Therapeutic Effect of Costunolide in Autoimmune Hepatitis: Network Pharmacology and Experimental Validation
Abstract
:1. Introduction
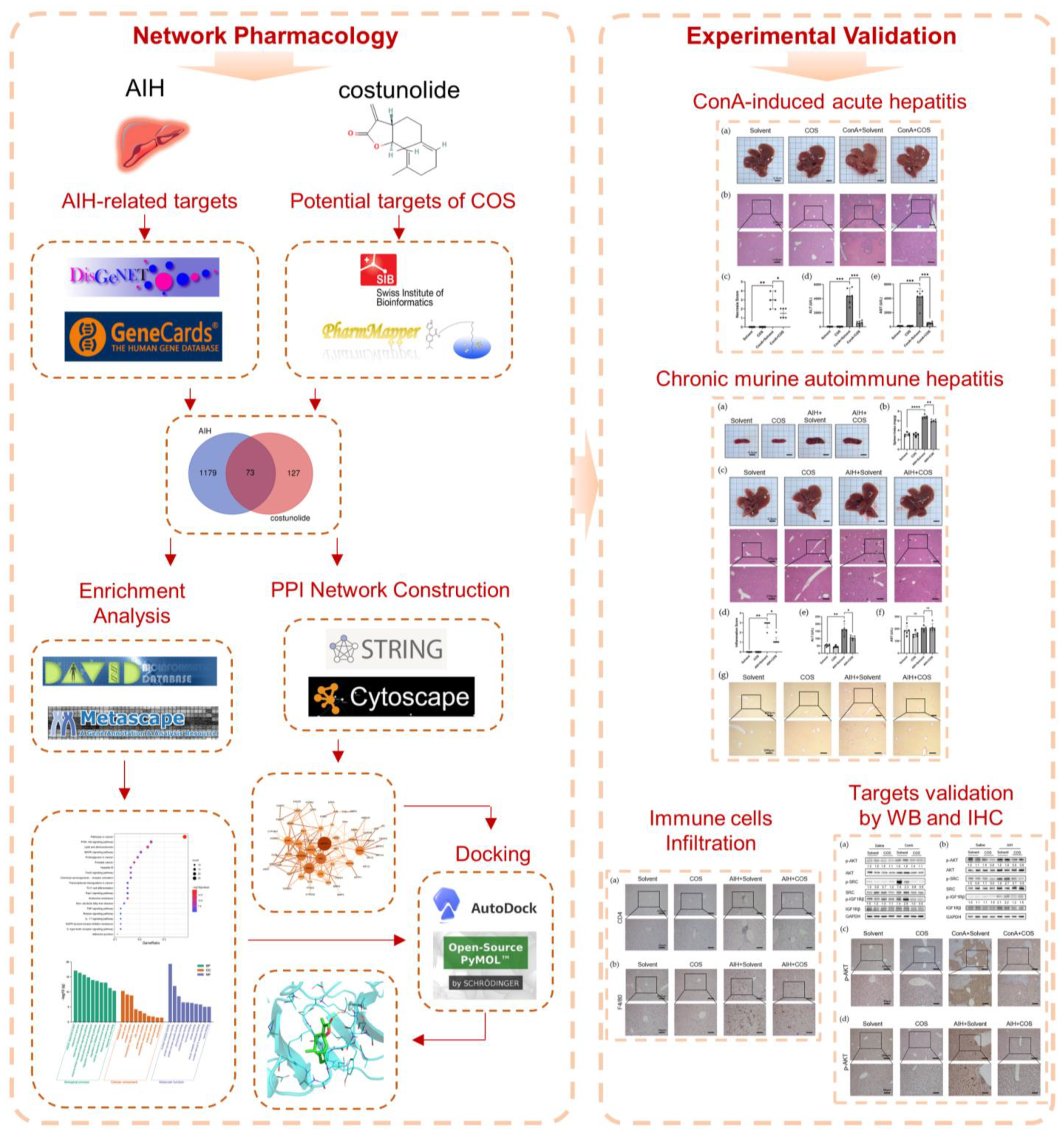
2. Results
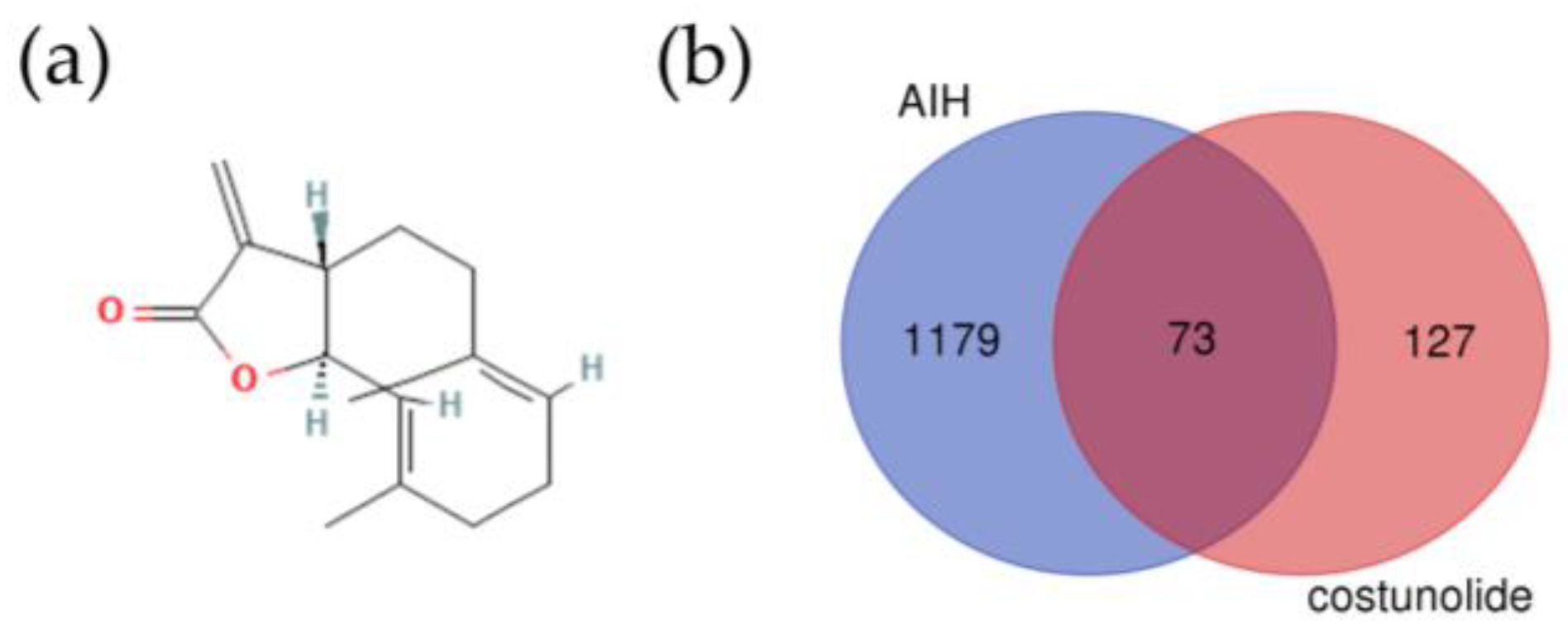
2.1. Identification of the Potential Targets of Costunolide in AIH
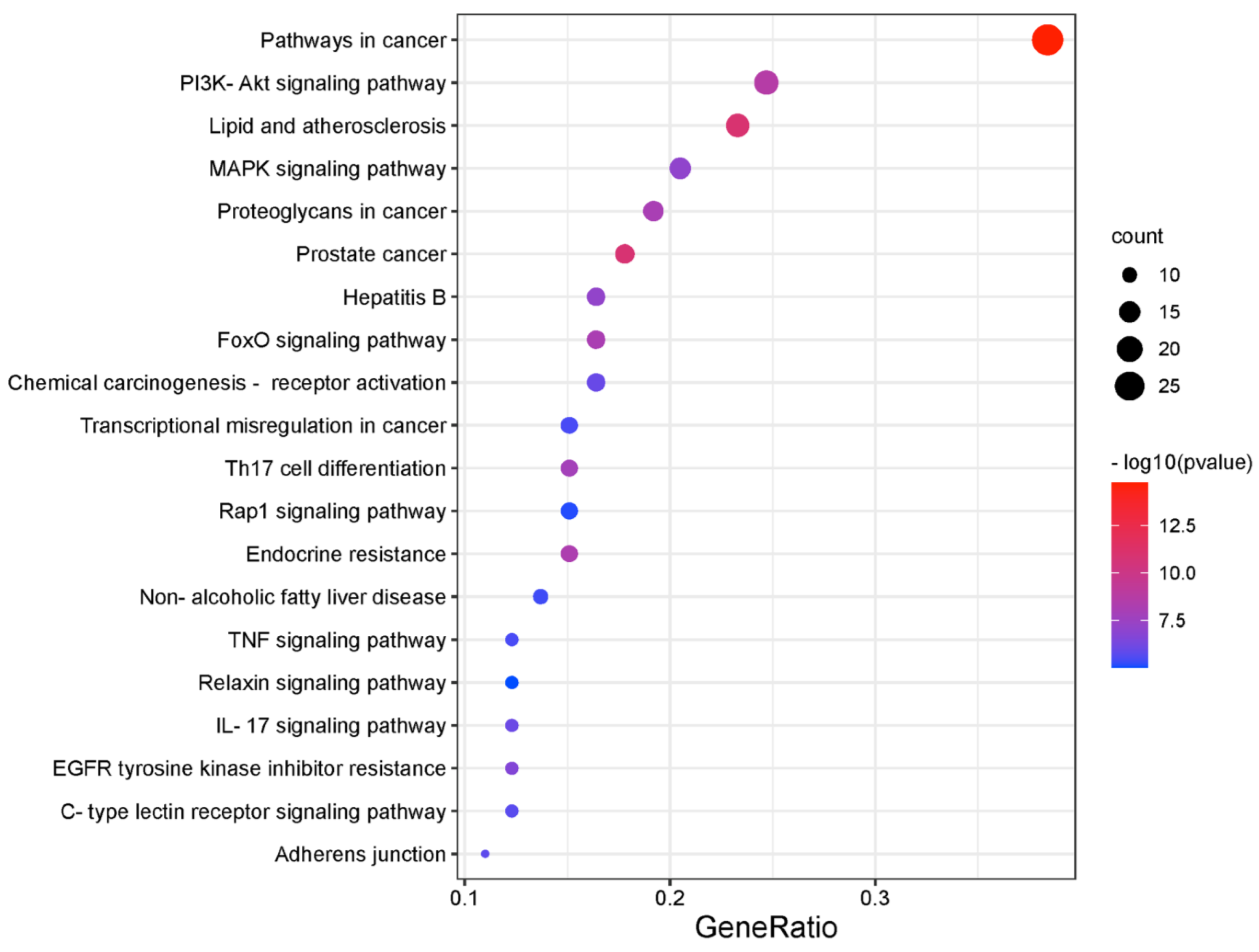
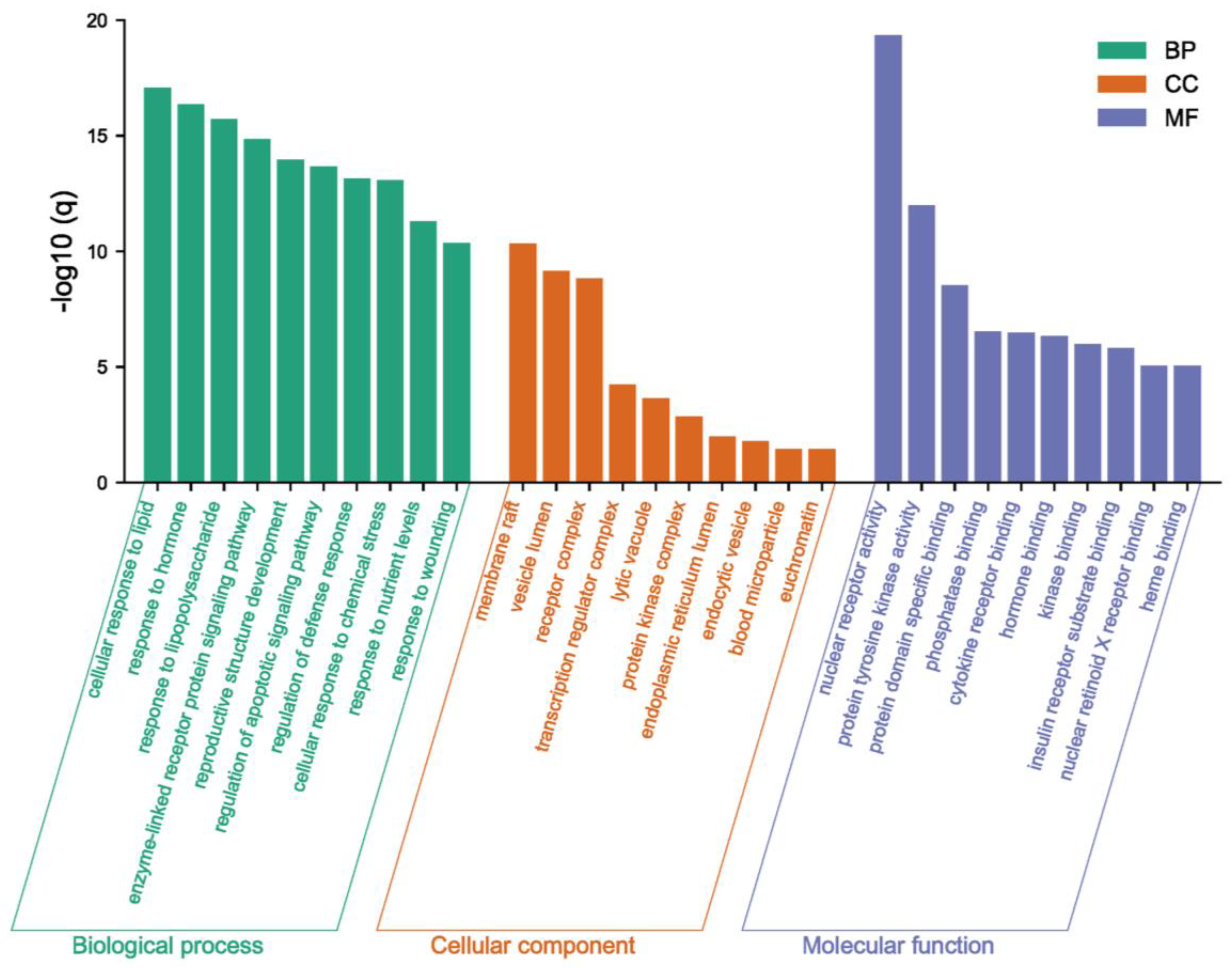
2.2. Recognition of Enriched Pathway of the Potential Targets of Costunolide in AIH
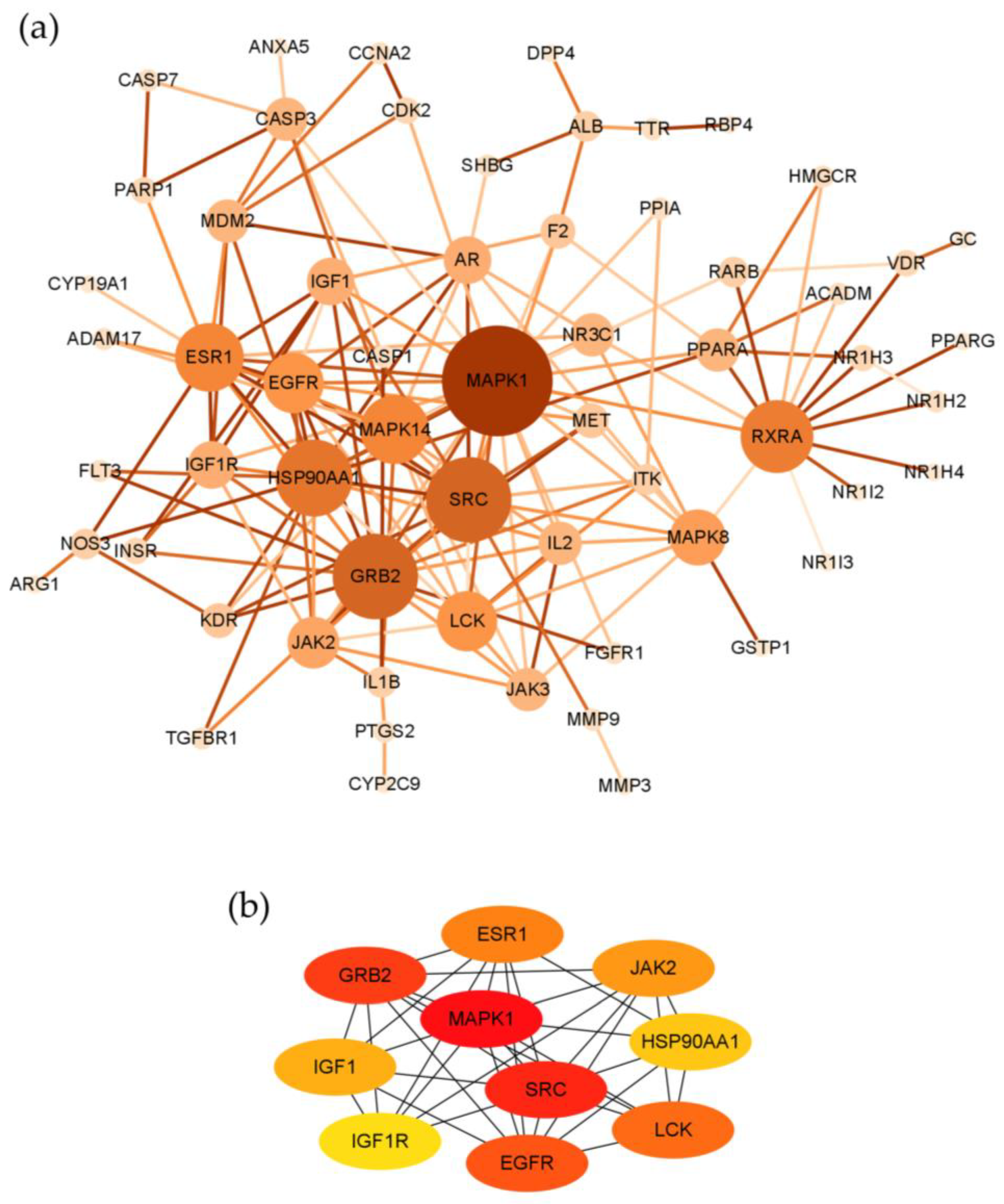
2.3. Topological Network Analysis of the Potential Targets of Costunolide in AIH
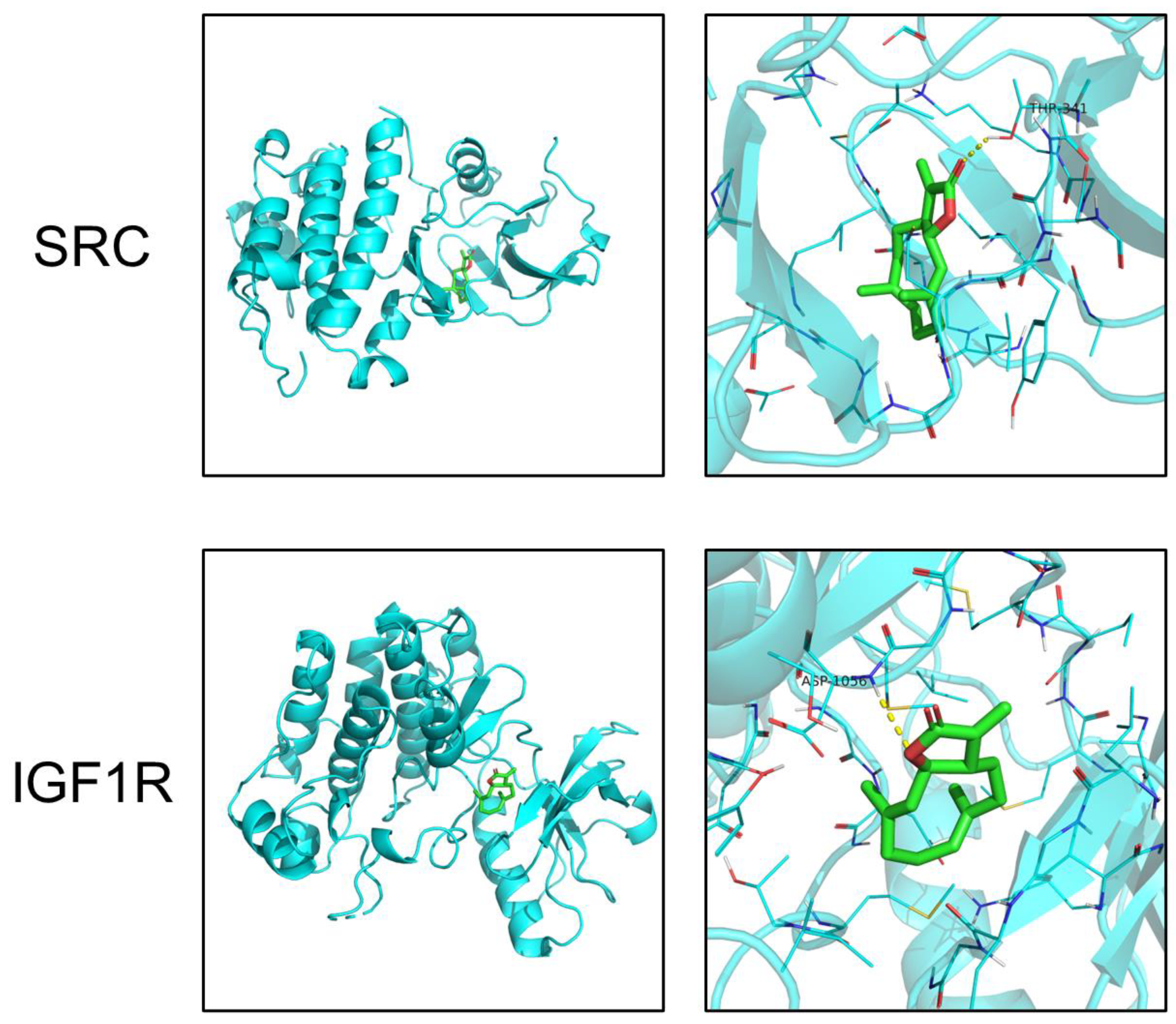
2.4. Molecular Docking
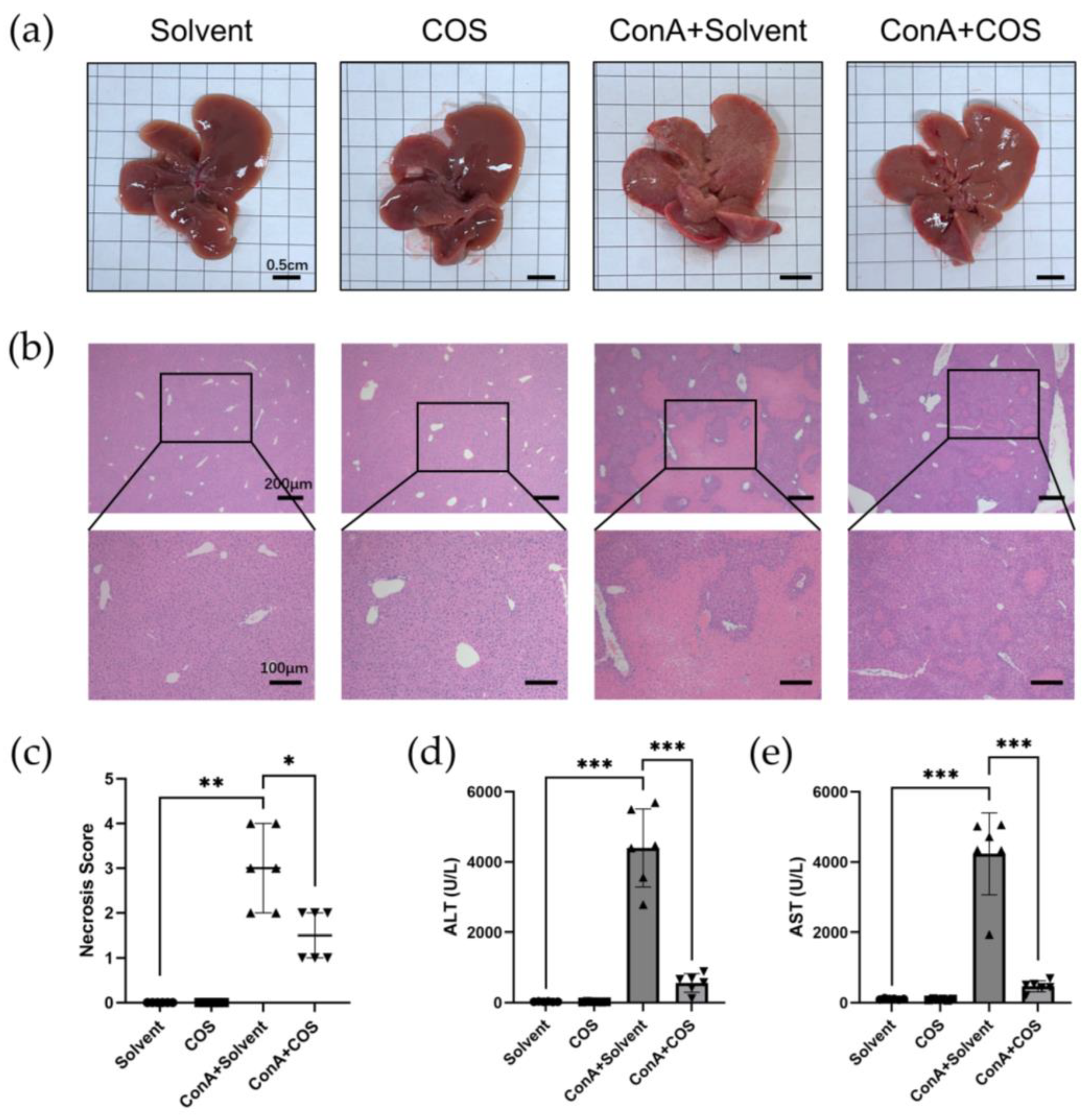
2.5. Costunolide Attenuated ConA-Induced Acute Hepatitis
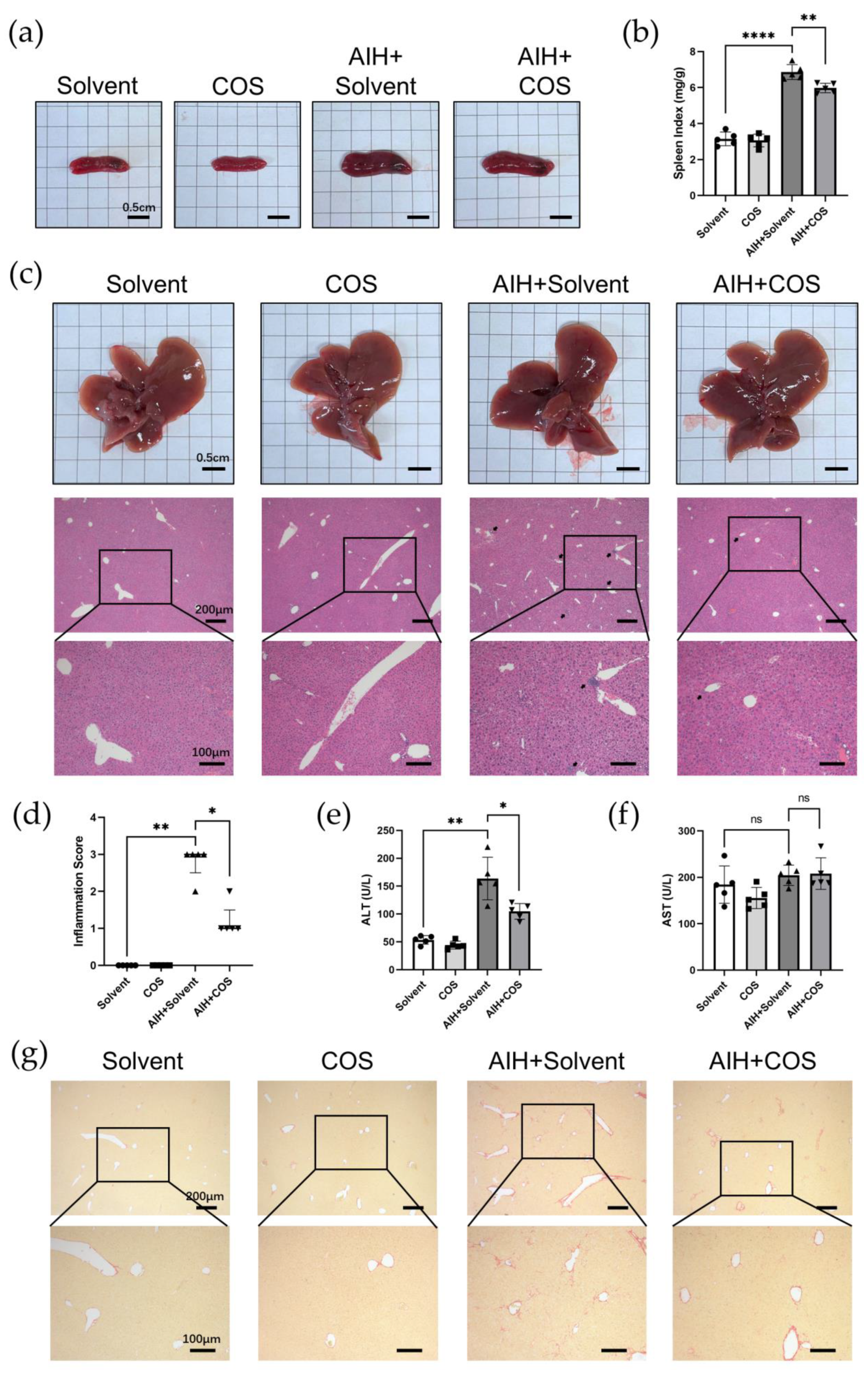
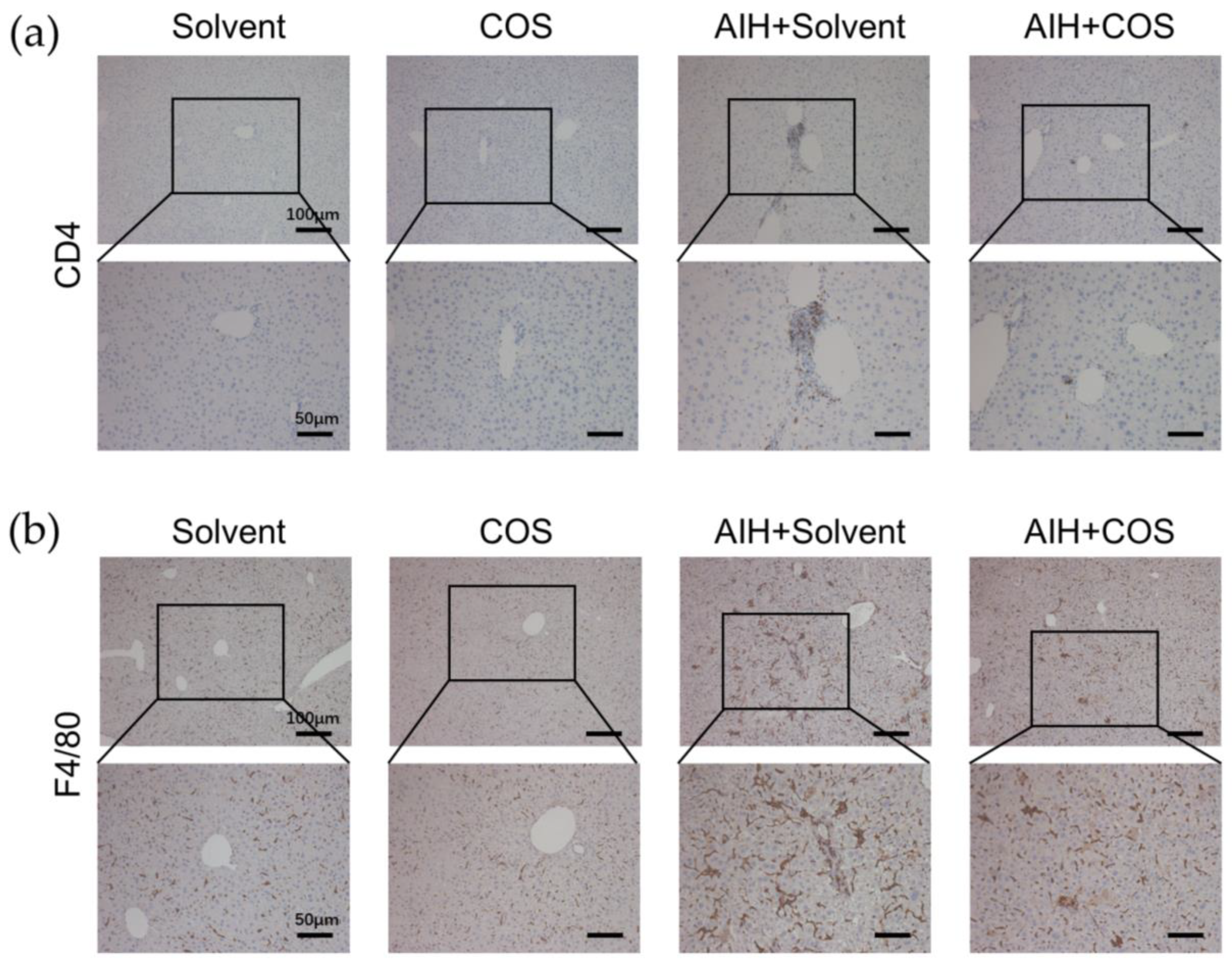
2.6. Costunolide Ameliorated Chronic Murine Autoimmune Hepatitis
2.7. Costunolide Inhibited the Activation of PI3K-AKT Pathway and Suppressed the Phosphorylation of SRC and IGF1R in AIH
3. Discussion
4. Materials and Methods
4.1. Chemical Information Collection and Targets Retrieval of Costunolide
4.2. Prediction of Potential Targets of Costunolide in AIH and Enrichment Analysis
4.3. Protein–Protein Interaction Target Network Construction and Visualization
4.4. Molecular Docking
4.5. Establishment of Two Experimental AIH Models and Administration of Costunolide
4.6. Histopathology, Immunohistochemistry, and Biochemical Analysis
4.7. Western Blot
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sirbe, C.; Simu, G.; Szabo, I.; Grama, A.; Pop, T.L. Pathogenesis of Autoimmune Hepatitis-Cellular and Molecular Mechanisms. Int. J. Mol. Sci. 2021, 22, 13578. [Google Scholar] [CrossRef] [PubMed]
- Terziroli Beretta-Piccoli, B.; Mieli-Vergani, G.; Vergani, D. Autoimmmune hepatitis. Cell. Mol. Immunol. 2022, 19, 158–176. [Google Scholar] [CrossRef] [PubMed]
