Anti-Tuberculosis Mur Inhibitors: Structural Insights and the Way Ahead for Development of Novel Agents
Abstract
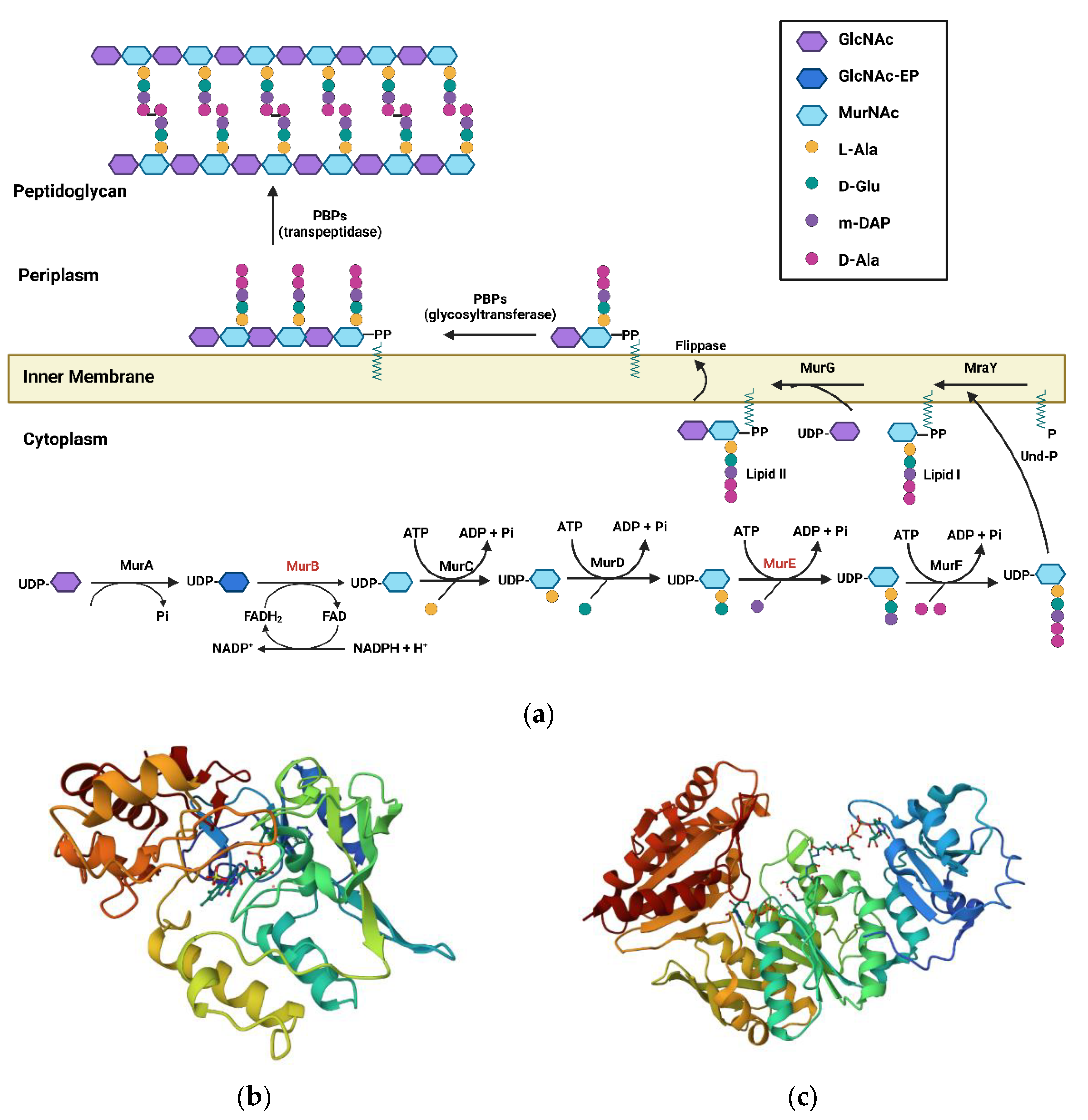
1. Introduction
1.1. MurB Enzyme
1.2. MurE Enzyme
2. Structural Classes of Mur Inhibitors
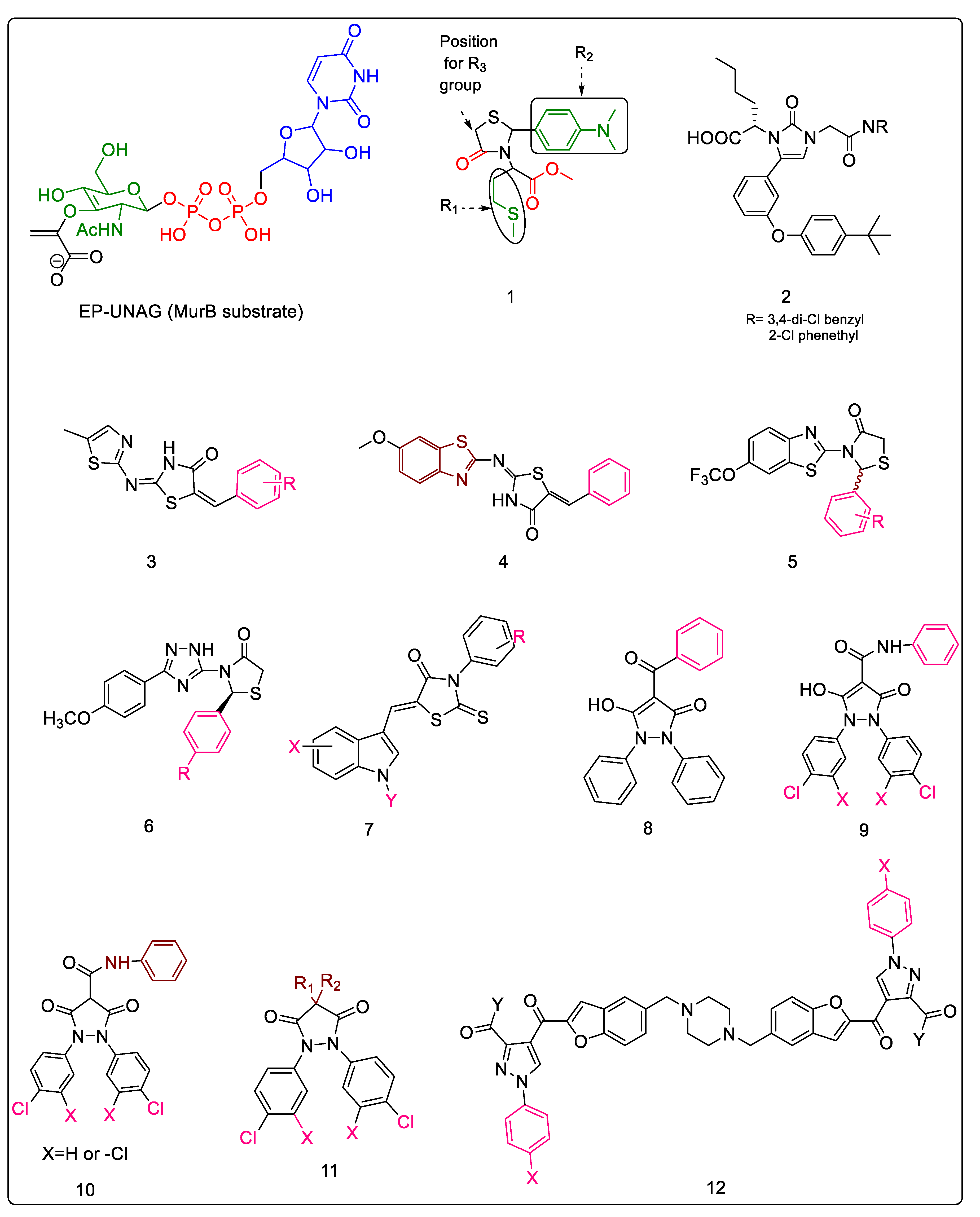
2.1. Mur B Inhibitors
2.1.1. 4-Thiazolidinones
2.1.2. Imidazolinone—A Thiazolidinone Bioisostere
2.1.3. Benzylidene Thiazolylimino Thiazolidinones
2.1.4. Benzylidene Benzothiazolo Thiazolidinones
2.1.5. Benzo[d]thiazole-Based Thiazolidinones
2.1.6. 1,2,4-Triazole-Based 4-Thiazolidinones
2.1.7. 5-Indolylmethylen-4-oxo-2-thioxothiazolidines
2.1.8. 3,5-Dioxopyrazolidinedione and Its Derivatives
2.1.9. Bis(pyrazole-benzofuran) Hybrids Possessing Piperazine Linkers
2.1.10. 1,2,3-Triazolyl Pyrazole Derivatives
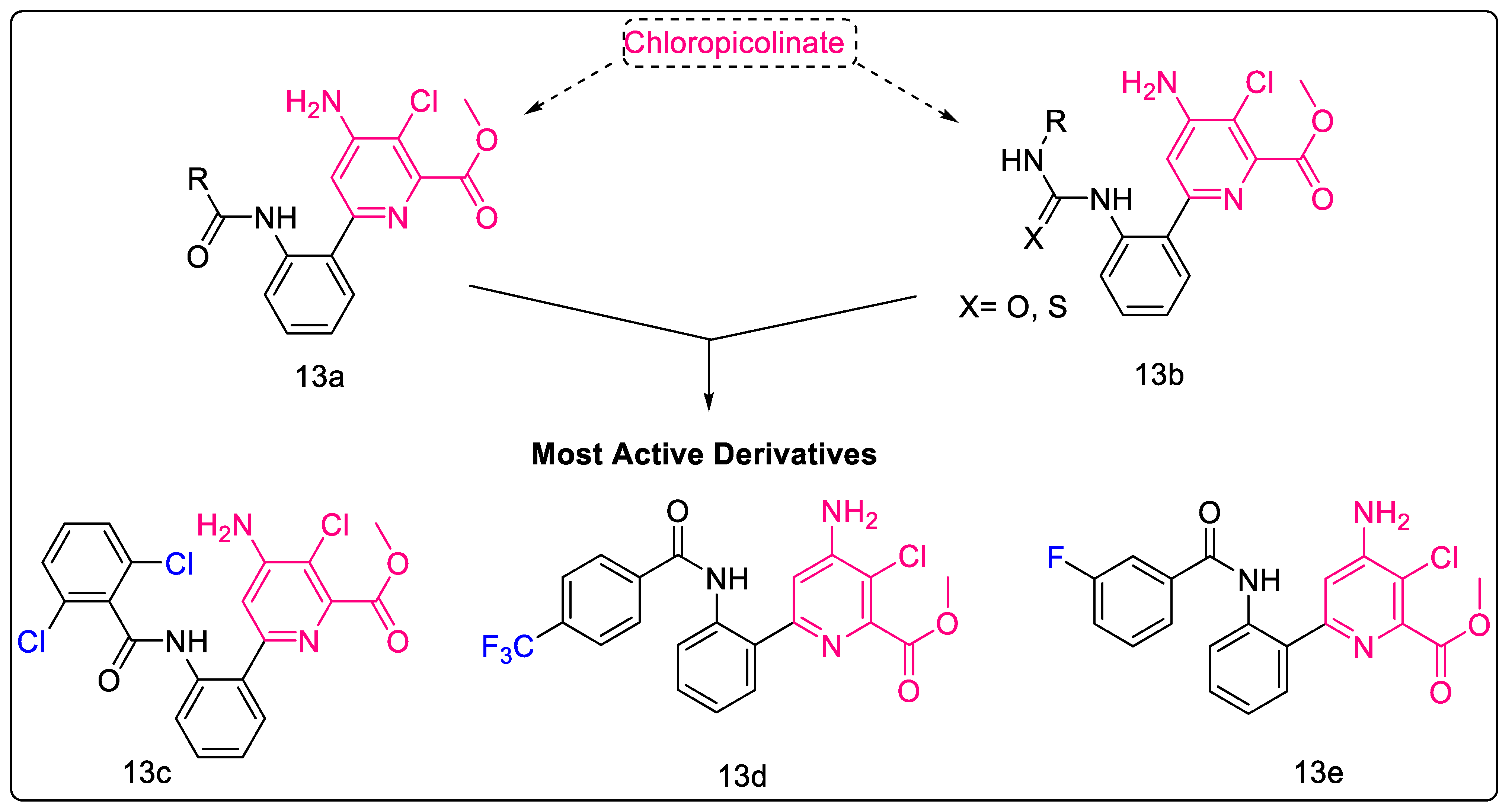
2.1.11. Chloropicolinate Amide, Urea, and Thiourea Derivatives
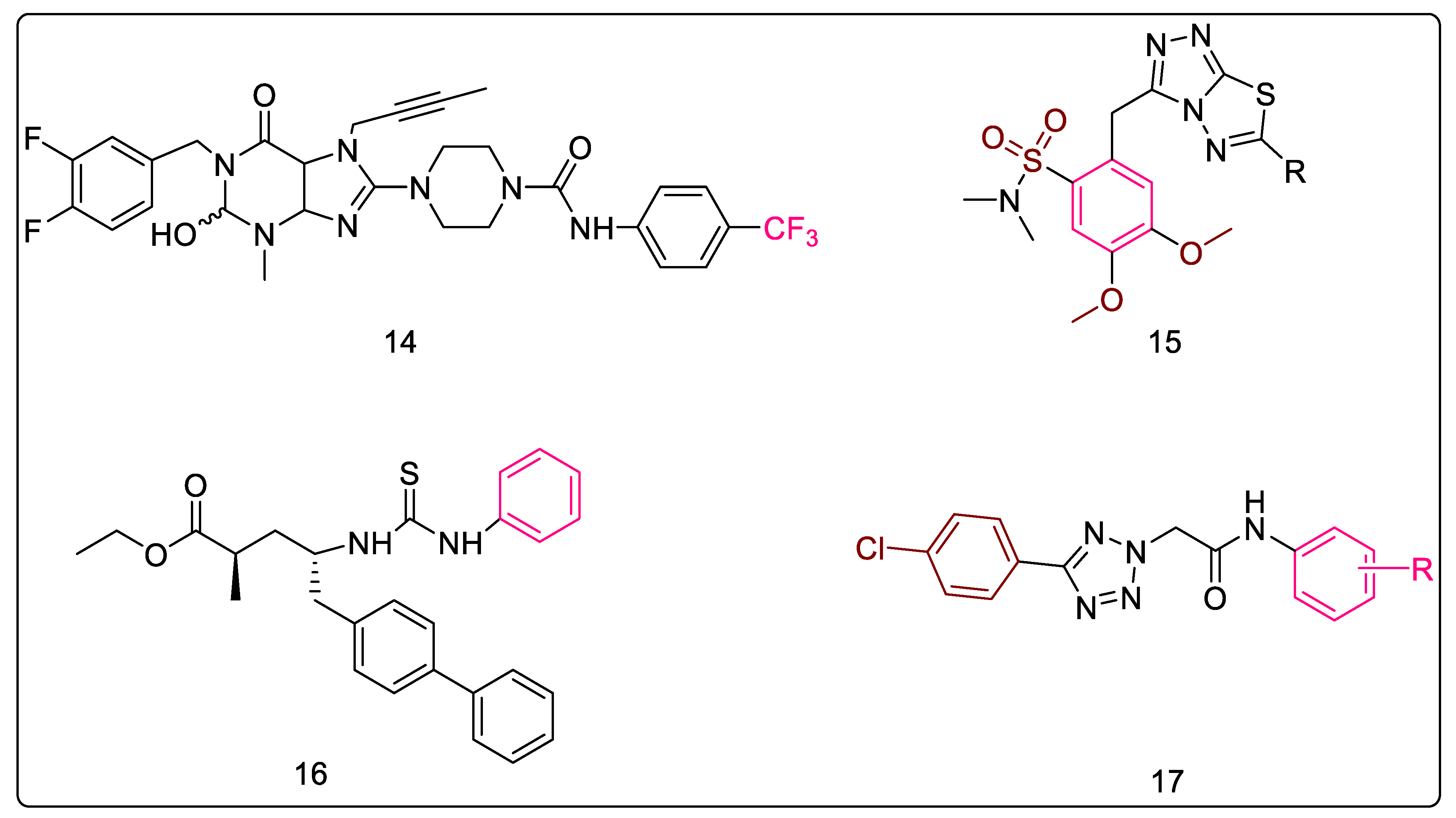
2.1.12. Purine-2,6-dione Linked Piperazine Derivatives
2.1.13. Triazolo-Thiadiazole Derivatives
2.1.14. Sacubitril-Based Urea and Thiourea Derivatives
2.1.15. 5-Substituted Tetrazol-2-yl Acetamides
2.1.16. Coumarins: MurB Inhibitors
