Virtual Screening of Benzimidazole Derivatives as Potential Triose Phosphate Isomerase Inhibitors with Biological Activity against Leishmania mexicana
Abstract
:1. Introduction
2. Results and Discussion
2.1. Binding Site Prediction and Molecular Docking of Control Ligands
2.2. LBSV in ZINC15
2.3. Leishmanicidal Activity
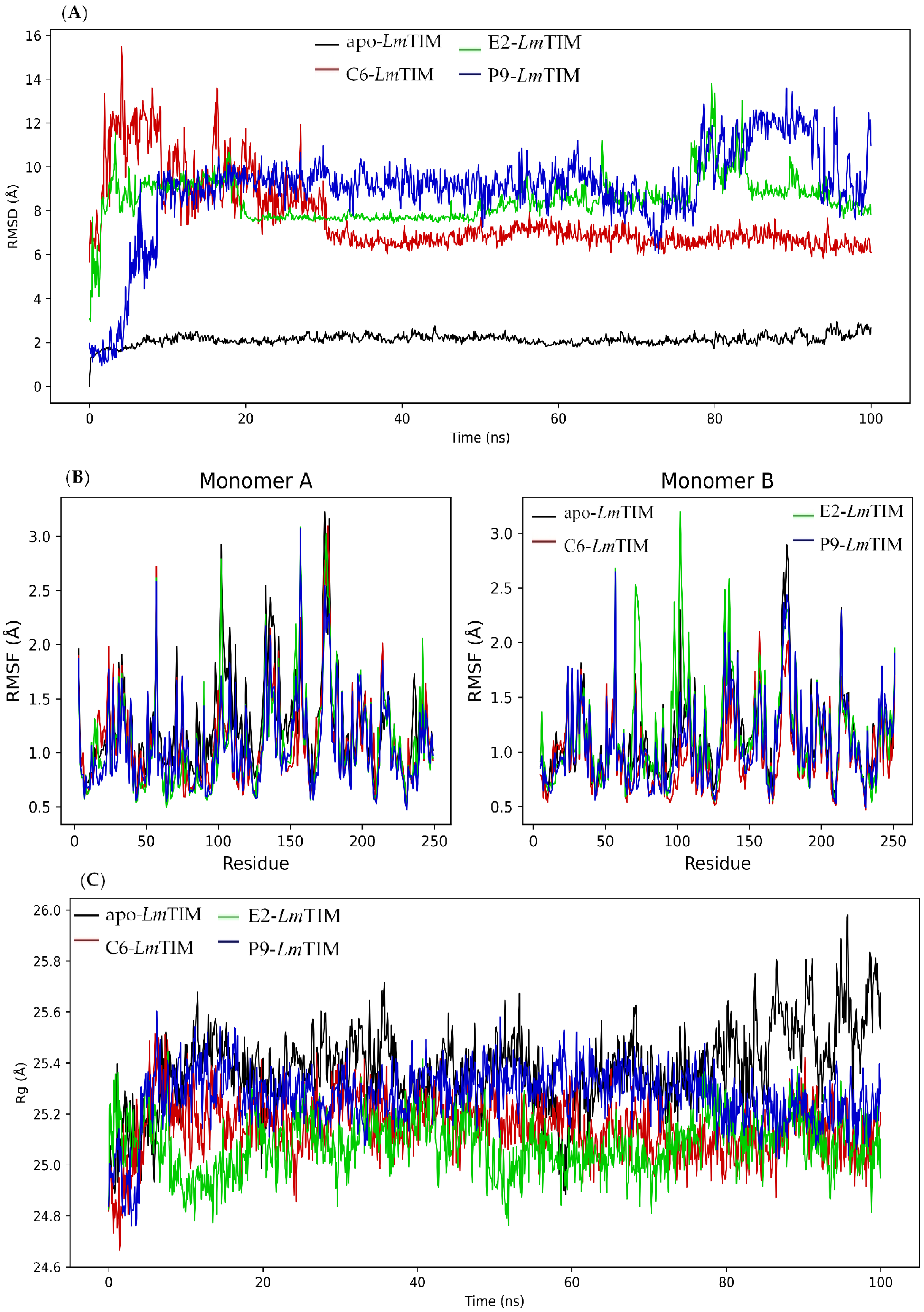
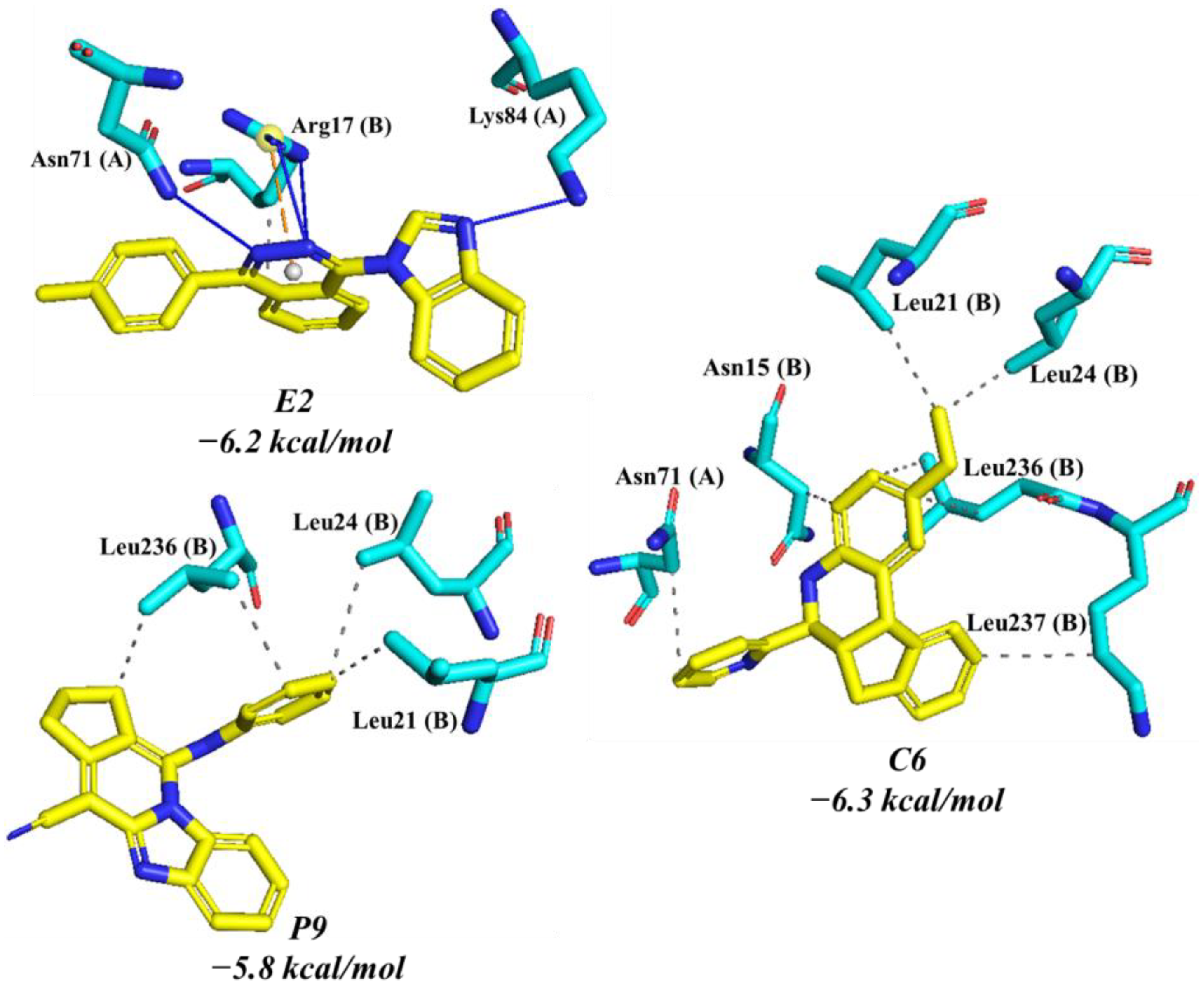
2.4. Molecular Dynamics Analysis
2.5. ADMET In Silico
2.6. Molecular Docking on HsTIM
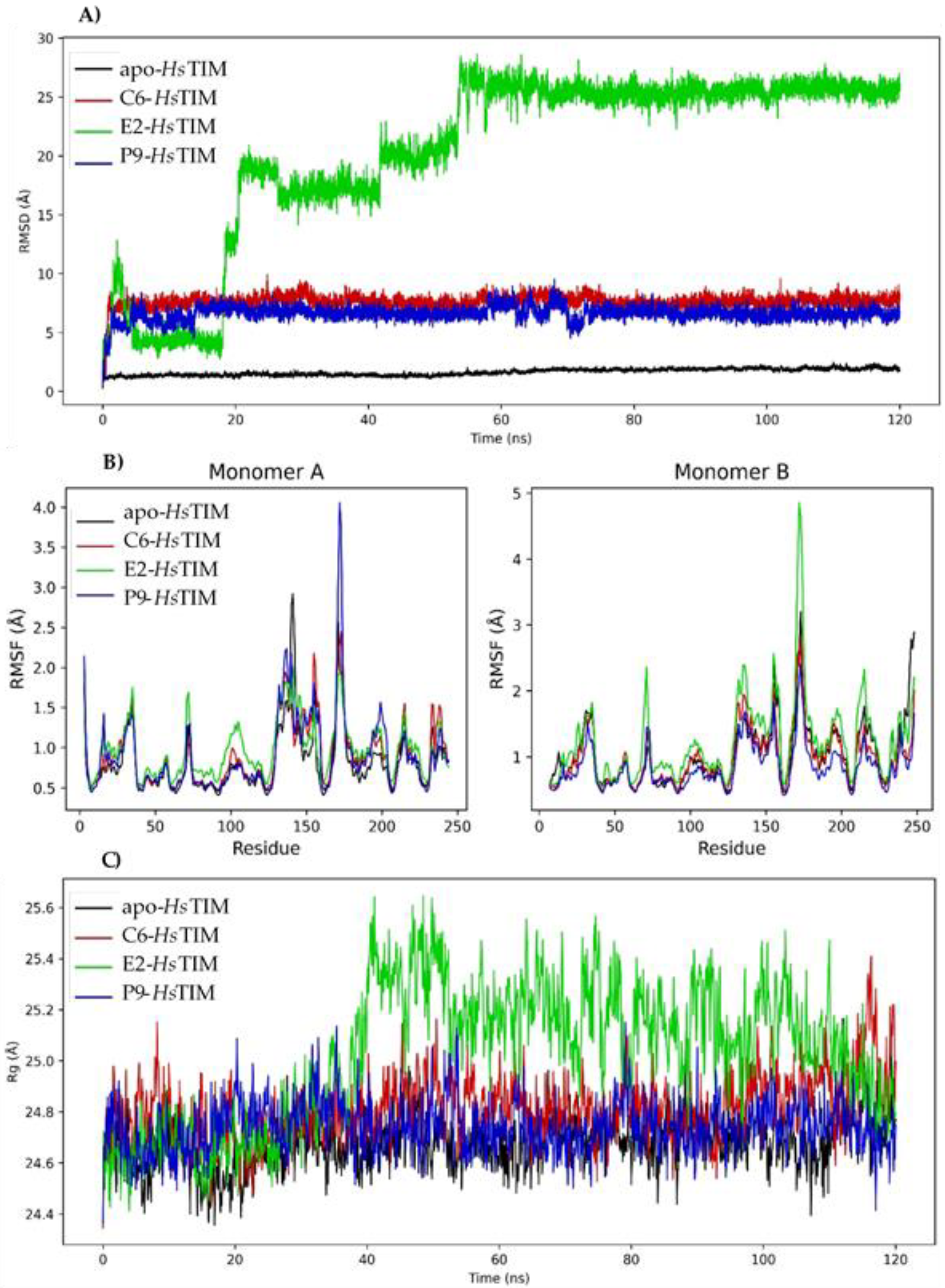
2.7. Molecular Dynamics Simulation on HsTIM
3. Materials and Methods
3.1. Control Compounds
3.2. Molecular Docking Analysis
3.3. Ligand-Based Virtual Screening
3.4. In Vitro Leishmanicidal Activity
3.5. Molecular Dynamics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abadías-Granado, I.; Diago, A.; Cerro, P.A.; Palma-Ruiz, A.M.; Gilaberte, Y. Cutaneous and Mucocutaneous Leishmaniasis. Actas Dermosifiliogr. 2021, 27, 601–618. [Google Scholar] [CrossRef]
- Zamora-Ledesma, S.; Hernández-Camacho, N.; Sánchez-Moreno, M.; Ruiz-Piña, H.; Villagrán-Herrera, M.E.; Marín-Sánchez, C.; Carrillo-Angeles, I.G.; Jones, R.W.; Camacho-Macías, B. Seropositivity for Trypanosoma cruzi and Leishmania mexicana in dogs from a metropolitan region of Central Mexico. Vet. Parasitol. Reg. Stud. Rep. 2020, 22, 100459. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization (PAHO). Cutaneous and Mucosal Leishmaniasis. Available online: https://www3.paho.org/hq/index.php?option=com_content&view=article&id=6417:2012-leishmaniasis-cutanea-mucosa&Itemid=39345&lang=en (accessed on 10 May 2022).
- Ulloa, J.L.; Spina, R.; Casasco, A.; Petray, P.B.; Martino, V.; Sosa, M.A.; Frank, F.M.; Muschietti, L.V. Germacranolide-type sesquiterpene lactones from Smallanthus sonchifolius with promising activity against Leishmania mexicana and Trypanosoma cruzi. Parasit. Vectors. 2017, 10, 567. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Control of the Leishmaniases. Report of a meeting of the WHO Expert Committee on the Control of Leishmaniases. WHO Technol. Rep. Ser. 2010, 949, 1–186. [Google Scholar]
- DNDi-Drugs for Neglected Diseases Initiative. Available online: http://www.dndi.org/diseases-projects/leishmaniasis/ (accessed on 31 May 2022).
- Matadamas-Martínez, F.; Hernández-Campos, A.; Téllez-Valencia, A.; Vázquez-Raygoza, A.; Comparán-Alarcón, S.; Yépez-Mulia, L.; Castillo, R. Leishmania mexicana Trypanothione Reductase Inhibitors: Computational and Biological Studies. Molecules 2019, 24, 3216. [Google Scholar] [CrossRef] [Green Version]
- De Sousa, L.R.; Wu, H.; Nebo, L.; Fernandes, J.B.; da Silva, M.F.; Kiefer, W.; Schirmeister, T.; Vieira, P.C. Natural products as inhibitors of recombinant cathepsin L of Leishmania mexicana. Exp. Parasitol. 2015, 156, 42–48. [Google Scholar] [CrossRef]