- Lohse, A.W.; Sebode, M.; Jorgensen, M.H.; Ytting, H.; Karlsen, T.H.; Kelly, D.; Manns, M.P.; Vesterhus, M.; European Reference Network on Hepatological Diseases; The International Autoimmune Hepatitis Group (IAIHG). Second-line and third-line therapy for autoimmune hepatitis: A position statement from the European Reference Network on Hepatological Diseases and the International Autoimmune Hepatitis Group. J. Hepatol. 2020, 73, 1496–1506. [Google Scholar] [CrossRef] [PubMed]
- Webb, G.J.; Hirschfield, G.M.; Krawitt, E.L.; Gershwin, M.E. Cellular and Molecular Mechanisms of Autoimmune Hepatitis. Annu. Rev. Pathol. 2018, 13, 247–292. [Google Scholar] [CrossRef]
- Liu, J.; Ma, Z.; Li, H.; Li, X. Chinese medicine in the treatment of autoimmune hepatitis: Progress and future opportunities. Anim. Model. Exp. Med. 2022, 5, 95–107. [Google Scholar] [CrossRef]
- Kim, D.Y.; Choi, B.Y. Costunolide-A Bioactive Sesquiterpene Lactone with Diverse Therapeutic Potential. Int. J. Mol. Sci. 2019, 20, 2926. [Google Scholar] [CrossRef] [Green Version]
- Kassuya, C.A.; Cremoneze, A.; Barros, L.F.; Simas, A.S.; Lapa Fda, R.; Mello-Silva, R.; Stefanello, M.E.; Zampronio, A.R. Antipyretic and anti-inflammatory properties of the ethanolic extract, dichloromethane fraction and costunolide from Magnolia ovata (Magnoliaceae). J. Ethnopharmacol. 2009, 124, 369–376. [Google Scholar] [CrossRef]
- Butturini, E.; Di Paola, R.; Suzuki, H.; Paterniti, I.; Ahmad, A.; Mariotto, S.; Cuzzocrea, S. Costunolide and Dehydrocostuslactone, two natural sesquiterpene lactones, ameliorate the inflammatory process associated to experimental pleurisy in mice. Eur. J. Pharmacol. 2014, 730, 107–115. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, Y.; Zhang, J.; Wang, L.; Jin, Z.; Huang, H.; Man, S.; Gao, W. Evaluation of protective effects of costunolide and dehydrocostuslactone on ethanol-induced gastric ulcer in mice based on multi-pathway regulation. Chem. Biol. Interact. 2016, 250, 68–77. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, D.; Li, M.; Wang, B. Costunolide ameliorates lipoteichoic acid-induced acute lung injury via attenuating MAPK signaling pathway. Int. Immunopharmacol. 2018, 61, 283–289. [Google Scholar] [CrossRef]
