Heparin Mimetics and Their Impact on Extracellular Matrix Protein Assemblies
Abstract
:1. Introduction
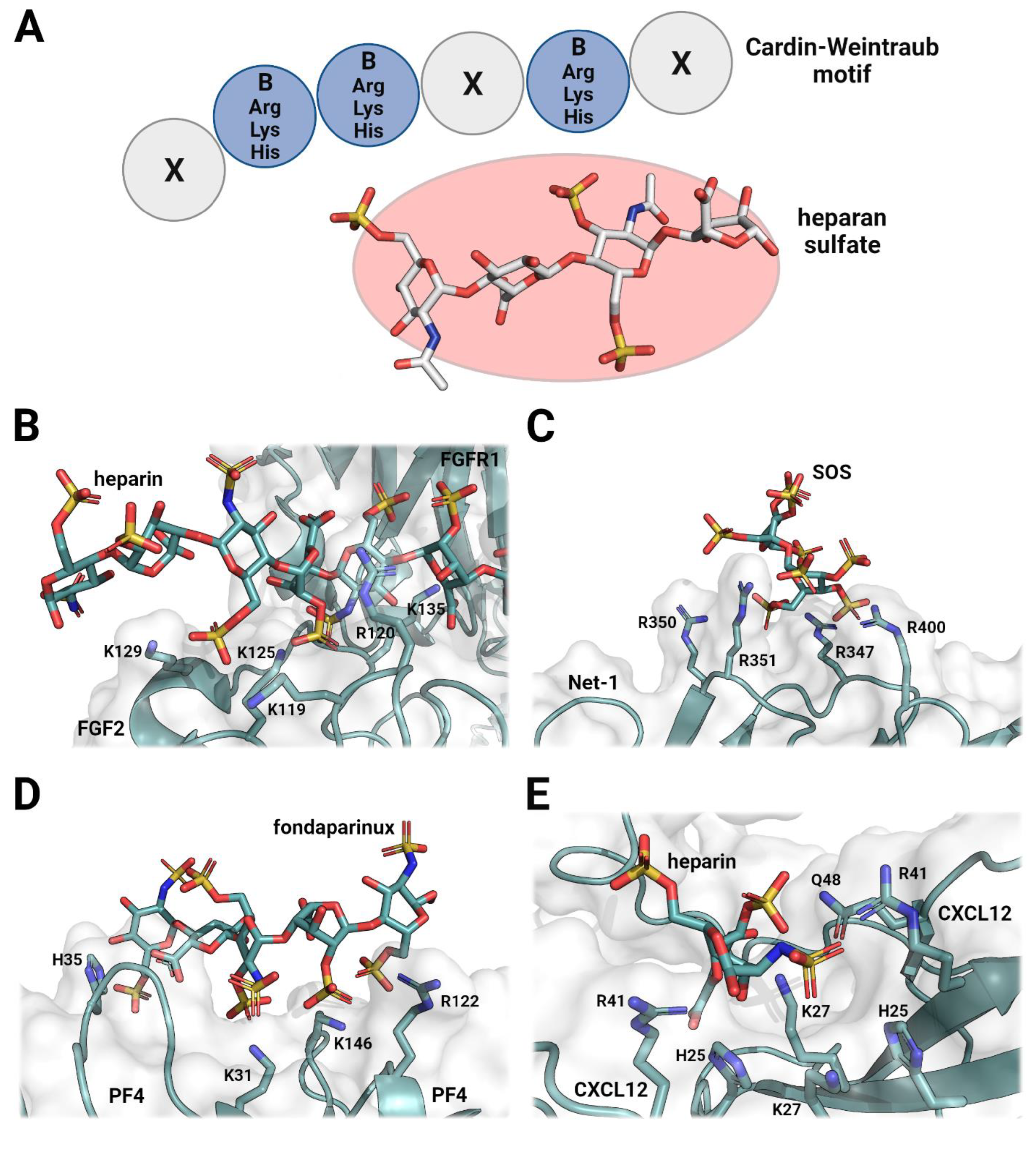
2. The Role of Heparan Sulfates in Cellular Signaling and Organization
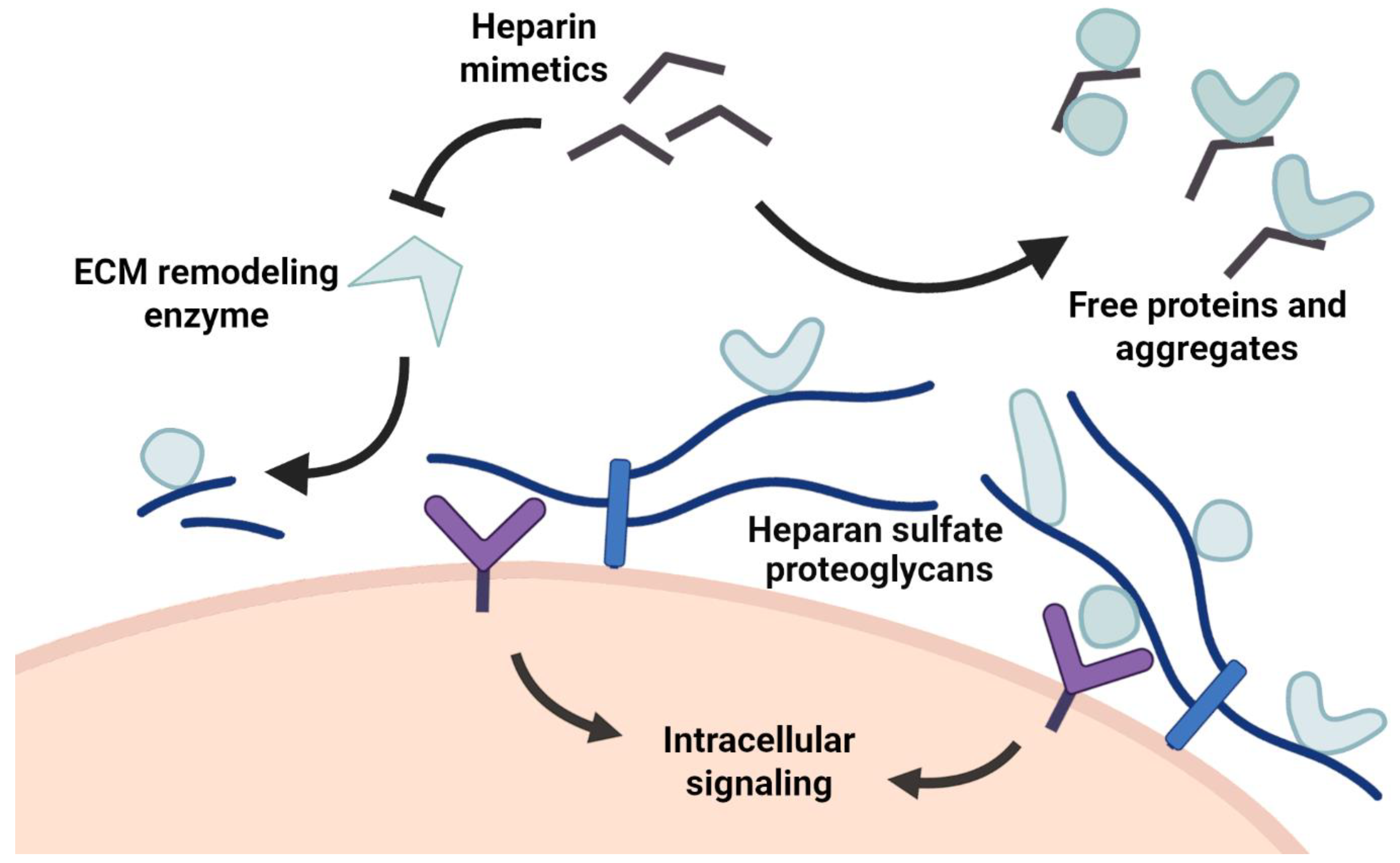
3. Heparin Mimetics Influence Extracellular Matrix Organization
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stickens, D.; Zak, B.M.; Rougier, N.; Esko, J.D.; Werb, Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development 2005, 132, 5055–5068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Wei, G.; Shi, Z.; Dryer, L.; Esko, J.D.; Wells, D.E.; Matzuk, M.M. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev. Biol. 2000, 224, 299–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iozzo, R.V.; Gubbiotti, M.A. Extracellular matrix: The driving force of mammalian diseases. Matrix Biol. 2018, 71–72, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Meneghetti, M.C.Z.; Hughes, A.; Rudd, T.; Nader, H.B.; Powell, A.K.; Yates, E.A.; Lima, M.A. Heparan sulfate and heparin interactions with proteins. J. R. Soc. Interface 2015, 12, 20150589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Kett, W.C.; Severin, I.C.; Agyekum, I.; Duan, J.; Amster, I.J.; Proudfoot, A.E.I.; Coombe, D.R.; Woods, R.J. The interaction of heparin tetrasaccharides with chemokine CCL5 is modulated by sulfation pattern and PH. J. Biol. Chem. 2015, 290, 15421–15436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Esko, J.D. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 2014, 83, 129–157. [Google Scholar] [CrossRef]
- Maaroufi, R.M.; Jozefowicz, M.; Tapon-Bretaudière, J.; Fischer, A.M. Mechanism of thrombin inhibition by antithrombin and heparin cofactor II in the presence of heparin. Biomaterials 1997, 18, 203–211. [Google Scholar] [CrossRef]
- Raitman, I.; Huang, M.L.; Williams, S.A.; Friedman, B.; Godula, K.; Schwarzbauer, J.E. Heparin-fibronectin interactions in the development of extracellular matrix insolubility. Matrix Biol. 2018, 67, 107–122. [Google Scholar] [CrossRef]
- Vallet, S.D.; Berthollier, C.; Ricard-Blum, S. The glycosaminoglycan interactome 2.0. Am. J. Physiol.-Cell Physiol. 2022, 322, C1271–C1278. [Google Scholar] [CrossRef]