2.2. Mixed Inhibitors
2.2.1. 5-Adamantan Thiadiazole-Based Thiazolidinones: MurA and MurB Inhibitors
2.2.2. Phenyl Thiazolyl Urea and Carbamate Derivatives: MurA and MurB Inhibitors
2.2.3. Pulvinones: MurA–MurD Inhibitors
2.2.4. 5′-Deoxy-5′-(4-substituted-1,2,3-triazol-1-yl)-uridine: MurE Inhibitor
3. MurE Inhibitors
3.1. Natural Products
3.2. Tetrahydoisoquinolines
3.3. N-Methyl-2-alkenyl-4-quinolones
3.4. 3-Bromo-4,5-dihydroisoxazole
3.5. Phosphinates as Dual MurD and MurE Inhibtors
3.6. 5-Benzylidenethiazolidin-4-one Derivatives as Dual MurC-MurF Inhibitors
3.7. D-Glutamate-based 5-benzylidenethiazolidin-4-one Derivatives as MurD and MurE Inhibitors
3.8. Phosphorylated Hydroxyethylamines: MurC–MurF Inhibitors
3.9. Furan-Containing Compounds as MurC–MurF Inhibitors
3.10. Benzene-1,3-dicarboxylic Acids as MurD and MurE Inhibitors
3.11. Peptidosulfonamides: MurD & MurE Inhibitors
3.12. Naphthyl Tetronic Acids as MurA–MurE Inhibitors
4. Repurposed Drugs
5. Conclusions
6. Opportunities and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Global Tuberculosis Report 2021; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2021.
