, 115403. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.R.; Ghanavatkar, C.W.; Mali, S.N.; Qureshi, S.I.; Chaudhari, H.K.; Sekar, N. Design, synthesis, antimicrobial activity, and computational studies of novel azo linked substituted benzimidazole, benzoxazole and benzothiazole derivatives. Comput. Biol. Chem. 2019, 78, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Sudha, V.; Chaturvedi, S.C. Docking Studies on Some Benzimidazole and Triazole Analogues as Antihypertensive Agents. J. Bioinform. 2018, 5, 37–39. [Google Scholar]
- Mohanty, S.K.; Khuntia, A.; Yellasubbaiah, N.; Ayyanna, C.; Sudha, B.N.; Harika, M.S. Design, synthesis of novel azo derivatives of benzimidazole as potent antibacterial and anti-tubercular agents. Beni-Suef Univ. J. Basic Appl. Sci. 2018, 7, 646–651. [Google Scholar] [CrossRef]
- AlMalki, W.H.; Shahid, I.; Abdalla, A.N.; Johargy, A.K.; Ahmed, M.; Hassan, S. Virological surveillance, molecular phylogeny, and evolutionary dynamics of hepatitis C virus subtypes 1a and 4a isolates in patients from Saudi Arabia. Saudi J. Biol. Sci. 2021, 28, 1664–1677. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Kumar, V.; Mal, M.; Houghton, P.J. Acetylcholinesterase inhibitors from plants. Phytomedicine 2007, 14, 289–300. [Google Scholar] [CrossRef]
- Nordberg, A.; Ballard, C.; Bullock, R.; Darreh-Shori, T.; Somogyi, M. A Review of Butyrylycholinesterase as a Therapeutic Target in the Treatment of Alzheimer’s Disease. Prim. Care Companion CNS Disord. 2013, 15, 26731. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, S.; Janelidze, S.; Quiroz, Y.T.; Zetterberg, H.; Lopera, F.; Stomrud, E.; Hansson, O. Discriminative accuracy of plasma phospho-tau217 for Alzheimer disease vs other neurodegenerative disorders. JAMA 2020, 324, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Hussein, E.M.; ElGuesmi, N.; Maji, T.K.; Jassas, R.S.; Alsimaree, A.A.; Altass, H.M.; Moussa, Z.; Pal, S.K.; Ahmed, S.A. Synthesis and photophysical properties of benzimidazoles grafted pyrazole-containing pyrene or fluorene moiety: A combined spectroscopic and computational study. J. Photochem. Photobiol. A Chem. 2021, 419, 113465. [Google Scholar] [CrossRef]
- Rahim, F.; Javed, M.T.; Ullah, H.; Wadood, A.; Taha, M.; Ashraf, M.; Khan, M.A.; Khan, F.; Mirza, S.; Khan, K.M. molecular docking, acetylcholinesterase and butyrylcholinesterase inhibitory potential of thiazole analogs as new inhibitors for Alzheimer disease. Bioorg. Chem. 2015, 62, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Sen, J.; Srivastava, S. Synthesis and antimicrobial activity of some new 2-[2”, 3”-dinitrophenyl]-3-(substituted aryl)-3, 3a-dihydrobenzimidazo [2, 1-a] pyrazolo [3, 4-d] thiazoles derivatives. Int. J. Theoret. Appl. Sci. 2009, 1, 41–47. [Google Scholar]
- Shojaei, P.; Mokhtari, B.; Ghorbanpoor, M. Synthesis, in vitro antifungal evaluation and docking studies of novel derivatives of imidazoles and benzimidazoles. Med. Chem. Res. 2019, 28, 1359–1367. [Google Scholar] [CrossRef]
- Skrzypek, A.; Matysiak, J.; Niewiadomy, A.; Bajda, M.; Szymanski, P. Synthesis and biological evaluation of 1, 3, 4-thiadiazole analogues as novel AChE and BuChE inhibitors. Eur. J. Med. Chem. 2013, 62, 311–319. [Google Scholar] [CrossRef]