- Suresh, S.; Bressi, J.C.; Kennedy, K.J.; Verlinde, C.L.; Gelb, M.H.; Hol, W.G. Conformational changes in Leishmania mexicana glyceraldehyde-3-phosphate dehydrogenase induced by designed inhibitors. J. Mol. Biol. 2001, 309, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Olivares-Illana, V.; Pérez-Montfort, R.; López-Calahorra, F.; Costas, M.; Rodríguez-Romero, A.; Tuena de Gómez-Puyou, M.; Gómez Puyou, A. Structural differences in triosephosphate isomerase from different species and discovery of a multi-trypanosomatid inhibitor. Biochemistry 2006, 45, 2556–2560. [Google Scholar] [CrossRef]
- Velanker, S.S.; Ray, S.S.; Gokhale, R.S.; Suma, S.; Balaram, H.; Balaram, P.; Murthy, M.R. Triosephosphate isomerase from Plasmodium falciparum: The crystal structure provides insights into antimalarial drug design. Structure 1997, 5, 751–761. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Raygoza, A.; Cano-González, L.; Velázquez-Martínez, I.; Trejo-Soto, P.; Castillo, R.; Hernández-Campos, A.; Hernández-Luis, F.; Oria-Hernández, J.; Castillo-Villanueva, A.; Avitia-Domínguez, C.; et al. Species-Specific Inactivation of Triosephosphate Isomerase from Trypanosoma brucei: Kinetic and Molecular Dynamics Studies. Molecules 2017, 22, 2055. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Puyou, A.; Saavedra-Lira, E.; Becker, I.; Zubillaga, R.A.; Rojo-Domínguez, A.; Perez-Montfort, R. Using evolutionary changes to achieve species-specific inhibition of enzyme action-studies with triosephosphate isomerase. Chem. Biol. 1995, 2, 847. [Google Scholar] [CrossRef] [Green Version]
- Sanabria-Ayala, V.; Belmont, I.; Abraham, L. Triosephosphate isomerase of Taenia solium (TTPI): Phage display and antibodies as tools for finding target regions to inhibit catalytic activity. Parasitol. Res. 2015, 114, 55. [Google Scholar] [CrossRef]
- Saramago, L.; Gomes, H.; Aguilera, E.; Cerecetto, H.; González, M.; Cabrera, M.; Alzugaray, M.; daSilva Vaz Junior, I.; Nunes da Fonseca, R.; Aguirre-López, B.; et al. Novel and Selective Rhipicephalus microplus Triosephosphate Isomerase Inhibitors with Acaricidal Activity. Vet. Sci. 2018, 5, 74. [Google Scholar] [CrossRef] [Green Version]
- Juárez-Saldivar, A.; Barbosa-Cabrera, E.; Lara-Ramírez, E.E.; Paz-González, A.D.; Martínez-Vázquez, A.V.; Bocanegra-García, V.; Palos, I.; Campillo, N.E.; Rivera, G. Virtual Screening of FDA-Approved Drugs against Triose Phosphate Isomerase from Entamoeba histolytica and Giardia lamblia Identifies Inhibitors of Their Trophozoite Growth Phase. Int. J. Mol. Sci. 2021, 22, 5943. [Google Scholar] [CrossRef]
- Azahar, N.H.; Ab Dullah, S.; Abdullah, R.; Ahmat, N.; Md Akim, A.; Ab Hamid, H. Mutagenic Study of Benzimidazole Derivatives with (+S9) and without (-S9) Metabolic Activation. Int. J. Mol. Sci. 2019, 20, 4324. [Google Scholar] [CrossRef] [Green Version]