- Zhang, Y.; Vilchèze, C.; Jacobs, W.R. Mechanisms of Drug Resistance in Mycobacterium tuberculosis. In Tuberculosis and the Tubercle Bacillus; Wiley Online Library: Hoboken, NJ, USA; ASM Press: Washington, DC, USA, 2004; pp. 115–140. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for Bad Bugs: Confronting the Challenges of Antibacterial Discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef]
- Macielag, M.J. Chemical Properties of Antimicrobials and Their Uniqueness. Antibiot. Discov. Dev. 2012, 9781461414, 793–820. [Google Scholar] [CrossRef]
- Dhiman, R.; Singh, R. Critical Review Recent Advances for Identification of New Scaffolds and Drug Targets for Mycobacterium tuberculosis. Life 2018, 70, 905–916. [Google Scholar] [CrossRef]
- El Zoeiby, A.; Sanschagrin, F.; Levesque, R.C. Structure and Function of the Mur Enzymes: Development of Novel Inhibitors. Mol. Microbiol. 2003, 47, 1–12. [Google Scholar] [CrossRef]
- Anishetty, S.; Pulimi, M.; Pennathur, G. Potential Drug Targets in Mycobacterium Tuberculosis through Metabolic Pathway Analysis. Comput. Biol. Chem. 2005, 29, 368–378. [Google Scholar] [CrossRef]
- Mdluli, K.; Spigelman, M. Novel Targets for Tuberculosis Drug Discovery. Curr. Opin. Pharmacol. 2006, 6, 459–467. [Google Scholar] [CrossRef]
- Lamichhane, G.; Freundlich, J.S.; Ekins, S.; Wickramaratne, N.; Nolan, S.T.; Bishai, W.R. Essential Metabolites of Mycobacterium Tuberculosis and Their Mimics. MBio 2011, 2, e00301–e00310. [Google Scholar] [CrossRef]
- Gaur, V.; Bera, S. Recent Developments on UDP-N-Acetylmuramoyl-L-Alanine-D-Gutamate Ligase (Mur D) Enzyme for Antimicrobial Drug Development: An Emphasis on in-Silico Approaches. Curr. Res. Pharmacol. Drug Discov. 2022, 3, 100137. [Google Scholar] [CrossRef]
- Isa, M.A. Homology Modeling and Molecular Dynamic Simulation of UDP-N-Acetylmuramoyl-l-Alanine-d-Glutamate Ligase (MurD) from Mycobacterium Tuberculosis H37Rv Using in Silico Approach. Comput. Biol. Chem. 2019, 78, 116–126. [Google Scholar] [CrossRef]
- Eniyan, K.; Bajpai, U. Mur Ligases as Potential Drug Targets in Mycobacterium Tuberculosis. A Review. DU J. Undergrad. Res. Innov. 2017, 3, 97–115. [Google Scholar]
- Catalão, M.J.; Filipe, S.R.; Pimentel, M. Revisiting Anti-Tuberculosis Therapeutic Strategies That Target the Peptidoglycan Structure and Synthesis. Front. Microbiol. 2019, 10, 190. [Google Scholar] [CrossRef]
- Moraes, G.L.; Gomes, G.C.; Monteiro De Sousa, P.R.; Alves, C.N.; Govender, T.; Kruger, H.G.; Maguire, G.E.M.; Lamichhane, G.; Lameira, J. Structural and Functional Features of Enzymes of Mycobacterium Tuberculosis Peptidoglycan Biosynthesis as Targets for Drug Development. Tuberculosis 2015, 95, 95–111. [Google Scholar] [CrossRef]
- Eniyan, K.; Rani, J.; Ramachandran, S.; Bhat, R.; Khan, I.A.; Bajpai, U. Screening of Antitubercular Compound Library Identifies Inhibitors of Mur Enzymes in Mycobacterium Tuberculosis. SLAS Discov. 2020, 25, 70–78. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network Pharmacology: The next Paradigm in Drug Discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Bouhss, A.; Mengin-Lecreulx, D.; Blanot, D.; Van Heijenoort, J.; Parquet, C. Invariant Amino Acids in the Mur Peptide Synthetases of Bacterial Peptidoglycan Synthesis and Their Modification by Site-Directed Mutagenesis in the UDP-MurNAc:L-Alanine Ligase from Escherichia Coli. Biochemistry 1997, 36, 11556–11563. [Google Scholar] [CrossRef]
- El Zoeiby, A.; Beaumont, M.; Dubuc, E.; Sanschagrin, F.; Voyer, N.; Levesque, R.C. Combinatorial Enzymatic Assay for the Screening of a New Class of Bacterial Cell Wall Inhibitors. Bioorg. Med. Chem. 2003, 11, 1583–1592. [Google Scholar] [CrossRef]
- Gordon, E.; Flouret, B.; Chantalat, L.; Van Heijenoort, J.; Mengin-Lecreulx, D.; Dideberg, O. Crystal Structure of UDP-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: Meso-Diaminopimelate Ligase from Escherichia Coli. J. Biol. Chem. 2001, 276, 10999–11006. [Google Scholar] [CrossRef]
- Bouhss, A.; Dementin, S.; Parquet, C.; Mengin-Lecreulx, D.; Bertrand, J.A.; Le Beller, D.; Dideberg, O.; Van Heijenoort, J.; Blanot, D. Role of the Ortholog and Paralog Amino Acid Invariants in the Active Site of the UDP-MurNAc-L-Alanine:D-Glutamate Ligase (MurD). Biochemistry 1999, 38, 12240–12247. [Google Scholar] [CrossRef]
- Silver, L.L. Does the Cell Wall of Bacteria Remain a Viable Source of Targets for Novel Antibiotics? Biochem. Pharmacol. 2006, 71, 996–1005. [Google Scholar] [CrossRef]
- Peters, J.U. Polypharmacology—Foe or Friend? J. Med. Chem. 2013, 56, 8955–8971. [Google Scholar] [CrossRef]
- Proschak, E.; Stark, H.; Merk, D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. J. Med. Chem. 2019, 62, 420–444. [Google Scholar] [CrossRef]
- Katz, A.; Caufield, C. Structure-Based Design Approaches to Cell Wall Biosynthesis Inhibitors. Curr. Pharm. Des. 2003, 9, 857–866. [Google Scholar] [CrossRef]
- Image Generated from the RCSB PDB (rcsb.org) of PDB ID 1MBT. Available online: https://www.rcsb.org/3d-view/1MBT/1 (accessed on 18 February 2023).
- Eniyan, K.; Dharavath, S.; Vijayan, R.; Bajpai, U.; Gourinath, S. Crystal Structure of UDP-N-Acetylglucosamine-Enolpyruvate Reductase (MurB) from Mycobacterium Tuberculosis. BBA-Proteins Proteom. 2018, 1866, 397–406. [Google Scholar] [CrossRef]
- Image Generated from the RCSB PDB (rcsb.org) of PDB ID 2XJA. Available online: https://www.rcsb.org/3d-view/2XJA/1 (accessed on 18 February 2023).
- Basavannacharya, C.; Moody, P.R.; Munshi, T.; Cronin, N.; Keep, N.H.; Bhakta, S. Essential Residues for the Enzyme Activity of ATP-Dependent MurE Ligase from Mycobacterium Tuberculosis. Protein Cell 2010, 1, 1011–1022. [Google Scholar] [CrossRef]