- Litov, L.; Petkov, P.; Rangelov, M.; Ilieva, N.; Lilkova, E.; Todorova, N.; Krachmarova, E.; Malinova, K.; Gospodinov, A.; Hristova, R.; et al. Molecular mechanism of the anti-inflammatory action of heparin. Int. J. Mol. Sci. 2021, 22, 10730. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, M.; Doherty, G.G.; See, N.W.; Gandhi, N.S.; Ferro, V. From cancer to COVID-19: A perspective on targeting heparan sulfate-protein interactions. Chem. Rec. 2021, 21, 3087–3101. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, R.A.; Aliter, K.F.; Kar, S.; Mottamal, M. Sulfonated nonsaccharide heparin mimetics are potent and noncompetitive inhibitors of human neutrophil elastase. ACS Omega 2021, 6, 12699–12710. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.; Coombe, D. Heparin mimetics: Their therapeutic potential. Pharmaceuticals 2017, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Afosah, D.K.; Al-Horani, R.A.; Sankaranarayanan, N.V.; Desai, U.R. Potent, selective, allosteric inhibition of human plasmin by sulfa non-saccharide glycosaminoglycan mimetics. J. Med. Chem. 2017, 60, 641–657. [Google Scholar] [CrossRef] [PubMed]
- Nahain, A.A.; Ignjatovic, V.; Monagle, P.; Tsanaktsidis, J.; Ferro, V. Heparin mimetics with anticoagulant activity. Med. Res. Rev. 2018, 38, 1582–1613. [Google Scholar] [CrossRef] [Green Version]
- Gockel, L.M.; Heyes, M.; Li, H.; Al Nahain, A.; Gorzelanny, C.; Schlesinger, M.; Holdenrieder, S.; Li, J.P.; Ferro, V.; Bendas, G. Inhibition of tumor-host cell interactions using synthetic heparin mimetics. ACS Appl. Mater. Interfaces 2021, 13, 7080–7093. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, S.P.; Li, J.Y.; Chen, P.C.; Lee, Y.Z.; Li, K.M.; Zarivach, R.; Sun, Y.J.; Sue, S.C. Integrative Model to coordinate the oligomerization and aggregation mechanisms of CCL5. J. Mol. Biol. 2020, 432, 1143–1157. [Google Scholar] [CrossRef]
- Heide, F.; Legare, S.; To, V.; Gupta, M.; Gabir, H.; Imhof, T.; Moya-Torres, A.; McDougall, M.; Meier, M.; Koch, M.; et al. Heparins mediate the multimer assembly of secreted noggin. Protein Sci. 2022, 31, 1–13. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Xu, Y.; Brandt, S.; Mandelkow, M.; Raschke, R.; Strobel, U.; Delcea, M.; Zhou, W.; Liu, J.; Greinacher, A. Characterization of the interaction between platelet factor 4 and homogeneous synthetic low molecular weight heparins. J. Thromb. Haemost. 2020, 18, 390–398. [Google Scholar] [CrossRef] [Green Version]
- Cardin, A.D.; Weintraub, H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler. Off. J. Am. Heart Assoc. Inc. 1989, 9, 21–32. [Google Scholar] [CrossRef]