- Tonelli, M.; Novelli, F.; Tasso, B.; Vazzana, I.; Sparatore, A.; Boido, V.; Sparatore, F.; La Colla, P.; Sanna, G.; Giliberti, G.; et al. Antiviral activity of benzimidazole derivatives. III. Novel anti-CVB-5, anti-RSV and anti-Sb-1 agents. Bioorg. Med. Chem. 2014, 22, 4893–4909. [Google Scholar] [CrossRef]
- Picanço, G.A.; de Lima, N.F.; Gomes, T.C.; Fraga, C.M.D.; Alves, D.S.M.M.; Castillo, R.; da Costa, T.L.; Lino-Junior, R.S.; Ambrosio, J.; Vinaud, M.C. Partial inhibition of the main energetic pathways and its metabolic consequences after in vivo treatment with benzimidazole derivatives in experimental neurocysticercosis. Parasitology 2019, 146, 1578–1582. [Google Scholar] [CrossRef]
- Sirim, M.M.; Krishna, V.S.; Sriram, D.M.; Unsal Tan, O. Novel benzimidazole-acrylonitrile hybrids and their derivatives: Design, synthesis and antimycobacterial activity. Eur. J. Med. Chem. 2020, 188, 112010. [Google Scholar] [CrossRef]
- Tonelli, M.; Gabriele, E.; Piazza, F.; Basilico, N.; Parapini, S.; Tasso, B.; Loddo, R.; Sparatore, F.; Sparatore, A. Benzimidazole derivatives endowed with potent antileishmanial activity. J. Enzyme Inhib. Med. Chem. 2018, 33, 210–226. [Google Scholar] [CrossRef] [Green Version]
- Babkov, D.A.; Zhukowskaya, O.N.; Borisov, A.V.; Babkova, V.A.; Sokolova, E.V.; Brigadirova, A.A.; Litvinov, R.A.; Kolodina, A.A.; Morkovnik, A.S.; Sochnev, V.; et al. Towards multi-target antidiabetic agents: Discovery of biphenyl-benzimidazole conjugates as AMPK activators. Bioorg. Med. Chem. Lett. 2019, 29, 2443–2447. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Ko, P.W.; Chang, Y.J.; Kapoor, M.; Liang, Y.C.; Chu, H.L.; Lin, H.H.; Horng, J.C.; Hsu, M.H. Design and Synthesis of Benzimidazole-Chalcone Derivatives as Potential Anticancer Agents. Molecules 2019, 24, 3259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Gómez, H.; Hernández-Núñez, E.; León-Rivera, I.; Guerrero-Alvarez, J.; Cedillo-Rivera, R.; Moo-Puc, R.; Argotte-Ramos, R.; Rodríguez-Gutiérrez, M.C.; Chan-Bacab, M.J.; Navarrete-Vázquez, G. Design, synthesis and in vitro antiprotozoal activity of benzimidazole-pentamidine hybrids. Bioorg. Med. Chem. Lett. 2008, 18, 3147–3151. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Meneses, R.; Castillo, R.; Hernández-Campos, A.; Maldonado-Rangel, A.; Matius-Ruiz, J.B.; Trejo-Soto, P.J.; Nogueda-Torres, B.; Dea-Ayuela, M.A.; Bolás-Fernández, F.; Méndez-Cuesta, C.; et al. In vitro activity of new N-benzyl-1H-benzimidazol-2-amine derivatives against cutaneous, mucocutaneous and visceral Leishmania species. Exp. Parasitol. 2018, 184, 82–89. [Google Scholar] [CrossRef]
- Hernández-Chinea, C.; Carbajo, E.; Sojo, F.; Arvelo, F.; Kouznetsov, V.V.; Romero-Bohórquez, A.R.; Romero, P.J. In Vitro Activity of Synthetic tetrahydroindeno[2,1-c]quinolines on Leishmania Mexicana. Parasitol. Int. 2015, 64, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef] [Green Version]