- Andres, C.J.; Bronson, J.J.; Andrea, S.V.D.; Deshpande, M.S.; Falk, P.J.; Grant-young, K.A.; Harte, W.E.; Ho, H.; Misco, P.F.; Robertson, J.G.; et al. 4-Thiazolidinones: Novel Inhibitors of the Bacterial Enzyme MurB. Bioorg. Med. Chem. Lett. 2000, 10, 715–717. [Google Scholar] [CrossRef]
- Babaoglu, K.; Page, M.A.; Jones, V.C.; McNeil, M.R.; Dong, C.; Naismith, J.H.; Lee, R.E. Novel Inhibitors of an Emerging Target in Mycobacterium Tuberculosis; Substituted Thiazolidinones as Inhibitors of DTDP-Rhamnose Synthesis. Bioorg. Med. Chem. Lett. 2003, 13, 3227–3230. [Google Scholar] [CrossRef]
- Bronson, J.J.; Denbleyker, K.L.; Falk, P.J.; Mate, R.A.; Ho, H.-T.; Pucci, M.J.; Snyder, L.B. Discovery of the First Antibacterial Small Molecule Inhibitors of MurB. Bioorg. Med. Chem. Lett. 2003, 13, 873–875. [Google Scholar] [CrossRef]
- Viegas-Junior, C.; Barreiro, E.J. Carlos Alberto Manssour Fraga Molecular Hybridization: A Useful Tool in the Design of New Drug Prototypes. Curr. Med. Chem. 2007, 14, 1829–1852. [Google Scholar] [CrossRef]
- Tratrat, C. Novel Thiazole-Based Thiazolidinones as Potent Anti-Infective Agents: In Silico PASS and Toxicity Prediction, Synthesis, Biological Evaluation and Molecular Modelling. Comb. Chem. High Throughput Screen. 2020, 23, 126–140. [Google Scholar] [CrossRef]
- Hardy, B.; Douglas, N.; Helma, C.; Rautenberg, M.; Jeliazkova, N.; Jeliazkov, V.; Nikolova, I.; Benigni, R.; Tcheremenskaia, O.; Kramer, S.; et al. Collaborative Development of Predictive Toxicology Applications. J. Cheminform. 2010, 2, 7. [Google Scholar] [CrossRef]
- Haroun, M.; Tratrat, C.; Kositzi, K.; Tsolaki, E.; Petrou, A.; Aldhubiab, B.; Attimarad, M.; Harsha, S.; Geronikaki, A.; Venugopala, K.N.; et al. New Benzothiazole-Based Thiazolidinones as Potent Antimicrobial Agents. Design, Synthesis and Biological Evaluation. Curr. Top. Med. Chem. 2018, 18, 75–87. [Google Scholar] [CrossRef]
- Drwal, M.N.; Banerjee, P.; Dunkel, M.; Wettig, M.R.; Preissner, R. ProTox: A Web Server for the in Silico Prediction of Rodent Oral Toxicity. Nucleic Acids Res. 2014, 42, W53–W58. [Google Scholar] [CrossRef]
- Haroun, M.; Tratrat, C.; Petrou, A.; Geronikaki, A.; Ivanov, M.; Ciric, A.; Sokovic, M. 2-Aryl-3-(6-Trifluoromethoxy)Benzo[d]Thiazole-Based Thiazolidinone Hybrids as Potential Anti-Infective Agents: Synthesis, Biological Evaluation and Molecular Docking Studies. Bioorg. Med. Chem. Lett. 2021, 32, 127718. [Google Scholar] [CrossRef]
- Ahmed, S.; Zayed, M.F.; El-Messery, S.M.; Al-Agamy, M.H.; Abdel-Rahman, H.M. Design, Synthesis, Antimicrobial Evaluation and Molecular Modeling Study of 1,2,4-Triazole-Based 4-Thiazolidinones. Molecules 2016, 21, 568. [Google Scholar] [CrossRef]
- Šink, R.; Barreteau, H.; Patin, D.; Mengin-Lecreulx, D.; Gobec, S.; Blanot, D. MurD Enzymes: Some Recent Developments. Biomol. Concepts 2013, 4, 539–556. [Google Scholar] [CrossRef]
- Horishny, V.; Geronikaki, A.; Kartsev, V.; Matiychuk, V.; Petrou, A.; Pogodin, P.; Poroikov, V.; Papadopoulou, T.A.; Vizirianakis, I.S.; Kostic, M.; et al. Synthesis, Biological Evaluation and Molecular Docking Studies of 5-Indolylmethylen-4-Oxo-2-Thioxothiazolidine Derivatives. Molecules 2022, 27, 1068. [Google Scholar] [CrossRef]
- Gilbert, A.M.; Failli, A.; Shumsky, J.; Yang, Y.; Severin, A.; Singh, G.; Hu, W.; Keeney, D.; Petersen, P.J.; Katz, A.H. Pyrazolidine-3,5-Diones and 5-Hydroxy-1 H -Pyrazol-3(2 H)-Ones, Inhibitors of UDP- N -Acetylenolpyruvyl Glucosamine Reductase. J. Med. Chem. 2006, 49, 6027–6036. [Google Scholar] [CrossRef]
- Yang, Y.; Severin, A.; Chopra, R.; Krishnamurthy, G.; Singh, G.; Hu, W.; Keeney, D.; Svenson, K.; Petersen, P.J.; Labthavikul, P.; et al. 3,5-Dioxopyrazolidines, Novel Inhibitors of UDP-N-Acetylenolpyruvylglucosamine Reductase (MurB) with Activity against Gram-Positive Bacteria. Antimicrob. Agents Chemother. 2006, 50, 8543. [Google Scholar] [CrossRef]
- Kutterer, K.M.K.; Davis, J.M.; Singh, G.; Yang, Y.; Hu, W.; Severin, A.; Rasmussen, B.A.; Krishnamurthy, G.; Failli, A.; Katz, A.H. 4-Alkyl and 4,4′-Dialkyl 1,2-Bis(4-Chlorophenyl)Pyrazolidine-3,5-Dione Derivatives as New Inhibitors of Bacterial Cell Wall Biosynthesis. Chem. Screen. Sci. 2005, 15, 2527–2531. [Google Scholar] [CrossRef]
- Mekky, A.E.M.; Sanad, S.M.H. Novel Bis(Pyrazole-Benzofuran) Hybrids Possessing Piperazine Linker: Synthesis of Potent Bacterial Biofilm and MurB Inhibitors. Bioorg. Chem. 2020, 102, 104094. [Google Scholar] [CrossRef]
- Bhat, M.; Nagaraja, G.K.; Kayarmar, R.; Peethamber, S.K.; Shafeeulla, R.M. Design, Synthesis and Characterization of New 1,2,3-Triazolyl Pyrazole Derivatives as Potential Antimicrobial Agents: Via a Vilsmeier-Haack Reaction Approach. RSC Adv. 2016, 6, 59375–59388. [Google Scholar] [CrossRef]
- Konduri, S.; Bhargavi, D.; Prashanth, J.; Krishna, V.S.; Sriram, D.; Rao, K.P. Design and Synthesis of “Chloropicolinate Amides and Urea Derivatives” as Novel Inhibitors for Mycobacterium Tuberculosis. ACS Omega 2021, 6, 1657–1667. [Google Scholar] [CrossRef]
- Tang, Z.; Ye, W.; Chen, H.; Kuang, X.; Guo, J.; Xiang, M.; Peng, C.; Chen, X.; Liu, H. Role of Purines in Regulation of Metabolic Reprogramming. Purinergic Signal. 2019, 15, 423–438. [Google Scholar] [CrossRef]
- Renuka, J.; Reddy, K.I.; Srihari, K.; Jeankumar, V.U.; Shravan, M.; Sridevi, J.P.; Yogeeswari, P.; Babu, K.S.; Sriram, D. Design, Synthesis, Biological Evaluation of Substituted Benzofurans as DNA GyraseB Inhibitors of Mycobacterium Tuberculosis. Bioorg. Med. Chem. 2014, 22, 4924–4934. [Google Scholar] [CrossRef]
- D’hooghe, M.; Mollet, K.; De Vreese, R.; Jonckers, T.H.M.; Dams, G.; De Kimpe, N. Design, Synthesis, and Antiviral Evaluation of Purine-β-Lactam and Purine-Aminopropanol Hybrids. J. Med. Chem. 2012, 55, 5637–5641. [Google Scholar] [CrossRef]