- Friedrich, U.; Blom, A.M.; Dahlbäck, B.; Villoutreix, B.O. Structural and energetic characteristics of the heparin-binding site in antithrombotic Protein, C. J. Biol. Chem. 2001, 276, 24122–24128. [Google Scholar] [CrossRef] [Green Version]
- Ricard-Blum, S.; Beraud, M.; Raynal, N.; Farndale, R.W.; Ruggiero, F. Structural requirements for heparin/heparan sulfate binding to type V collagen. J. Biol. Chem. 2006, 281, 25195–25204. [Google Scholar] [CrossRef] [Green Version]
- Billings, P.C.; Yang, E.; Mundy, C.; Pacifici, M. Domains with highest heparan sulfate–binding affinity reside at opposite ends in BMP2/4 versus BMP5/6/7: Implications for function. J. Biol. Chem. 2018, 293, 14371–14383. [Google Scholar] [CrossRef] [Green Version]
- Sandoval, D.R.; Gomez Toledo, A.; Painter, C.D.; Tota, E.M.; Sheikh, M.O.; West, A.M.V.; Frank, M.M.; Wells, L.; Xu, D.; Bicknell, R.; et al. Proteomics-based screening of the endothelial heparan sulfate interactome reveals that C-Type lectin 14a (CLEC14A) is a heparin-binding protein. J. Biol. Chem. 2020, 295, 2804–2821. [Google Scholar] [CrossRef] [Green Version]
- Schlessinger, J.; Plotnikov, A.N.; Ibrahimi, O.A.; Eliseenkova, A.V.; Yeh, B.K.; Yayon, A.; Linhardt, R.J.; Mohammadi, M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in fgfr binding and dimerization. Mol. Cell 2000, 6, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.C.; Schwab, R.A.; El Omari, K.; Bishop, B.; Iverson, E.J.; Malinauskas, T.; Dubey, R.; Qian, M.; Covey, D.F.; Gilbert, R.J.C.; et al. Hedgehog-interacting protein is a multimodal antagonist of hedgehog signalling. Nat. Commun. 2021, 12, 7171. [Google Scholar] [CrossRef] [PubMed]
- Paine-Saunders, S.; Viviano, B.L.; Economides, A.N.; Saunders, S. Heparan sulfate proteoglycans retain noggin at the cell surface. A potential mechanism for shaping bone morphogenetic protein gradients. J. Biol. Chem. 2002, 277, 2089–2096. [Google Scholar] [CrossRef] [Green Version]
- Proudfoot, A.E.I.; Handel, T.M.; Johnson, Z.; Lau, E.K.; LiWang, P.; Clark-Lewis, I.; Borlat, F.; Wells, T.N.C.; Kosco-Vilbois, M.H. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. USA 2003, 100, 1885–1890. [Google Scholar] [CrossRef] [Green Version]
- Kogut, M.M.; Marcisz, M.; Samsonov, S.A. Modeling glycosaminoglycan–protein complexes. Curr. Opin. Struct. Biol. 2022, 73. [Google Scholar] [CrossRef] [PubMed]