- Xie, F.; Zhang, H.; Zheng, C.; Shen, X.F. Costunolide improved dextran sulfate sodium-induced acute ulcerative colitis in mice through NF-kappaB, STAT1/3, and Akt signaling pathways. Int. Immunopharmacol. 2020, 84, 106567. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Xing, Y.; Dong, D.; Hu, Y.; Chen, Q.; Zhai, L.; Hu, L.; Zhang, Y. Costunolide ameliorates colitis via specific inhibition of HIF1alpha/glycolysis-mediated Th17 differentiation. Int. Immunopharmacol. 2021, 97, 107688. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Yi, M.; Wang, R.; Huang, Y.; Chen, M. Protective Effects of Costunolide Against D-Galactosamine and Lipopolysaccharide-Induced Acute Liver Injury in Mice. Front. Pharmacol. 2018, 9, 1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, J.; Zhan, H.; Meng, F.; Wang, G.; Huang, D.; Liao, Z.; Chen, M. Costunolide protects against alcohol-induced liver injury by regulating gut microbiota, oxidative stress and attenuating inflammation in vivo and in vitro. Phytother. Res. 2022, 36, 1268–1283. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.X.; Liu, H.T.; Zhang, N.; Niu, W.X.; Lu, Z.N.; Bao, Y.Y.; Huang, R.; Yu, D.K.; Shao, R.G.; He, H.W. Costunolide represses hepatic fibrosis through WW domain-containing protein 2-mediated Notch3 degradation. Br. J. Pharmacol. 2020, 177, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- To, K.I.; Zhu, Z.X.; Wang, Y.N.; Li, G.A.; Sun, Y.M.; Li, Y.; Jin, Y.H. Integrative network pharmacology and experimental verification to reveal the anti-inflammatory mechanism of ginsenoside Rh4. Front. Pharmacol. 2022, 13, 953871. [Google Scholar] [CrossRef]
- Wang, C.R.; Chen, H.W.; Li, Y.; Zhou, M.Y.; Wong, V.K.; Jiang, Z.H.; Zhang, W. Network Pharmacology Exploration Reveals Anti-Apoptosis as a Common Therapeutic Mechanism for Non-Alcoholic Fatty Liver Disease Treated with Blueberry Leaf Polyphenols. Nutrients 2021, 13, 4060. [Google Scholar] [CrossRef]