- Kuo, T.C.; Li, L.W.; Pan, S.H.; Fang, J.M.; Liu, J.H.; Cheng, T.J.; Wang, C.J.; Hung, P.F.; Chen, H.Y.; Hong, T.M.; et al. Purine-Type Compounds Induce Microtubule Fragmentation and Lung Cancer Cell Death through Interaction with Katanin. J. Med. Chem. 2016, 59, 8521–8534. [Google Scholar] [CrossRef]
- Konduri, S.; Prashanth, J.; Krishna, V.S.; Sriram, D.; Behera, J.N.; Siegel, D.; Rao, K.P. Design and Synthesis of Purine Connected Piperazine Derivatives as Novel Inhibitors of Mycobacterium Tuberculosis. Bioorg. Med. Chem. Lett. 2020, 30, 127512. [Google Scholar] [CrossRef]
- Kamoutsis, C.; Fesatidou, M.; Petrou, A.; Geronikaki, A.; Poroikov, V.; Ivanov, M.; Soković, M.; Ćirić, A.; Carazo, A.; Mladěnka, P. Triazolo Based-Thiadiazole Derivatives. Synthesis, Biological Evaluation and Molecular Docking Studies. Antibiotics 2021, 10, 804. [Google Scholar] [CrossRef]
- Halama, A.; Zapadlo, M. Synthesis, Isolation, and Analysis of Stereoisomers of Sacubitril. Org. Process Res. Dev. 2019, 23, 102–107. [Google Scholar] [CrossRef]
- Ksander, G.M.; Ghai, R.D.; deJesus, R.; Diefenbacher, C.G.; Yuan, A.; Berry, C.; Sakane, Y.; Trapani, A. Dicarboxylic Acid Dipeptide Neutral Endopeptidase Inhibitors. J. Med. Chem. 1995, 38, 1689–1700. [Google Scholar] [CrossRef]
- Novel Drugs Summary 2015. Available online: https://www.Fda.Gov/Drugs/New-Drugs-Fda-Cders-New-Molecular-Entities-and-New-Therapeutic-Biological-Products/Novel-Drugs-Summary-2015 (accessed on 6 December 2022).
- Konduri, S.; Pogaku, V.; Prashanth, J.; Siva Krishna, V.; Sriram, D.; Basavoju, S.; Behera, J.N.; Prabhakara Rao, K. Sacubitril-Based Urea and Thiourea Derivatives as Novel Inhibitors for Anti-Tubercular against Dormant Tuberculosis. ChemistrySelect 2021, 6, 3869–3874. [Google Scholar] [CrossRef]
- Hrast, M.; Jukič, M.; Patin, D.; Tod, J.; Dowson, C.G.; Roper, D.I.; Barreteau, H.; Gobec, S. In Silico Identification, Synthesis and Biological Evaluation of Novel Tetrazole Inhibitors of MurB. Chem. Biol. Drug. Des. 2018, 91, 1101–1112. [Google Scholar] [CrossRef]
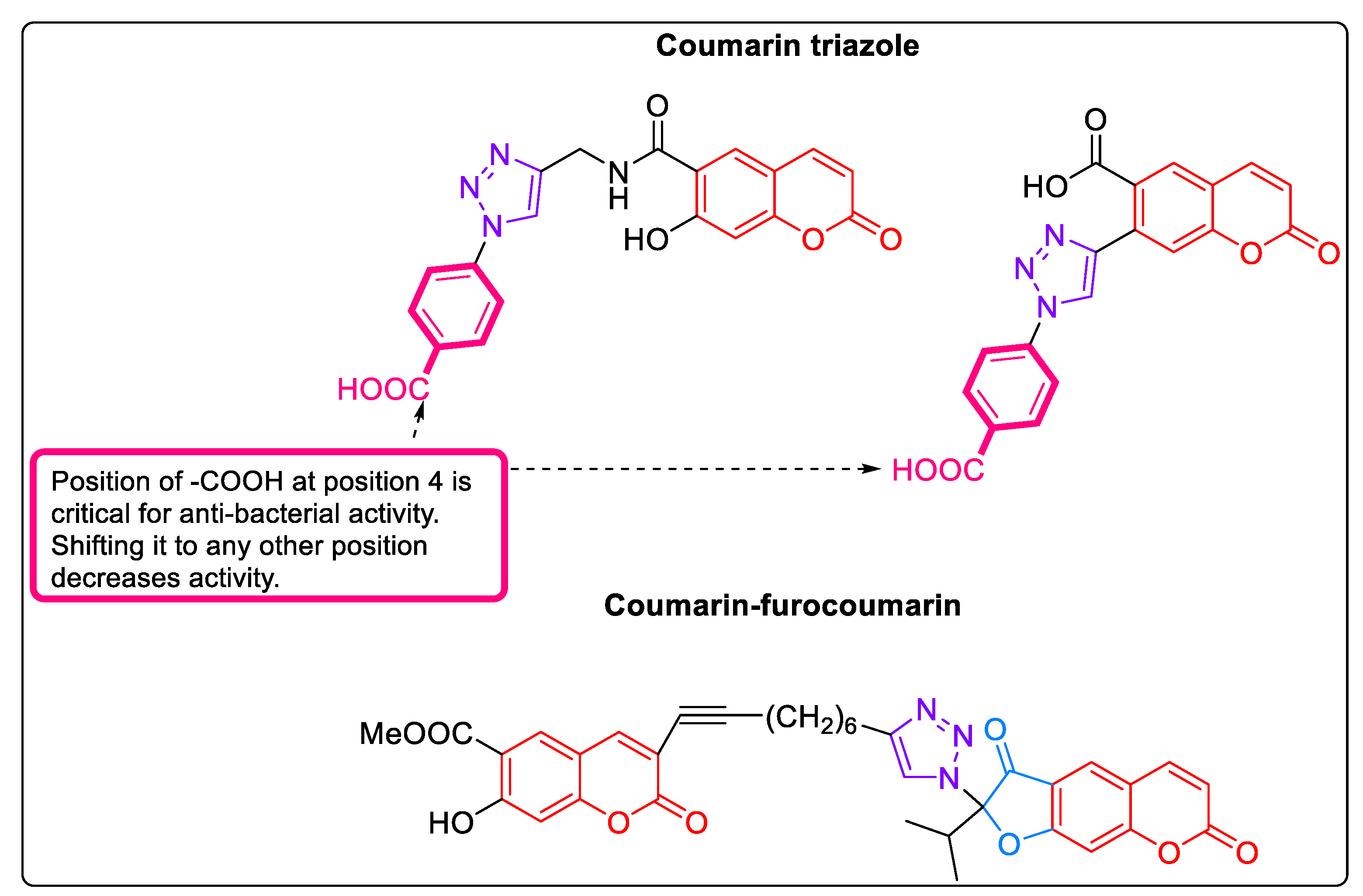
- Lipeeva, A.V.; Zakharov, D.O.; Burova, L.G.; Frolova, T.S.; Baev, D.S.; Shirokikh, I.V.; Evstropov, A.N.; Sinitsyna, O.I.; Tolsikova, T.G.; Shults, E.E. Design, Synthesis and Antibacterial Activity of Coumarin-1,2,3-Triazole Hybrids Obtained from Natural Furocoumarin Peucedanin. Molecules 2019, 24, 2126. [Google Scholar] [CrossRef]
- Fesatidou, M.; Zagaliotis, P.; Camoutsis, C.; Petrou, A.; Eleftheriou, P.; Tratrat, C.; Haroun, M.; Geronikaki, A.; Ciric, A.; Sokovic, M. 5-Adamantan Thiadiazole-Based Thiazolidinones as Antimicrobial Agents. Design, Synthesis, Molecular Docking and Evaluation. Bioorg. Med. Chem. 2018, 26, 4664–4676. [Google Scholar] [CrossRef]
- Proctor, R.A.; Dalal, S.C.; Kahl, B.; Brar, D.; Peters, G.; Nichols, W.W. Two Diarylurea Electron Transport Inhibitors Reduce Staphylococcus Aureus Hemolytic Activity and Protect Cultured Endothelial Cells from Lysis. Antimicrob. Agents Chemother. 2002, 46, 2333–2336. [Google Scholar] [CrossRef]
- Beaver, D.J.; Roman, D.P.; Stoffel, P.J. The Preparation and Bacteriostatic Activity of Substituted Ureas. J. Am. Chem. Soc. 1957, 79, 1236–1245. [Google Scholar] [CrossRef]
- Francisco, G.D.; Li, Z.; Albright, J.D.; Eudy, N.H.; Katz, A.H.; Petersen, P.J.; Labthavikul, P.; Singh, G.; Yang, Y.; Rasmussen, B.A.; et al. Phenyl Thiazolyl Urea and Carbamate Derivatives as New Inhibitors of Bacterial Cell-Wall Biosynthesis. Bioorg. Med. Chem. Lett. 2004, 14, 235–238. [Google Scholar] [CrossRef]
- Antane, S.; Caufield, C.E.; Hu, W.; Keeney, D.; Labthavikul, P.; Morris, K.; Naughton, S.M.; Petersen, P.J.; Rasmussen, B.A.; Singh, G.; et al. Pulvinones as Bacterial Cell Wall Biosynthesis Inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 176–180. [Google Scholar] [CrossRef]
- Hervin, V.; Arora, R.; Rani, J.; Ramchandran, S.; Bajpai, U.; Agrofoglio, L.A.; Roy, V. Design and Synthesis of Various 5′-Deoxy-5′-(4-Substituted-1,2,3-Triazol-1-Yl)-Uridine Analogues as Inhibitors of Mycobacterium Tuberculosis Mur Ligases. Molecules 2020, 25, 4953. [Google Scholar] [CrossRef]
- Gutierrez-Lugo, M.T.; Bewley, C.A. Natural Products, Small Molecules, and Genetics in Tuberculosis Drug Development. J. Med. Chem. 2008, 51, 2606–2612. [Google Scholar] [CrossRef]
- Osman, K.; Evangelopoulos, D.; Basavannacharya, C.; Gupta, A.; McHugh, T.D.; Bhakta, S.; Gibbons, S. An Antibacterial from Hypericum Acmosepalum Inhibits ATP-Dependent MurE Ligase from Mycobacterium Tuberculosis. Int. J. Antimicrob. Agents 2012, 39, 124–129. [Google Scholar] [CrossRef]
- Guzman, J.D.; Gupta, A.; Evangelopoulos, D.; Basavannacharya, C.; Pabon, L.C.; Plazas, E.A.; Muñoz, D.R.; Delgado, W.A.; Cuca, L.E.; Ribon, W.; et al. Anti-Tubercular Screening of Natural Products from Colombian Plants: 3-Methoxynordomesticine, an Inhibitor of MurE Ligase of Mycobacterium Tuberculosis. J. Antimicrob. Chemother. 2010, 65, 2101–2107. [Google Scholar] [CrossRef]
- Guzman, J.D.; Pesnot, T.; Barrera, D.A.; Davies, H.M.; McMahon, E.; Evangelopoulos, D.; Mortazavi, P.N.; Munshi, T.; Maitra, A.; Lamming, E.D.; et al. Tetrahydroisoquinolines Affect the Whole-Cell Phenotype of Mycobacterium Tuberculosis by Inhibiting the ATP-Dependent MurE Ligase. J. Antimicrob. Chemother. 2015, 70, 1691–1703. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X. DNA Gyrase, Topoisomerase IV, and the 4-Quinolones. Microbiol. Mol. Biol. Rev. 1997, 61, 377–392. [Google Scholar] [CrossRef]
- Guzman, J.D.; Wube, A.; Evangelopoulos, D.; Gupta, A.; Hüfner, A.; Basavannacharya, C.; Rahman, M.M.; Thomaschitz, C.; Bauer, R.; McHugh, T.D.; et al. Interaction of N-Methyl-2-Alkenyl-4-Quinolones with ATP-Dependent MurE Ligase of Mycobacterium Tuberculosis: Antibacterial Activity, Molecular Docking and Inhibition Kinetics. J. Antimicrob. Chemother. 2011, 66, 1766–1772. [Google Scholar] [CrossRef]