- Zagris, N. Extracellular matrix in development of the early embryo. Micron 2001, 32, 427–438. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; del Río Hernández, A. Role of extracellular matrix in development and cancer progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier, M.; Gupta, M.; Akgül, S.; McDougall, M.; Imhof, T.; Nikodemus, D.; Reuten, R.; Moya-Torres, A.; To, V.; Ferens, F.; et al. The dynamic nature of Netrin-1 and the structural basis for glycosaminoglycan fragment-induced filament formation. Nat. Commun. 2023, 14, 1226. [Google Scholar] [CrossRef]
- Cai, Z.; Yarovoi, S.V.; Zhu, Z.; Rauova, L.; Hayes, V.; Lebedeva, T.; Liu, Q.; Poncz, M.; Arepally, G.; Cines, D.B.; et al. Atomic description of the immune complex involved in heparin-induced thrombocytopenia. Nat. Commun. 2015, 6, 8277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, J.W.; Cho, Y.; Sachpatzidis, A.; Fan, C.; Hodsdon, M.E.; Lolis, E. Structural and functional basis of CXCL12 (Stromal Cell-Derived Factor-1α) binding to heparin. J. Biol. Chem. 2007, 282, 10018–10027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crijns, H.; Adyns, L.; Ganseman, E.; Cambier, S.; Vandekerckhove, E.; Pörtner, N.; Vanbrabant, L.; Struyf, S.; Gerlza, T.; Kungl, A.; et al. Affinity and specificity for binding to glycosaminoglycans can be tuned by adapting peptide length and sequence. Int. J. Mol. Sci. 2022, 23, 447. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Proudfoot, A.E.I. The biological relevance of chemokine–proteoglycan interactions. Biochem. Soc. Trans. 2006, 34, 422–426. [Google Scholar] [CrossRef] [Green Version]
- Jägers, C.; Roelink, H. Association of sonic hedgehog with the extracellular matrix requires its zinc-coordination center. BMC Mol. Cell Biol. 2021, 22, 1–18. [Google Scholar] [CrossRef]
- Nesterenko, A.M.; Orlov, E.E.; Ermakova, G.V.; Ivanov, I.A.; Semenyuk, P.I.; Orlov, V.N.; Martynova, N.Y.; Zaraisky, A.G. Affinity of the heparin binding motif of noggin1 to heparan sulfate and its visualization in the embryonic tissues. Biochem. Biophys. Res. Commun. 2015, 468, 331–336. [Google Scholar] [CrossRef]
- Graf, F.; Horn, P.; Ho, A.D.; Boutros, M.; Maercker, C. The extracellular matrix proteins type I collagen, type III collagen, fibronectin, and laminin 421 stimulate migration of cancer cells. FASEB J. 2021, 35, e21692. [Google Scholar] [CrossRef]