- Li, X.; Wen, H.; Zhang, Y.; Liu, A.; Zhang, X.; Fu, M.; Pan, Y.; Xu, J.; Zhang, J. DPHB, a diarylheptane from Alpinia officinarum Hance, ameliorates insulin resistance: A network pharmacology and in vitro study. Front. Pharmacol. 2022, 13, 956812. [Google Scholar] [CrossRef]
- Xu, Z.; Lin, S.; Gong, J.; Feng, P.; Cao, Y.; Li, Q.; Jiang, Y.; You, Y.; Tong, Y.; Wang, P. Exploring the Protective Effects and Mechanism of Crocetin From Saffron Against NAFLD by Network Pharmacology and Experimental Validation. Front. Med. 2021, 8, 681391. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yan, W.; Feng, Z.; Gao, Y.; Zhang, L.; Feng, X.; Tian, D. Plasma proteomic analysis of autoimmune hepatitis in an improved AIH mouse model. J. Transl. Med. 2020, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Lv, T.; Li, M.; Zeng, N.; Zhang, J.; Li, S.; Chen, S.; Zhang, C.; Shan, S.; Duan, W.; Wang, Q.; et al. Systematic review and meta-analysis on the incidence and prevalence of autoimmune hepatitis in Asian, European, and American population. J. Gastroenterol. Hepatol. 2019, 34, 1676–1684. [Google Scholar] [CrossRef]
- Huang, H.; Park, S.; Zhang, H.; Park, S.; Kwon, W.; Kim, E.; Zhang, X.; Jang, S.; Yoon, D.; Choi, S.K.; et al. Targeting AKT with costunolide suppresses the growth of colorectal cancer cells and induces apoptosis in vitro and in vivo. J. Exp. Clin. Cancer Res. 2021, 40, 114. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Antonucci, L.; Yamachika, S.; Zhang, Z.; Taniguchi, K.; Umemura, A.; Hatzivassiliou, G.; Roose-Girma, M.; Reina-Campos, M.; Duran, A.; et al. NRF2 activates growth factor genes and downstream AKT signaling to induce mouse and human hepatomegaly. J. Hepatol. 2020, 72, 1182–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, R.; Tang, J.; Zhang, X.; He, Y.; Yu, Z.; Chen, S.; Xia, J.; Lin, J.; Ou, Q. CCN1 Promotes Inflammation by Inducing IL-6 Production via alpha6beta1/PI3K/Akt/NF-kappaB Pathway in Autoimmune Hepatitis. Front. Immunol. 2022, 13, 810671. [Google Scholar] [CrossRef]