- Jöst, C.; Nitsche, C.; Scholz, T.; Roux, L.; Klein, C.D. Promiscuity and Selectivity in Covalent Enzyme Inhibition: A Systematic Study of Electrophilic Fragments. J. Med. Chem. 2014, 57, 7590–7599. [Google Scholar] [CrossRef]
- Zeng, B.; Wong, K.K.; Pompliano, D.L.; Reddy, S.; Tanner, M.E. A Phosphinate Inhibitor of the Meso -Diaminopimelic Acid-Adding Enzyme (MurE) of Peptidoglycan Biosynthesis. J. Org. Chem. 1998, 63, 10081–10085. [Google Scholar] [CrossRef]
- Štrancar, K.; Boniface, A.; Blanot, D.; Gobec, S. Phosphinate Inhibitors of UDP-N-Acetylmuramoyl-L-Alanyl-D-Glutamate: L-Lysine Ligase (MurE). Arch. Pharm. 2007, 340, 127–134. [Google Scholar] [CrossRef]
- Tomašić, T.; Kovač, A.; Simčič, M.; Blanot, D.; Grdadolnik, S.G.; Gobec, S.; Kikelj, D.; Peterlin Mašič, L. Novel 2-Thioxothiazolidin-4-One Inhibitors of Bacterial MurD Ligase Targeting D-Glu- and Diphosphate-Binding Sites. Eur. J. Med. Chem. 2011, 46, 3964–3975. [Google Scholar] [CrossRef]
- Tomašić, T.; Zidar, N.; Kovač, A.; Turk, S.; Simčič, M.; Blanot, D.; Müller-Premru, M.; Filipič, M.; Grdadolnik, S.G.; Zega, A.; et al. 5-Benzylidenethiazolidin-4-Ones as Multitarget Inhibitors of Bacterial Mur Ligases. ChemMedChem 2010, 5, 286–295. [Google Scholar] [CrossRef]
- Tomašić, T.; Šink, R.; Zidar, N.; Fic, A.; Contreras-Martel, C.; Dessen, A.; Patin, D.; Blanot, D.; Müller-Premru, M.; Gobec, S.; et al. Dual Inhibitor of MurD and MurE Ligases from Escherichia Coli and Staphylococcus Aureus. ACS Med. Chem. Lett. 2012, 3, 626–630. [Google Scholar] [CrossRef]
- Humljan, J.; Kotnik, M.; Boniface, A.; Šolmajer, T.; Urleb, U.; Blanot, D.; Gobec, S. A New Approach towards Peptidosulfonamides: Synthesis of Potential Inhibitors of Bacterial Peptidoglycan Biosynthesis Enzymes MurD and MurE. Tetrahedron 2006, 62, 10980–10988. [Google Scholar] [CrossRef]
- Sova, M.; Kovač, A.; Turk, S.; Hrast, M.; Blanot, D.; Gobec, S. Phosphorylated Hydroxyethylamines as Novel Inhibitors of the Bacterial Cell Wall Biosynthesis Enzymes MurC to MurF. Bioorg. Chem. 2009, 37, 217–222. [Google Scholar] [CrossRef]
- Sova, M.; Čadež, G.; Turk, S.; Majce, V.; Polanc, S.; Batson, S.; Lloyd, A.J.; Roper, D.I.; Fishwick, C.W.G.; Gobec, S. Design and Synthesis of New Hydroxyethylamines as Inhibitors of D-Alanyl-D-Lactate Ligase (VanA) and D-Alanyl-D-Alanine Ligase (DdlB). Bioorg. Med. Chem. Lett. 2009, 19, 1376–1379. [Google Scholar] [CrossRef]
- Perdih, A.; Hrast, M.; Pureber, K.; Barreteau, H.; Grdadolnik, S.G.; Kocjan, D.; Gobec, S.; Solmajer, T.; Wolber, G. Furan-Based Benzene Mono- and Dicarboxylic Acid Derivatives as Multiple Inhibitors of the Bacterial Mur Ligases (MurC–MurF): Experimental and Computational Characterization. J. Comput. Aided Mol. Des. 2015, 29, 541–560. [Google Scholar] [CrossRef]
- Sinko, W.; Wang, Y.; Zhu, W.; Zhang, Y.; Feixas, F.; Cox, C.L.; Mitchell, D.A.; Oldfield, E.; McCammon, J.A. Undecaprenyl Diphosphate Synthase Inhibitors: Antibacterial Drug Leads. J. Med. Chem. 2014, 57, 5693–5701. [Google Scholar] [CrossRef]
- Brvar, M.; Perdih, A.; Hodnik, V.; Renko, M.; Anderluh, G.; Jerala, R.; Solmajer, T. In Silico Discovery and Biophysical Evaluation of Novel 5-(2-Hydroxybenzylidene) Rhodanine Inhibitors of DNA Gyrase B. Bioorg. Med. Chem. 2012, 20, 2572–2580. [Google Scholar] [CrossRef]
- Xu, Y.; Brenning, B.; Clifford, A.; Vollmer, D.; Bearss, J.; Jones, C.; Mccarthy, V.; Shi, C.; Wolfe, B.; Aavula, B.; et al. Discovery of Novel Putative Inhibitors of UDP-GlcNAc 2-Epimerase as Potent Antibacterial Agents. ACS Med. Chem. Lett. 2013, 4, 1142–1147. [Google Scholar] [CrossRef]
- Tomasic, T.; Masic, L. Rhodanine as a Privileged Scaffold in Drug Discovery. Curr. Med. Chem. 2009, 16, 1596–1629. [Google Scholar] [CrossRef]
- Mariner, K.R.; Trowbridge, R.; Agarwal, A.K.; Miller, K.; O’Neill, A.J.; Fishwick, C.W.G.; Chopra, I. Furanyl-Rhodanines Are Unattractive Drug Candidates for Development as Inhibitors of Bacterial RNA Polymerase. Antimicrob. Agents Chemother. 2010, 54, 4506–4509. [Google Scholar] [CrossRef]
- Tomašić, T.; Peterlin Mašič, L. Rhodanine as a Scaffold in Drug Discovery: A Critical Review of Its Biological Activities and Mechanisms of Target Modulation. Expert Opin. Drug Discov. 2012, 7, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.; Walters, M.A. Chemistry: Chemical Con Artists Foil Drug Discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A. Structure, Function and Dynamics in the Mur Family of Bacterial Cell Wall Ligases. J. Mol. Biol. 2006, 362, 640–655. [Google Scholar] [CrossRef]
- Perdih, A.; Kovač, A.; Wolber, G.; Blanot, D.; Gobec, S.; Solmajer, T. Discovery of Novel Benzene 1,3-Dicarboxylic Acid Inhibitors of Bacterial MurD and MurE Ligases by Structure-Based Virtual Screening Approach. Bioorg. Med. Chem. Lett. 2009, 19, 2668–2673. [Google Scholar] [CrossRef]
- Perdih, A.; Hrast, M.; Barreteau, H.; Gobec, S.; Wolber, G.; Solmajer, T. Benzene-1,3-Dicarboxylic Acid 2,5-Dimethylpyrrole Derivatives as Multiple Inhibitors of Bacterial Mur Ligases (MurC-MurF). Bioorg. Med. Chem. 2014, 22, 4124–4134. [Google Scholar] [CrossRef]
- Obreza, A.; Gobec, S. Recent Advances in Design, Synthesis and Biological Activity of Aminoalkylsulfonates and Sulfonamidopeptides. Curr. Med. Chem. 2004, 11, 3263–3278. [Google Scholar] [CrossRef]
- Mansour, T.S.; Caufield, C.E.; Rasmussen, B.; Chopra, R.; Krishnamurthy, G.; Morris, K.M.; Svenson, K.; Bard, J.; Smeltzer, C.; Naughton, S.; et al. Naphthyl Tetronic Acids as Multi-Target Inhibitors of Bacterial Peptidoglycan Biosynthesis. ChemMedChem 2007, 2, 1414–1417. [Google Scholar] [CrossRef]
- Andronis, C.; Sharma, A.; Virvilis, V.; Deftereos, S.; Persidis, A. Literature Mining, Ontologies and Information Visualization for Drug Repurposing. Brief. Bioinform. 2011, 12, 357–368. [Google Scholar] [CrossRef]