- Taha, M.; Rahim, F.; Zaman, K.; Selvaraj, M.; Uddin, N.; Farooq, R.K.; Khan, K.M. Synthesis, α-glycosidase inhibitory potential and molecular docking study of benzimidazole derivatives. Bioorg. Chem. 2020, 95, 103555. [Google Scholar] [CrossRef]
- Yiannopoulou, K.G.; Papageorgiou, S.G. Current and future treatments in Alzheimer disease: An update. J. Cent. Nerv. Syst. Dis. 2020, 12, 1179573520907397. [Google Scholar] [CrossRef]
- Zawawi, N.K.N.A.; Taha, M.; Ahmat, N.; Ismail, N.H.; Wadood, A.; Rahim, F. Synthesis, molecular docking studies of hybrid benzimidazole as a-glucosidase inhibitor. Bioorg. Chem. 2017, 70, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Zueva, I.; Dias, J.; Lushchekina, S.; Semenov, V.; Mukhamedyarov, M.; Pashirova, T.; Petrov, K. New evidence for dual binding site inhibitors of acetylcholinesterase as improved drugs for treatment of Alzheimer’s disease. Neuropharmacology 2019, 155, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Shivanika, C.; Kumar, D.; Ragunathan, V.; Tiwari, P.; Sumitha, A. Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J. Biomol. Struct. Dyn. 2020, 1, 1–27. [Google Scholar]
- Ajmal, A.; Ali, Y.; Khan, A.; Wadood, A.; Rehman, A.U. Identification of novel peptide inhibitors for the KRas-G12C variant to prevent oncogenic signaling. J. Biomol. Struct. Dynam. 2022, 1–10. [Google Scholar] [CrossRef]
- Jarvis, T.R.; Chughtai, B.; Kaplan, S.A. Testosterone and benign prostatic hyperplasia. Asian J. Androl. 2015, 17, 212. [Google Scholar] [PubMed]
- ML, E.U.; LG, D.T. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J. Numerical integration of a System with Constraints: Of the Cartesian Equations of Motion Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef]
- Gotz, A.W.; Williamson, M.J.; Xu, D.; Poole, D.; Le Grand, S.; Walker, R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs 1. Generalized born. J. Chem. Theory Comput. 2012, 8, 1542–1555. [Google Scholar] [CrossRef]



| S.No | Weight | logP | H-donor | H-acceptor |
|---|---|---|---|---|
| Compound 3 | 293.15 | 4.11 | 1 | 2 |
| Compound 10 | 309.15 | 3.65 | 2 | 3 |
| Compound 11 | 293.15 | 4.14 | 1 | 2 |
| Reference | 380.51 | 1.04 | 1 | 3 |
| Complex | vdW | EEL | ESURF | EGB | ΔG TOTAL |
|---|---|---|---|---|---|
| Compound3_AChE | −59.8288 | −10.1721 | −7.8161 | 5.5700 | −56.4166 |
| Compound10_AChE | −57.0777 | −2.0551 | −6.1095 | 12.6979 | −52.5444 |
| Donepezil_ache | −55.1897 | −4.2318 | −5.7198 | 14.9143 | −50.6618 |
| Compound3_BuChE | −41.5357 | −2.6287 | −4.2604 | 10.9467 | −37.4781 |
| Compound10_BuChE | −35.5181 | −5.0815 | −3.4819 | 12.2147 | −31.8668 |
| Donepezil_BuChE | −33.1783 | −2.1543 | −3.1661 | 11.2146 | −30.1013 |
| Category | Property with Unit | Compound’s ID | |||
|---|---|---|---|---|---|
| 3 | 10 | 11 | Donepezil | ||
| Absorption | Water solubility (log mol/L) | −4.959 | −3.705 | −8.852 | −6.483 |
| Intestinal absorption (%) | 92.4 | 90.359 | 86.645 | 88.561 | |
| Skin permeability(log Kp) | −0.823 | −1.672 | −2.178 | −2.728 | |
| Distribution | VDss (human) (log L/kg) | 0.487 | 0.196 | 0.803 | 0.943 |
| BBB permeability | 0.458 | 0.21 | 0.453 | 0.514 | |
| CNS permeability (log PS) | −1.394 | −1.394 | −0.914 | −0.97 | |
| Metabolism | CYP2D6 substrate | No | No | No | No |
| CYP3A4 substrate | No | No | Yes | Yes | |
| CYP1A2 inhibitor | Yes | Yes | Yes | No | |
| CYP2C19 inhibitor | No | No | Yes | No | |
| CYP2C9 inhibitor | No | No | Yes | No | |