- Xiang, M.; Liu, T.; Tan, W.; Ren, H.; Li, H.; Liu, J.; Cao, H.; Cheng, Q.; Liu, X.; Zhu, H.; et al. Effects of kinsenoside, a potential immunosuppressive drug for autoimmune hepatitis, on dendritic cells/CD8(+) T cells communication in mice. Hepatology 2016, 64, 2135–2150. [Google Scholar] [CrossRef]
- Morales-Ruiz, M.; Santel, A.; Ribera, J.; Jimenez, W. The Role of Akt in Chronic Liver Disease and Liver Regeneration. Semin. Liver Dis. 2017, 37, 11–16. [Google Scholar] [CrossRef]
- Haxhinasto, S.; Mathis, D.; Benoist, C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 2008, 205, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgoffe, G.M.; Pollizzi, K.N.; Waickman, A.T.; Heikamp, E.; Meyers, D.J.; Horton, M.R.; Xiao, B.; Worley, P.F.; Powell, J.D. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 2011, 12, 295–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, K.; Yin, N.; Peng, M.; Stamatiades, E.G.; Chhangawala, S.; Shyu, A.; Li, P.; Zhang, X.; Do, M.H.; Capistrano, K.J.; et al. Glycolytic ATP fuels phosphoinositide 3-kinase signaling to support effector T helper 17 cell responses. Immunity 2021, 54, 976–987.e7. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Yin, N.; Peng, M.; Stamatiades, E.G.; Shyu, A.; Li, P.; Zhang, X.; Do, M.H.; Wang, Z.; Capistrano, K.J.; et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 2021, 371, 405–410. [Google Scholar] [CrossRef]
- Herrero-Sanchez, M.C.; Rodriguez-Serrano, C.; Almeida, J.; San Segundo, L.; Inoges, S.; Santos-Briz, A.; Garcia-Brinon, J.; Corchete, L.A.; San Miguel, J.F.; Del Canizo, C.; et al. Targeting of PI3K/AKT/mTOR pathway to inhibit T cell activation and prevent graft-versus-host disease development. J. Hematol. Oncol. 2016, 9, 113. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, M.J.; Tsantikos, E.; Kong, A.M.; Vanhaesebroeck, B.; Tarlinton, D.M.; Hibbs, M.L. Attenuation of phosphoinositide 3-kinase delta signaling restrains autoimmune disease. J. Autoimmun. 2012, 38, 381–391. [Google Scholar] [CrossRef]