- Breznik, M.; Ge, Y.; Bluck, J.P.; Briem, H.; Hahn, D.F.; Christ, C.D.; Mortier, J.; Mobley, D.L.; Meier, K. Prioritizing Small Sets of Molecules for Synthesis through in-silico Tools: A Comparison of Common Ranking Methods. Chem. Med. Chem. 2023, 18, e202200425. [Google Scholar] [CrossRef]
- Espinoza-Fonseca, L.M.; Trujillo-Ferrara, J.G. Toward a rational design of selective multi-trypanosomatid inhibitors: A computational docking study. Bioorg. Med. Chem. Lett. 2006, 16, 6288–6292. [Google Scholar] [CrossRef]
- Velázquez-López, J.M.; Hernández-Campos, A.; Yépez-Mulia, L.; Téllez-Valencia, A.; Flores-Carrillo, P.; Nieto-Meneses, R.; Castillo, R. Synthesis and trypanocidal activity of novel benzimidazole derivatives. Bioorg. Med. Chem. Lett. 2016, 26, 4377–4381. [Google Scholar] [CrossRef]
- Kurkcuoglu, Z.; Ural, G.; Demet Akten, E.; Doruker, P. Blind Dockings of Benzothiazoles to Multiple Receptor Confor-mations of Triosephosphate Isomerase from Trypanosoma cruzi and Human. Mol. Inform. 2011, 30, 986–995. [Google Scholar] [CrossRef]
- Téllez-Valencia, A.; Avila-Ríos, S.; Pérez-Montfort, R.; Rodríguez-Romero, A.; de Gómez-Puyou, M.T.; López-Calahorra, F.; Gómez-Puyou, A. Highly Specific Inactivation of Triosephosphate Isomerase from Trypanosoma Cruzi. Biochem. Biophys. Res. Commun. 2002, 295, 958–963. [Google Scholar] [CrossRef]
- Romo-Mancillas, A.; Téllez-Valencia, A.; Yépez-Mulia, L.; Hernández-Luis, F.; Hernández-Campo, A.; Castillo, R. The design and inhibitory profile of new benzimidazole derivatives against triosephosphate isomerase from Trypanosoma cruzi: A problem of residue motility. J. Mol. Graph Model. 2011, 30, 90–99. [Google Scholar] [CrossRef]
- Bansal, Y.; Silakari, O. The therapeutic journey of benzimidazoles: A review. Bioorg Med. Chem. 2012, 20, 6208–6236. [Google Scholar] [CrossRef]
- Beltran-Hortelano, I.; Alcolea, V.; Font, M.; Pérez-Silanes, S. The role of imidazole and benzimidazole heterocycles in Chagas disease: A review. Eur. J. Med. Chem. 2020, 206, 112692. [Google Scholar] [CrossRef]
- De Vivo, M.; Masetti, M.; Bottegoni, G.; Cavalli, A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 2016, 59, 4035–4061. [Google Scholar] [CrossRef]
- Arooj, M.; Shehadi, I.; Nassab, C.N.; Mohamed, A.A. Physicochemical stability study of protein-benzoic acid complexes using molecular dynamics simulations. Amino Acids 2020, 52, 1353–1362. [Google Scholar] [CrossRef]
- Espinosa-Bustos, C.; Ortiz-Pérez, M.; González-González, A.; Zarate, A.M.; Rivera, G.; Belmont-Díaz, J.A.; Saavedra, E.; Cuellar, M.A.; Vázquez, K.; Salas, C.O. New Amino Naphthoquinone Derivatives as Anti-Trypanosoma cruzi Agents Targeting Trypanothione Reductase. Pharmaceutics 2022, 14, 1121. [Google Scholar] [CrossRef]
- Vázquez-Jiménez, L.K.; Juárez-Saldivar, A.; Gómez-Escobedo, R.; Delgado-Maldonado, T.; Méndez-Álvarez, D.; Palos, I.; Bandyopadhyay, D.; Gaona-Lopez, C.; Ortiz-Pérez, E.; Nogueda-Torres, B.; et al. Ligand-Based Virtual Screening and Molecular Docking of Benzimidazoles as Potential Inhibitors of Triosephosphate Isomerase Identified New Trypanocidal Agents. Int. J. Mol. Sci. 2022, 23, 10047. [Google Scholar] [CrossRef]