- Artinger, M.; Gerken, O.J.; Purvanov, V.; Legler, D.F. Distinct fates of chemokine and surrogate molecule gradients: Consequences for CCR7-guided dendritic cell migration. Front. Immunol. 2022, 13, 913366. [Google Scholar] [CrossRef] [PubMed]
- Serafini, T.; Colamarino, S.A.; Leonardo, E.D.; Wang, H.; Beddington, R.; Skarnes, W.C.; Tessier-Lavigne, M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell 1996, 87, 1001–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segerer, S.; Johnson, Z.; Rek, A.; Baltus, T.; von Hundelshausen, P.; Kungl, A.J.; Proudfoot, A.E.I.; Weber, C.; Nelson, P.J. The basic residue cluster 55KKWVR59 in CCL5 is required for in vivo biologic function. Mol. Immunol. 2009, 46, 2533–2538. [Google Scholar] [CrossRef] [PubMed]
- Cain, S.A.; McGovern, A.; Baldwin, A.K.; Baldock, C.; Kielty, C.M. Fibrillin-1 mutations causing weill-marchesani syndrome and acromicric and geleophysic dysplasias disrupt heparan sulfate interactions. PLoS ONE 2012, 7, e48634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, S.; Namba, K.; Mutai, H.; Usui, S.; Miyanaga, Y.; Kaneko, H.; Matsunaga, T. A mutation in the heparin-binding site of noggin as a novel mechanism of proximal symphalangism and conductive hearing loss. Biochem. Biophys. Res. Commun. 2014, 447, 496–502. [Google Scholar] [CrossRef]
- Schwaninger, R.; Rentsch, C.A.; Wetterwald, A.; Van Der Horst, G.; Van Bezooijen, R.L.; Van Der Pluijm, G.; Löwik, C.W.G.M.; Ackermann, K.; Pyerin, W.; Hamdy, F.C.; et al. Lack of noggin expression by cancer cells is a determinant of the osteoblast response in bone metastases. Am. J. Pathol. 2007, 170, 160–175. [Google Scholar] [CrossRef] [Green Version]
- Feeley, B.T.; Krenek, L.; Liu, N.; Hsu, W.K.; Gamradt, S.C.; Schwarz, E.M.; Huard, J.; Lieberman, J.R. Overexpression of noggin inhibits bmp-mediated growth of osteolytic prostate cancer lesions. Bone 2006, 38, 154–166. [Google Scholar] [CrossRef]
- Ouahoud, S.; Hardwick, J.C.H.; Hawinkels, L.J.A.C. Extracellular bmp antagonists, multifaceted orchestrators in the tumor and its microenvironment. Int. J. Mol. Sci. 2020, 21, 3888. [Google Scholar] [CrossRef]
- Dyer, D.P.; Salanga, C.L.; Volkman, B.F.; Kawamura, T.; Handel, T.M. The dependence of chemokine-glycosaminoglycan interactions on chemokine oligomerization. Glycobiology 2015, 26, 312–326. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Davari, P.; Takada, Y.K.; Takada, Y. Stromal cell-derived factor-1 (CXCL12) activates integrins by direct binding to an allosteric ligand-binding site (site 2) of integrins without CXCR4. Biochem. J. 2018, 475, 723–732. [Google Scholar] [CrossRef]