- Camps, M.; Ruckle, T.; Ji, H.; Ardissone, V.; Rintelen, F.; Shaw, J.; Ferrandi, C.; Chabert, C.; Gillieron, C.; Francon, B.; et al. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat. Med. 2005, 11, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, R.; Gu, T.; Ma, F.; Laster, K.V.; Li, X.; Liu, K.; Lee, M.H.; Dong, Z. Costunolide is a dual inhibitor of MEK1 and AKT1/2 that overcomes osimertinib resistance in lung cancer. Mol. Cancer 2022, 21, 193. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Men, R.; Wang, H.; Shen, M.; Wang, T.; Ye, T.; Luo, X.; Yang, L. Methylprednisolone Decreases Mitochondria-Mediated Apoptosis and Autophagy Dysfunction in Hepatocytes of Experimental Autoimmune Hepatitis Model via the Akt/mTOR Signaling. Front. Pharmacol. 2019, 10, 1189. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.; Messmer, M.; Bayer, M.; Pfeilschifter, J.M.; Hintermann, E.; Christen, U. Non-alcoholic fatty liver disease (NAFLD) potentiates autoimmune hepatitis in the CYP2D6 mouse model. J. Autoimmun. 2016, 69, 51–58. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 2015, 94, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Bai, Y.; Zhu, X.; Lin, L.; Zhao, L.; Wu, X.; Buch, S.; Wang, L.; Chao, J.; Yao, H. IL-17A induces MIP-1alpha expression in primary astrocytes via Src/MAPK/PI3K/NF-kB pathways: Implications for multiple sclerosis. J. Neuroimmune Pharmacol. 2014, 9, 629–641. [Google Scholar] [CrossRef]
- Hao, M.; Sun, J.; Zhang, Y.; Zhang, D.; Han, J.; Zhang, J.; Qiao, H. Exploring the Role of SRC in Extraocular Muscle Fibrosis of the Graves’ Ophthalmopathy. Front. Bioeng. Biotechnol. 2020, 8, 392. [Google Scholar] [CrossRef]
- Behnen, M.; Leschczyk, C.; Moller, S.; Batel, T.; Klinger, M.; Solbach, W.; Laskay, T. Immobilized immune complexes induce neutrophil extracellular trap release by human neutrophil granulocytes via FcgammaRIIIB and Mac-1. J. Immunol. 2014, 193, 1954–1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef] [PubMed]