- Méndez-Álvarez, D.; Herrera-Mayorga, V.; Juárez-Saldivar, A.; Paz-González, A.D.; Ortiz-Pérez, E.; Bandyopadhyay, D.; Pérez-Sánchez, H.; Rivera, G. Ligand-based virtual screening, molecular docking, and molecular dynamics of eugenol analogs as potential acetylcholinesterase inhibitors with biological activity against Spodoptera frugiperda. Mol. Divers. 2022, 26, 2025–2037. [Google Scholar] [CrossRef]
- Collier, T.A.; Piggot, T.J.; Allison, J.R. Molecular Dynamics Simulation of Proteins. Methods Mol. Biol. 2020, 2073, 311–327. [Google Scholar] [CrossRef]
- Fatriansyah, J.F.; Rizqillah, R.K.; Yandi, M.Y.; Fadilah, Sahlan, M. Molecular docking and dynamics studies on propolis sulabiroin-A as a potential inhibitor of SARS-CoV-2. J. King Saud Univ. Sci. 2022, 34, 101707. [Google Scholar] [CrossRef]
- Wierenga, R.K.; Noble, M.E. Comparison of the refined crystal structures of liganded and unliganded chicken, yeast and trypanosomal triosephosphate isomerase. J. Mol. Biol. 1992, 224, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Evaluation of green tea polyphenols as novel corona virus (SARS-CoV-2) main protease (Mpro) inhibitors-an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2021, 39, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Kirschner, K.N. Principal component and clustering analysis on molecular dynamics data of the ribosomal L11·23S subdomain. J. Mol. Model. 2013, 19, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Singh, S. Dynamics of heroin molecule inside the lipid membrane: A molecular dynamics study. J. Mol. Model. 2019, 25, 121. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Mollazadeh, S.; Sahebkar, A.; Hadizadeh, F.; Behravan, J.; Arabzadeh, S. Structural and functional aspects of P-glycoprotein and its inhibitors. Life Sci. 2018, 214, 118–123. [Google Scholar] [CrossRef]
- Waghray, D.; Zhang, Q. Inhibit or Evade Multidrug Resistance P-Glycoprotein in Cancer Treatment. J. Med. Chem. 2018, 61, 5108–5121. [Google Scholar] [CrossRef]
- Berman, H.M.; Kleywegt, G.J.; Nakamura, H.; Markley, J.L. The future of the Protein Data Bank. Biopolymers 2013, 99, 218–222. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Fährrolfes, R.; Bietz, S.; Flachsenberg, F.; Meyder, A.; Nittinger, E.; Otto, T.; Volkamer, A.; Rarey, M. ProteinsPlus: A web portal for structure analysis of macromolecules. Nucleic Acids Res. 2017, 45, W337–W343. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Téllez-Valencia, A.; Olivares-Illana, V.; Hernandez-Santoyo, A.; Perez-Montfort, R.; Costas, M.; Rodriguez-Romero, A.; López-Calahorra, F.; de Gómez-Puyou, M.T.; Gómez-Puyou, A. Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface. J. Mol. Biol. 2004, 341, 1355–1365. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15--Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Joachim-Haupt, V.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic. Acids. Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Abraham, A.; Pedregosa, F.; Eickenberg, M.; Gervais, P.; Mueller, A.; Kossaifi, J.; Gramfort, A.; Thirion, B.; Varoquaux, G. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 2014, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- López-López, E.; Naveja, J.J.; Medina-Franco, J.L. DataWarrior: An evaluation of the open-source drug discovery tool. Expert. Opin. Drug. Discov. 2019, 14, 335–341. [Google Scholar] [CrossRef]
- Muñoz, V.; Moretti, C.; Sauvain, M.; Caron, C.; Porzel, A.; Massiot, G.; Richard, B.; Le Men-Olivier, L. Isolation of bis-indole alkaloids with antileishmanial and antibacterial activities from Peschiera van heurkii (Syn. Tabernamontana van heurkii). Planta Med. 1994, 60, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Inchausti, A.; Yaluff, G.; Rojas de Arias, A.; Torres, S.; Ferreira, M.E.; Nakayama, H.; Schinini, A.; Lorenzen, K.; Anke, T.; Fournet, A. Leishmanicidal and trypanocidal activity of extracts and secondary metabolites from Besidiomycetes. Phytotherapy Res. 1997, 11, 193–197. [Google Scholar] [CrossRef]
- Van-Der-Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Sousa-da-Silva, A.W.; Wim Vranken, F. ACPYPE AnteChamber PYthon Parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [Green Version]

| Chemical Structure of Control Ligands | |
|---|---|
 C1 DS= −7.2 Kcal/mol 1 IC50 = 0.71 μM [24] HI: Ile69B, Ala70B, Lys71B, Glu105B, Ile109B; HB: Gln112B, Ala70B, Lys71B, Lys113B; π-S: Phe75B, Tyr102A, Tyr103A |  C2 DS= −7.8 Kcal/mol 1 IC50 = 0.36 μM [24] HI: Ile69B, Ala70B, Lys71B, Glu105B, Ile109B; HB: Gln112B, Ala70B, Lys71B, Lys113B; π-S: Phe75B, Tyr103A |
 C3 DS= −7.3 Kcal/mol 1 IC50 = 2.62 μM, 2 IC50 = 0.28 μM [25] HI: Ile69B, Ala70B, Lys71B, Glu105B, Ile109B, Phe75B; HB: Gln112B, Lys113B; π-S: Tyr103(A) |  C4 DS= −7.1 Kcal/mol 1 IC50 = 3.21 μM, 2 IC50 = 0.26 μM [25] HI: Ile69B, Lys71B, Glu105B, Ile109B; HB: Tyr103A π-S: Phe75B, Tyr103A |
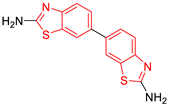 C5 DS= −7.6 Kcal/mol 70% enzyme inhibition at 100 µM [10] HI: Glu105B, Ile109B; HB: Tyr103A |  C6 DS= −9.2 Kcal/mol 1 IC50 = 1.02 μg/mL, Cell line J774 = 187.0 μg/mL; SI = 183.3 [26] HI: Ile69B, Phe75B, Ala70B, Tyr102A, Tyr103A, Ile109B, Glu105B |
| Group | Lead Compound | Group | Lead Compound |
|---|---|---|---|
| 1 (17 compounds) | 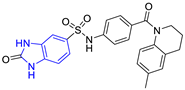 ZINC000030028937 (P1) DS= −9.6 Kcal/mol HI: Ala70B, Ile109B, Tyr102A, Ile69B, Lys71B; HB: Tyr103A, Lys71B, Ser72B, Ala74B; π-S: Phe75B, Tyr103A | 6 (11 compounds) | 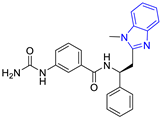 ZINC000010116378 (P6) DS= −9.3 Kcal/mol HI: Ile109B, Glu116B, Ala70B, Gln112B, Ile69B, Tyr102A, Phe75B; HB: Lys113B, Gln112B, Glu105B, Arg99B, Asn64B, Glu78B; π-S: Tyr103A, Tyr102A |