- Rani, J.; Silla, Y.; Borah, K.; Ramachandran, S.; Bajpai, U. Repurposing of FDA-Approved Drugs to Target MurB and MurE Enzymes in Mycobacterium Tuberculosis. J. Biomol. Struct. Dyn. 2020, 38, 2521–2532. [Google Scholar] [CrossRef]
- Haller, L.; Sossouhounto, R.; Coulibaly, I.M.; Dosso, M.; Kone, M.; Adom, H.; Meyer, U.A.; Betschart, B.; Wenk, M.; Haefeli, W.E.; et al. Isoniazid plus Sulphadoxine-Pyrimethamine Can Reduce Morbidity of HIV-Positive Patients Treated for Tuberculosis in Africa: A Controlled Clinical Trial. Chemotherapy 1999, 45, 452–465. [Google Scholar] [CrossRef]
- Ghofrani, H.A.; Osterloh, I.H.; Grimminger, F. Sildenafil: From Angina to Erectile Dysfunction to Pulmonary Hypertension and Beyond. Nat. Rev. Drug Discov. 2006, 5, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Shen Lee, B.; Toyos, M.; Karpecki, P.; Schiffbauer, J.; Sheppard, J. Selective Pharmacologic Therapies for Dry Eye Disease Treatment: Efficacy, Tolerability, and Safety Data Review from Preclinical Studies and Pivotal Trials. Ophthalmol. Ther. 2022, 11, 1333–1369. [Google Scholar] [CrossRef] [PubMed]
- Bruning, J.B.; Murillo, A.C.; Chacon, O.; Barletta, R.G.; Sacchettini, J.C. Structure of the Mycobacterium Tuberculosis D-Alanine:D-Alanine Ligase, a Target of the Antituberculosis Drug D-Cycloserine. Antimicrob. Agents Chemother. 2011, 55, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Eniyan, K.; Kumar, A.; Rayasam, G.V.; Perdih, A.; Bajpai, U. Development of a One-Pot Assay for Screening and Identification of Mur Pathway Inhibitors in Mycobacterium Tuberculosis. Sci. Rep. 2016, 6, 35134. [Google Scholar] [CrossRef]
- Hrast, M.; Rožman, K.; Ogris, I.; Škedelj, V.; Patin, D.; Sova, M.; Barreteau, H.; Gobec, S.; Grdadolnik, S.G.; Zega, A. Evaluation of the Published Kinase Inhibitor Set to Identify Multiple Inhibitors of Bacterial ATP-Dependent Mur Ligases. J. Enzyme Inhib. Med. Chem. 2019, 34, 1010–1017. [Google Scholar] [CrossRef]
- Hrast, M.; Frlan, R.; Knez, D.; Zdovc, I.; Barreteau, H.; Gobec, S. Mur Ligases Inhibitors with Azastilbene Scaffold: Expanding the Structure–Activity Relationship. Bioorg. Med. Chem. Lett. 2021, 40, 127966. [Google Scholar] [CrossRef]
- Kumar, V.; Saravanan, P.; Arvind, A.; Mohan, C.G. Identification of Hotspot Regions of MurB Oxidoreductase Enzyme Using Homology Modeling, Molecular Dynamics and Molecular Docking Techniques. J. Mol. Model. 2011, 17, 939–953. [Google Scholar] [CrossRef]



| R Group | Interactions with MurB |
|---|---|
| 4-Cl | Most potent compound against all tested strains (including mycobacterium). -NH- of the triazole and oxygen of methoxy form H-bond. Phenyl ring attached to thiazolidinone forms cationic-arene interaction. |
| 4-NO2 | Forms a triad of H-bonding interactions where -NH- of the triazole and carbonyl of the thiazolidinone are involved. Phenyl ring attached to thiazolidinone forms cationic-arene interaction. |
| 4-F | Forms triad of H-bonding interactions, as the -NO2 group above does. |
| 4-CH3 | Forms H-bond via the carbonyl group of the thiazolidinone ring. |
| 4-OCH3 | Oxygen of methoxy is involved in H-bonding interaction. Phenyl ring attached to thiazolidinone forms cationic-arene interaction. |
| X | Y | R | Effect on Activity |
|---|---|---|---|
| H | CH3 | 3-OH | Beneficial for anti-bacterial activity. The carbonyl oxygen forms an H-bond with Ser288 (a crucial interaction for inhibition, as this residue takes part in the proton transfer at the second stage of peptidoglycan synthesis). The hydroxy group forms H-bonds with active site residues. This compound also has a slight effect on biofilm inhibition. |
| 5-OCH3 | H | 3-COOH | Both methoxy and carboxylic acid groups are important for activity. Replacement of the carboxyl group with hydroxyl or removal of methoxy group led to decreases in activity. |
| 6-OCH3 | H | 4-OH; 3-COOH | These substituents are beneficial for activity. Flipping the order of the R- groups, and thereby generating a 3-OH, 4-COOH analog, led to decrease in activity. |
| Sr.no | Class ID | Class | Figure | Compound | MIC/IC50 (Organisms) |
|---|---|---|---|---|---|
| 1 | 2.1.1 | 4-thiazolidinones [29,30] | Figure 2-1 | 2-(2-(4-(4-(tert-butyl)phenoxy)phenyl)-4-oxothiazolidin-3-yl)hexanoic acid | 7.7 μM (EC) |
| 2 | 2.1.2 | Imidazolinone- a thiazolidinone bioisostere [31] | Figure 2-2 | (S)-2-(5-(3-(4-(tert-butyl)phenoxy)phenyl)-3-(2-((3,4-dichlorobenzyl)amino)-2-oxoethyl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl)hexanoic acid | 15 μM (EC) |
| 3 | 2.1.3 | Benzylidene thiazolylimino thiazolidinones [32,33,34] | Figure 2-3 | 2-(5-methylthiazol-2-ylimino)-5-(3-nitrobenzyliden) thiazolidin-4-one | 43.3 ± 0.03 μM (EC) |
| 4 | 2.1.4 | Benzylidene benzothiazolo thiazolidinones [34,35,36] | Figure 2-4 | 5-((Z)-3-chlorobenzylidene)-2-((6-methoxybenzo[d]thiazol-2-yl)imino)thiazolidin-4-one | 0.18 ± 0.06 μM (EC) |
| 5 | 2.1.5 | Benzo[d]thiazole-based thiazolidinones [37] | Figure 2-5 | 2-(2,3-dichlorophenyl)-3-(6-(trifluoromethoxy)benzo[d]thiazol-2-yl)thiazolidin-4-one | 0.12 ± 0.001 mg/mL (EC) |
| 6 | 2.1.6 | 1,2,4-Triazole-based 4-thiazolidinones [38] | Figure 2-6 | 2-(4-Chlorophenyl)-3-(5-(4-methoxyphenyl)-2H-1,2,4-triazol-3-yl)thiazolidin-4-one | 16 μM (EC) |
| 7 | 2.1.7 | 5-Indolylmethylen-4-oxo-2-thioxothiazolidines [39,40] | Figure 2-7 | (Z)-3-(3-Hydroxyphenyl)-5-(1-methyl-1H-indol-3-ylmethylene)-2-thioxothiazolidin-4-one | 12.28 ± 0.1 μM (EC) |
| 8 | 2.1.8 | 3,5-Dioxopyrazolidinedione and its derivatives [41,42,43] | Figure 2-8, 9, 10, 11 | 4-(4-Butoxybenzoyl)-1,2-bis(4-chlorophenyl)-5-hydroxy-1,2-dihydropyrazol-3-one | 4.5 μM (EC) |
| 9 | 2.1.9 | Bis(pyrazole-benzofuran) hybrids possessing piperazine linker [44] | Figure 2-12 | 1,4-Bis[((2-(3-acetyl-1-(4-nitrophenyl)-1H-pyrazole-4-yl)carbonyl)benzofuran-5-yl)methyl]piperazine | 3.1 µM (EC) |