- DiToro, D.; Harbour, S.N.; Bando, J.K.; Benavides, G.; Witte, S.; Laufer, V.A.; Moseley, C.; Singer, J.R.; Frey, B.; Turner, H.; et al. Insulin-Like Growth Factors Are Key Regulators of T Helper 17 Regulatory T Cell Balance in Autoimmunity. Immunity 2020, 52, 650–667.e10. [Google Scholar] [CrossRef]
- Pritchard, J.; Tsui, S.; Horst, N.; Cruikshank, W.W.; Smith, T.J. Synovial fibroblasts from patients with rheumatoid arthritis, like fibroblasts from Graves’ disease, express high levels of IL-16 when treated with Igs against insulin-like growth factor-1 receptor. J. Immunol. 2004, 173, 3564–3569. [Google Scholar] [CrossRef] [Green Version]
- Douglas, R.S.; Gianoukakis, A.G.; Kamat, S.; Smith, T.J. Aberrant expression of the insulin-like growth factor-1 receptor by T cells from patients with Graves’ disease may carry functional consequences for disease pathogenesis. J. Immunol. 2007, 178, 3281–3287. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.Y.; Qiu, H.Y.; Zhang, M.D.; Hou, J.J.; Zhou, M.L.; Wu, Y. Lenalidomide attenuates IMQ-induced inflammation in a mouse model of psoriasis. Biomed. Pharmacother. 2022, 156, 113883. [Google Scholar] [CrossRef]
- Pae, H.O.; Jeong, G.S.; Kim, H.S.; Woo, W.H.; Rhew, H.Y.; Kim, H.S.; Sohn, D.H.; Kim, Y.C.; Chung, H.T. Costunolide inhibits production of tumor necrosis factor-alpha and interleukin-6 by inducing heme oxygenase-1 in RAW264.7 macrophages. Inflamm. Res. 2007, 56, 520–526. [Google Scholar] [CrossRef]
- Park, E.; Song, J.H.; Kim, M.S.; Park, S.H.; Kim, T.S. Costunolide, a sesquiterpene lactone, inhibits the differentiation of pro-inflammatory CD4(+) T cells through the modulation of mitogen-activated protein kinases. Int. Immunopharmacol. 2016, 40, 508–516. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines for the management of patients with decompensated cirrhosis. J. Hepatol. 2018, 69, 406–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, X.; Wang, X.; Niu, B.; Meng, Y.; He, H.; Wang, Y.; Li, G. Costunolide Loaded in pH-Responsive Mesoporous Silica Nanoparticles for Increased Stability and an Enhanced Anti-Fibrotic Effect. Pharmaceuticals 2021, 14, 951. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [Green Version]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Pinero, J.; Ramirez-Anguita, J.M.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [Green Version]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature: Singapore, 2021; pp. 27–56. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System; Version 1.8; Schrodinger, LLC.: New York, NY, USA, 2015.
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janet, H. Methods of Blood Collection in the Mouse. Lab. Animal 2000, 29, 47–53. [Google Scholar]

| MW | AlogP | Hdon | Hacc | OB (%) | Caco-2 (nm/s) | BBB | DL | FASA- | TPSA | RBN |
|---|---|---|---|---|---|---|---|---|---|---|
| 232.35 | 4.02 | 0 | 2 | 29.07 | 1.28 | 1.42 | 0.11 | 0.36 | 26.30 | 0 |
| Target | PDB ID | Binding Energy (kcal/mol) |
|---|---|---|
| SRC | 6E6E | −7.7 |
| IGF1R | 3NW7 | −5.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Nie, S.; Wang, S.; Wang, H.; Gong, J.; Yan, W.; Tian, D.; Liu, M. Therapeutic Effect of Costunolide in Autoimmune Hepatitis: Network Pharmacology and Experimental Validation. Pharmaceuticals 2023, 16, 316. https://doi.org/10.3390/ph16020316
Huang Z, Nie S, Wang S, Wang H, Gong J, Yan W, Tian D, Liu M. Therapeutic Effect of Costunolide in Autoimmune Hepatitis: Network Pharmacology and Experimental Validation. Pharmaceuticals. 2023; 16(2):316. https://doi.org/10.3390/ph16020316
Chicago/Turabian StyleHuang, Zheng, Shangshu Nie, Shuhui Wang, Han Wang, Jin Gong, Wei Yan, Dean Tian, and Mei Liu. 2023. "Therapeutic Effect of Costunolide in Autoimmune Hepatitis: Network Pharmacology and Experimental Validation" Pharmaceuticals 16, no. 2: 316. https://doi.org/10.3390/ph16020316
APA StyleHuang, Z., Nie, S., Wang, S., Wang, H., Gong, J., Yan, W., Tian, D., & Liu, M. (2023). Therapeutic Effect of Costunolide in Autoimmune Hepatitis: Network Pharmacology and Experimental Validation. Pharmaceuticals, 16(2), 316. https://doi.org/10.3390/ph16020316