| 2 (24 compounds) | 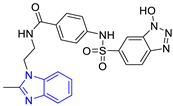 ZINC000010125646 (P2) DS= −9.6 Kcal/mol HI: Tyr103A, Tyr102A, Glu116B, Gln112B, Ile69B, HB: Ala70B, Lys113B; HB: Lys113B, Glu105B, Arg99B, Asn67B, Ala70B; π-S: Tyr103A, Tyr102A | 7 (22 compounds) |  ZINC000000182545 (P7) DS= −9.4 Kcal/mol HI: Tyr102A, Ile69B, Phe75B, Pys71B, Ala70B, Ile109B, Glu105B; HB: Tyr103A; π-S: Tyr103A |
| 3 (17 compounds) | 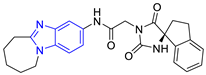 ZINC000030011443 (P3) DS= −9.6 Kcal/mol HI: Lys71B, Ile109B, Glu105B, Ile69B, Ala70B; HB: Lys71B, Gln112B, Ala70B; π-S: Tyr103A | 8 (9 compounds) |  ZINC000030009197 (P8) DS= −9.8 Kcal/mol HI: Ile109B, Glu105B, Ile69B, Phe75B; HB: Tyr103A, Gln112B, Tyr102A; π-S: Tyr103A |
| 4 (14 compounds) |  ZINC000010109617 (P4) DS= −10.3 Kcal/mol HI: Tyr103A, Ile109B, Phe75B, Ala70B, Glu116B, Gln112B, Lys71B, Ile69B; HB: Ala70B, Gln112B, Glu105B, Arg99B, Tyr103B, Lys113B | 9 (23 compounds) |  ZINC000000183176 (P9) DS= −9.4 Kcal/mol HI: Lys71B, Gln112B, Glu116B, Ala70B, Lys113B, Tyr103A, Tyr102A, Phe75B; HB: Gln112B, Lys113B; π-S: Phe75B |
| 5 (19 compounds) | 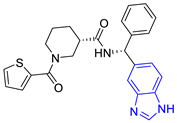 ZINC000040058994 (P5) DS= −9.4 Kcal/mol HI: Ile109B, Ala70B, Gln112B, Phe75B, Ile69B, Tyr102A, Tyr103A; HB: Lys113B, Tyr103B, Glu105B, Arg99B, Ala70B; π-S: Tyr103A | 10 (19 compounds) |  ZINC000030011418 (P10) DS= −9.7 Kcal/mol HI: Lys113B, Ala70B, Phe75B, Tyr102A, Ile69B, Ile109B, Tyr103A; HB: Lys113B, Ala70B, Lys71B, Gln112B, Glu105B; π-S: Tyr102A |
| Group | Lead Compound | Group | Lead Compound |
|---|---|---|---|
| 1 (20 compounds) | 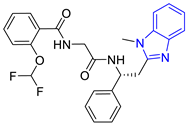 ZINC000010116253 (E1) DS= −10.8 Kcal/mol HI: Ile69B, Ile109B, Lys71B, Phe75B, Tyr102A; HB: Gln112B; π-S: Tyr103A | 6 (4 compounds) |  ZINC000000086631 (E6) DS= −9.0 Kcal/mol HI: Ile69B, Ile109B, Phe75B, Glu105B, Tyr102A; HB: Tyr102A; π-S: Tyr103A |
| 2 (26 compounds) |  ZINC000000087857 (E2) DS= −9.4 Kcal/mol HI: Ile69B, Ile109B, Phe75B, Tyr103A, Gln112B, Glu116B; HB: Gln112B | 7 (1 compound) | 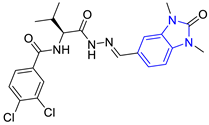 ZINC000010000495 (E7) DS= −9.7 Kcal/mol HI: Ile69B, Ala70B, Ile109B, Phe75B, Tyr102A, Tyr103A, Gln112B; HB: Ala70B, Tyr103A, Lys113B, Lys71B |
| 3 (102 compounds) |  ZINC000000018961 (E3) DS= −9.3 Kcal/mol HI: Ile69B, Ala70B, Ile109B; HB: Ala70B, Gln112B; π-S: Phe75B, Tyr103A | 8 (4 compounds) |  ZINC000010126510 (E8) DS= −9.5 Kcal/mol HI: Ile69B, Ile109B, Phe75B, Tyr102A, Tyr103A; HB: Gln112B, Tyr103A, Tyr102A; π-S: Phe75B, Tyr102A |
| 4 (4 compounds) | 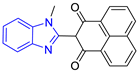 ZINC000000134117 (E4) DS= −9.3 Kcal/mol HI: Ala70B, Ile109B, Lys71B, Phe75B, Glu105B; HB: Gln112B, Lys113B; π-S: Tyr103A | 9 (3 compounds) | 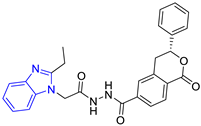 ZINC000010054868 (E9) DS= −9.4 Kcal/mol HI: Ile69B, Ile109B, Tyr103A; HB: Tyr102A; π-S: Phe75B; SB: Lys113B |