| 10 | 2.1.10 | 1,2,3-triazolyl pyrazole derivatives [45] | Figure 3 | 3-(1-(2,3-dichloro-6-methyl-5-(trifluoromethyl)phenyl)-5-methyl-1H-1,2,3-triazol-4-yl)-1-(phthalazin-1-yl)-1H-pyrazole-4-carbaldehyde | 10 ± 0.3 μg/mL (EC) |
| 11 | 2.1.11 | Chloropicolinate amide, urea and thiourea derivatives [46] | Figure 4 | Methyl4-amino-3-chloro-6-(2-(3-fluorobenzamido)phenyl)picolinate | 7.86 μM (Mtb) |
| 12 | 2.1.12 | Purine-2,6-dione linked piperazine derivatives [47,48,49,50,51] | Figure 5-14 | 4-(7-(but-2-yn-1-yl)-1-(3,4-difluorobenzyl)-3-methyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)-N-(4-(trifluoromethyl)phenyl)piperazine-1-carboxamide | 5.08 ± 0.4 μM (Mtb) |
| 13 | 2.1.13 | Triazolo-Thiadiazole derivatives [52] | Figure 5-15 | 4,5-dimethoxy-N,N-dimethyl-2-((6-phenyl-2l2,4l4-[1,2,4]triazolo [3,4-b][1,3,4]thiadiazol-3-yl)methyl)benzenesulfonamide | 5 μg/mL (EC) |
| 14 | 2.1.14 | Sacubitril-based urea and thiourea derivatives [53,54,55,56] | Figure 5-16 | Ethyl (2R,4S)-5-([1,1′-biphenyl]-4-yl)-2-methyl-4-(3-(4-nitrophenyl)thioureido)pentanoate and Ethyl (2R,4S)-5-([1,1′-biphenyl]-4-yl)-4-(3-(3,4-dichlorophenyl)thioureido)-2-methylpentanoate | 6.25 μg/mL (EC) |
| 15 | 2.1.15 | 5-substituted tetrazol-2-yl acetamides [57] | Figure 5-17 | 2-(5-(4-chlorophenyl)-2H-tetrazol-2-yl)-N-phenylacetamide | 25 ± 3 μM (EC) |
| 16 | 2.1.16 | Coumarins [58] | Figure 6 | Methyl 3-(8-(1-(2-ethyl-3,7-dioxo-2,3-dihydro-7H-furo [3,2-g]chromen-2-yl)-1H-1,2,3-triazol-4-yl)oct-1-yn-1-yl)-7-hydroxy-2-oxo-2H-chromene-6-carboxylate | 68.75 ± 11.97 μM (SA) |
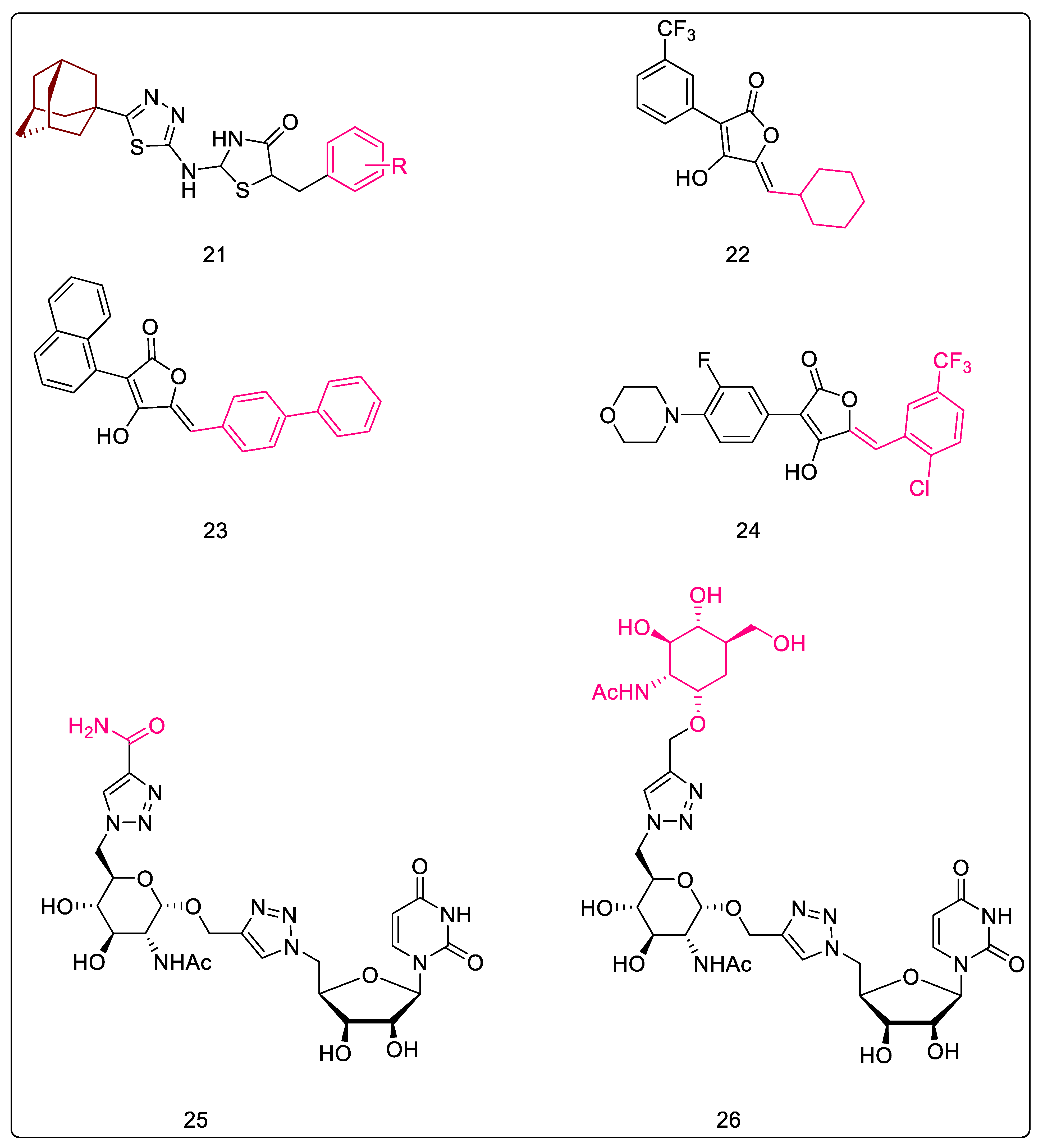
| 17 | 2.2.1 | 5-Adamantan thiadiazole-based thiazolidinones [59] | Figure 7-21 | 2-((5-((3R,5R,7R)-adamantan-1-yl)-1,3,4-thiadiazol-2-yl)amino)-5-(4-nitrobenzyl)thiazolidin-4-one | 0.022 μM (EC) |
| 18 | 2.2.2 | Phenyl thiazolyl urea and carbamate derivatives [60,61,62] | - | 1-(4-(tert-butyl)-5-cyanothiazol-2-yl)-3-(3,4-dichlorophenyl)urea | 6.2 μg/mL (EC); 2.8 μg/mL (SA) |
| 19 | 2.2.3 | Pulvinones [63] | Figure 7-22, 23, 24 | 5-(2-chloro-5-(trifluoromethyl)benzylidene)-3-(3-fluoro-4-morpholinophenyl)-4-hydroxyfuran-2(5H)-one | 1 μg/mL (EC) |
| 20 | 2.2.4 | 5′-deoxy-5′-(4-substituted-1,2,3-triazol-1-yl)-uridine [64] | Figure 7-25, 26 | N-((2S,3R,4R,5S,6R)-6-((4-((((1S,2R,3R,4R,5R)-2-acetamido-3,4-dihydroxy-5-(hydroxymethyl)cyclohexyl)oxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)-2-((1-(((2R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)-1H-1,2,3-triazol-4-yl)methoxy)-4,5-dihydroxytetrahydro-2H-pyran-3-yl)acetamide | ≥50% inhibition at 100 µM (EC) |
| 21 | 3.1 | Natural products [65,66,67] | - | 3-methoxynordomesticine | 57 ± 14 µM (Mtb) |
| 22 | 3.2 | Tetrahydoisoquinolines [68] | - | 1-(benzo[d][1,3]dioxol-5-ylmethyl)-8-methoxy-5-methyl-1,2,3,4-tetrahydroisoquinoline | <111 μM (Mtb) |
| 23 | 3.3 | N-methyl-2-alkenyl-4-quinolones [69,70] | Figure 8-27 | (Z)-1-methyl-2-(tetradec-5-en-1-yl)quinolin-4(1H)-one | 36 ± 16 μM (Mtb) |
| 24 | 3.4 | 3-bromo-4,5-dihydroisoxazole [71] | Figure 8-28 | 3-bromo-N-(4-nitrophenyl)-4,5-dihydroisoxazole-5-carboxamide | 44% Inhibition (EC) |
| 25 | 3.5 | Phosphinates as dual MurD and MurE inhibitors [72,73] | Figure 8-29 | 2-((hydroxy(1-((S)-2-methyl-3-(((4-nitrophenyl)methyl)sulfonamido)propanamido)ethyl)phosphoryl)methyl)pentanedioic acid | 13% RA (EC) |
| 26 | 3.6 | 5-benzylidenethiazolidin-4-one derivatives as dual MurC–MurF inhibitors [29,74,75] | Figure 9 | 2-thioxo-5-(2,3,4-trihydroxybenzylidene)thiazolidin-4-one | 6 μM (EC) |
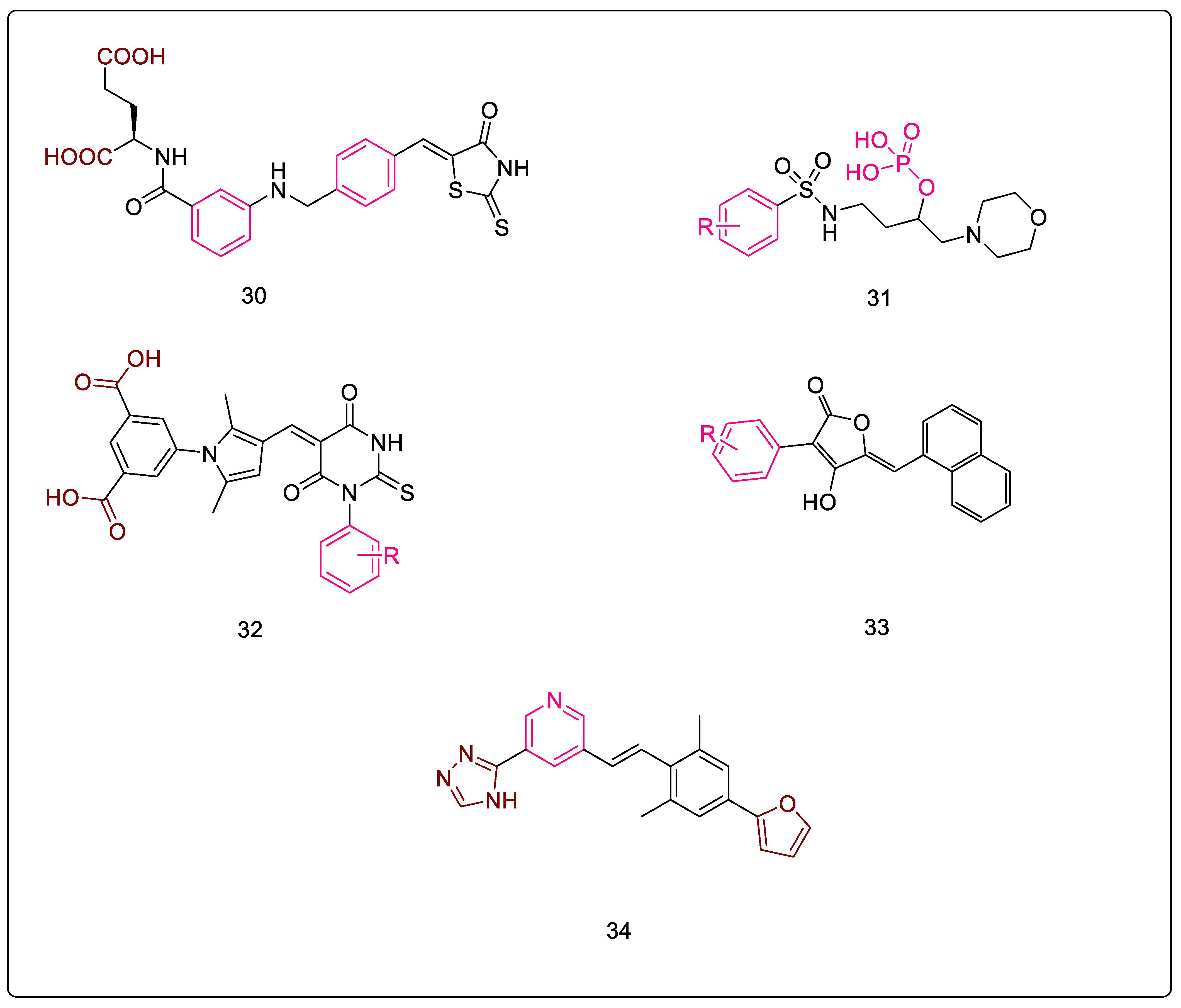
| 27 | 3.7 | D-gluatamate-based 5-benzylidenethiazolidin-4-one derivatives as MurD and MurE inhibitors [76] | Figure 10-30 | (3-((4-((4-oxo-2-thioxothiazolidin-5-ylidene)methyl)benzyl)amino)benzoyl)-D-glutamic acid | 180 μM (EC); 17 μM (SA) |
| 28 | 3.8 | Phosphorylated hydroxyethylamines: MurCMurF inhibitors [77,78,79] | Figure 10-31 | 4-((4-methoxyphenyl)sulfonamido)-1-morpholinobutan-2-yl dihydrogen phosphate | 6 μM (SA) |
| 29 | 3.9 | Furan containing compounds as MurC–MurF inhibitors [80,81,82,83,84,85,86,87,88] | Figure 10-34 | (E)-5-(5-((3-(benzo[d][1,3]dioxol-5-yl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)furan-2-yl)isophthalic acid | 32–94 μM (SA) |
| 30 | 3.10 | Benzene-1,3-dicarboxylic acids as MurD and MurE inhibitors [89,90] | Figure 10-32 | 5-(5-((3-(benzo[d][1,3]dioxol-5-yl)-4-oxo-2-thioxothiazolidin-5-ylidene)methyl)furan-2-yl)isophthalic acid | 32 Μm (EC) |
| 31 | 3.11 | Peptidosulfonamides: MurD and MurE inhibitors [91] | ((-2-([1,1′-biphenyl]-4-sulfonamido)propyl)sulfonyl)-D-glutamic acid | 12 μM (EC) | |
| 32 | 3.12 | Naphthyl tetronic acids as MurA–MurE inhibitors [92] | Figure 10-33 | 3-(3,5-dichlorophenyl)-4-hydroxy-5-(naphthalen-1-ylmethyl)furan-2(5H)-one | 16 μM (EC) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehta, K.; Khambete, M.; Abhyankar, A.; Omri, A. Anti-Tuberculosis Mur Inhibitors: Structural Insights and the Way Ahead for Development of Novel Agents. Pharmaceuticals 2023, 16, 377. https://doi.org/10.3390/ph16030377
Mehta K, Khambete M, Abhyankar A, Omri A. Anti-Tuberculosis Mur Inhibitors: Structural Insights and the Way Ahead for Development of Novel Agents. Pharmaceuticals. 2023; 16(3):377. https://doi.org/10.3390/ph16030377
Chicago/Turabian StyleMehta, Kunal, Mihir Khambete, Arundhati Abhyankar, and Abdelwahab Omri. 2023. "Anti-Tuberculosis Mur Inhibitors: Structural Insights and the Way Ahead for Development of Novel Agents" Pharmaceuticals 16, no. 3: 377. https://doi.org/10.3390/ph16030377
APA StyleMehta, K., Khambete, M., Abhyankar, A., & Omri, A. (2023). Anti-Tuberculosis Mur Inhibitors: Structural Insights and the Way Ahead for Development of Novel Agents. Pharmaceuticals, 16(3), 377. https://doi.org/10.3390/ph16030377