| Excretion | Total Clearance(log ml/min/kg) | 0.281 | 0.225 | −0.294 | 0.685 |
| Renal OCT2 substrate | No | No | No | No | |
| Toxicity | AMES toxicity | No | No | No | No |
| Max. tolerated dose (human) (log mg/kg/day) | 0.888 | 0.702 | 0.81 | 0.404 | |
| Oral Rat Acute Toxicity (LD50) (mol/kg) | 2.16 | 2.326 | 2.405 | 3.401 | |
| Hepatotoxicity | No | No | No | No | |
| Skin Sensitisation | Yes | Yes | No | No | |
| Code No. | R | AChE IC50 (µM ± SEM) | BuChE IC50 (µM ± SEM) |
|---|---|---|---|
| 1 |  | 2.10 ± 0.10 | 1.60 ± 0.10 |
| 2 |  | 5.10 ± 0.10 | 5.90 ± 0.10 |
| 3 | 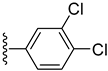 | 0.050 ± 0.001 | 0.080 ± 0.001 |
| 4 |  | 3.40 ± 0.10 | 3.60 ± 0.10 |
| 5 | 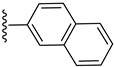 | 25.30 ± 0.40 | 25.80 ± 0.40 |
| 6 |  | 4.10 ± 0.10 | 5.10 ± 0.20 |
| 7 |  | 5.80 ± 0.10 | 5.90 ± 0.10 |
| 8 | 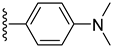 | 6.80 ± 0.20 | 4.70 ± 0.10 |
| 9 | 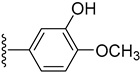 | 2.30 ± 0.10 | 2.10 ± 0.10 |
| 10 | 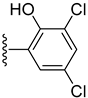 | 0.10 ± 0.001 | 0.40 ± 0.001 |
| 11 | 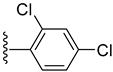 | 0.20 ± 0.001 | 0.30 ± 0.001 |
| 12 | 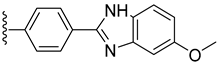 | 5.10 ± 0.10 | 5.80 ± 0.10 |
| 13 | 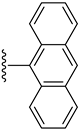 | N. A. | N. A. |
| 14 |  | N. A. | N. A. |
| 15 | 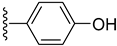 | 0.80 ± 0.001 | 0.90 ± 0.001 |
| 16 | 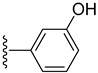 | 1.30 ± 0.10 | 2.10 ± 0.10 |
| 17 | 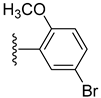 | 6.10 ± 0.10 | 3.90 ± 0.10 |
| 18 | 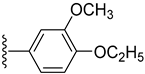 | 11.30 ± 0.30 | 9.80 ± 0.30 |
| 19 |  | 9.20 ± 0.20 | 6.80 ± 0.20 |
| 20 | 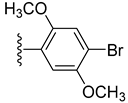 | N. A. | 15.20 ± 0.30 |
| 21 |  | 2.10 ± 0.10 | 2.70 ± 0.10 |
| Donepezil | 0.016 ± 0.12 | 0.30 ± 0.010 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adalat, B.; Rahim, F.; Rehman, W.; Ali, Z.; Rasheed, L.; Khan, Y.; Farghaly, T.A.; Shams, S.; Taha, M.; Wadood, A.; et al. Biologically Potent Benzimidazole-Based-Substituted Benzaldehyde Derivatives as Potent Inhibitors for Alzheimer’s Disease along with Molecular Docking Study. Pharmaceuticals 2023, 16, 208. https://doi.org/10.3390/ph16020208
Adalat B, Rahim F, Rehman W, Ali Z, Rasheed L, Khan Y, Farghaly TA, Shams S, Taha M, Wadood A, et al. Biologically Potent Benzimidazole-Based-Substituted Benzaldehyde Derivatives as Potent Inhibitors for Alzheimer’s Disease along with Molecular Docking Study. Pharmaceuticals. 2023; 16(2):208. https://doi.org/10.3390/ph16020208
Chicago/Turabian StyleAdalat, Bushra, Fazal Rahim, Wajid Rehman, Zarshad Ali, Liaqat Rasheed, Yousaf Khan, Thoraya A. Farghaly, Sulaiman Shams, Muhammad Taha, Abdul Wadood, and et al. 2023. "Biologically Potent Benzimidazole-Based-Substituted Benzaldehyde Derivatives as Potent Inhibitors for Alzheimer’s Disease along with Molecular Docking Study" Pharmaceuticals 16, no. 2: 208. https://doi.org/10.3390/ph16020208
APA StyleAdalat, B., Rahim, F., Rehman, W., Ali, Z., Rasheed, L., Khan, Y., Farghaly, T. A., Shams, S., Taha, M., Wadood, A., Shah, S. A. A., & Abdellatif, M. H. (2023). Biologically Potent Benzimidazole-Based-Substituted Benzaldehyde Derivatives as Potent Inhibitors for Alzheimer’s Disease along with Molecular Docking Study. Pharmaceuticals, 16(2), 208. https://doi.org/10.3390/ph16020208