| 5 (6 compounds) |  ZINC000010116541 (E5) DS= −9.2 Kcal/mol HI: Ile69B, Lys71B, Glu105B; HB: Gln112B; π-S: Phe75B, Tyr103A, Tyr102A | 10 (2 compounds) | 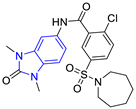 ZINC000020062149 (E10) DS= −9.1 Kcal/mol HI: Ile69B, Lys71B, Phe75B, Glu105B, Tyr103A; HB: Gln112B; π-S: Phe75B; Halogen: Lys71B |
| Compound | Promastigotes IC50 (µM) | |
|---|---|---|
| P9 | 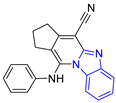 | >148.63 |
| E2 |  | 4.04 ± 0.39 |
| Pentamidine | 2.23 ± 0.79 | |
| Physicochemical Properties | Compounds | Pharmacokinetic Properties | Compounds | ||
|---|---|---|---|---|---|
| P9 | E2 | P9 | E2 | ||
| MW (g/mol) < 500 | 324.38 | 336.39 | Permeability BBB | Yes | Yes |
| Rotatable bonds < 10 | 2 | 2 | GI Absorption | Elevated | Elevated |
| Hydrogen bond acceptors < 10 | 2 | 3 | P-gp subtrate | Yes | Yes |
| Hydrogen bond donors < 5 | 1 | 0 | CYP1A2 inhibitor | Yes | Yes |
| TPSA (Å2) < 140 | 53.12 | 43.60 | CYP2C19 inhibitor | Yes | Yes |
| Log P < 5 | 3.97 | 4.31 | CYP2C9 inhibitor | Yes | No |
| Log S | Moderately soluble | Moderately soluble | CYP2D6 inhibitor | Yes | No |
| Hepatotoxicity | Inactive | Inactive | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez-Jiménez, L.K.; Juárez-Saldivar, A.; Chan-Bacab, M.J.; Delgado-Maldonado, T.; González-Morales, L.D.; Palos, I.; Ortiz-Pérez, E.; Lara-Ramírez, E.E.; Ramírez-Moreno, E.; Rivera, G. Virtual Screening of Benzimidazole Derivatives as Potential Triose Phosphate Isomerase Inhibitors with Biological Activity against Leishmania mexicana. Pharmaceuticals 2023, 16, 390. https://doi.org/10.3390/ph16030390
Vázquez-Jiménez LK, Juárez-Saldivar A, Chan-Bacab MJ, Delgado-Maldonado T, González-Morales LD, Palos I, Ortiz-Pérez E, Lara-Ramírez EE, Ramírez-Moreno E, Rivera G. Virtual Screening of Benzimidazole Derivatives as Potential Triose Phosphate Isomerase Inhibitors with Biological Activity against Leishmania mexicana. Pharmaceuticals. 2023; 16(3):390. https://doi.org/10.3390/ph16030390
Chicago/Turabian StyleVázquez-Jiménez, Lenci K., Alfredo Juárez-Saldivar, Manuel J. Chan-Bacab, Timoteo Delgado-Maldonado, Luis D. González-Morales, Isidro Palos, Eyra Ortiz-Pérez, Edgar E. Lara-Ramírez, Esther Ramírez-Moreno, and Gildardo Rivera. 2023. "Virtual Screening of Benzimidazole Derivatives as Potential Triose Phosphate Isomerase Inhibitors with Biological Activity against Leishmania mexicana" Pharmaceuticals 16, no. 3: 390. https://doi.org/10.3390/ph16030390
APA StyleVázquez-Jiménez, L. K., Juárez-Saldivar, A., Chan-Bacab, M. J., Delgado-Maldonado, T., González-Morales, L. D., Palos, I., Ortiz-Pérez, E., Lara-Ramírez, E. E., Ramírez-Moreno, E., & Rivera, G. (2023). Virtual Screening of Benzimidazole Derivatives as Potential Triose Phosphate Isomerase Inhibitors with Biological Activity against Leishmania mexicana. Pharmaceuticals, 16(3), 390. https://doi.org/10.3390/ph16030390