- Mayfosh, A.J.; Nguyen, T.K.; Hulett, M.D. The heparanase regulatory network in health and disease. Int. J. Mol. Sci. 2021, 22, 11096. [Google Scholar] [CrossRef]
- Bhattacharya, U.; Gutter-Kapon, L.; Kan, T.; Boyango, I.; Barash, U.; Yang, S.M.; Liu, J.J.; Gross-Cohen, M.; Sanderson, R.D.; Shaked, Y.; et al. Heparanase and chemotherapy synergize to drive macrophage activation and enhance tumor growth. Cancer Res. 2020, 80, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Barash, U.; Zohar, Y.; Wildbaum, G.; Beider, K.; Nagler, A.; Karin, N.; Ilan, N.; Vlodavsky, I. heparanase enhances myeloma progression via CXCL10 downregulation. Leukemia 2014, 28, 2178–2187. [Google Scholar] [CrossRef] [PubMed]
- Jayatilleke, K.M.; Hulett, M.D. Heparanase and the hallmarks of cancer. J. Transl. Med. 2020, 18, 453. [Google Scholar] [CrossRef] [PubMed]
- Coombe, D.R.; Gandhi, N.S. Heparanase: A challenging cancer drug target. Front. Oncol. 2019, 9, 1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauch, U. Extracellular matrix components associated with remodeling processes in brain. Cell. Mol. Life Sci. 2004, 61, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- Méneret, A.; Franz, E.A.; Trouillard, O.; Oliver, T.C.; Zagar, Y.; Robertson, S.P.; Welniarz, Q.; Gardner, R.J.M.; Gallea, C.; Srour, M.; et al. Mutations in the Netrin-1 gene cause congenital mirror movements. J. Clin. Investig. 2017, 127, 3923–3936. [Google Scholar] [CrossRef] [PubMed]
- Loka, R.S.; Song, Z.; Sletten, E.T.; Kayal, Y.; Vlodavsky, I.; Zhang, K.; Nguyen, H.M. Heparan sulfate mimicking glycopolymer prevents pancreatic β cell destruction and suppresses inflammatory cytokine expression in islets under the challenge of upregulated heparanase. ACS Chem. Biol. 2022, 17, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Feng, J.; Rao, Y.; Xu, Z.; Zu, J.; Wang, H.; Zhang, Z.; Chen, H. Artificial extracellular matrix composed of heparin-mimicking polymers for efficient anticoagulation and promotion of endothelial cell proliferation. ACS Appl. Mater. Interfaces 2022, 14, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Al-Horani, R.A.; Abdelfadiel, E.I.; Afosah, D.K.; Morla, S.; Sistla, J.C.; Mohammed, B.; Martin, E.J.; Sakagami, M.; Brophy, D.F.; Desai, U.R. A synthetic heparin mimetic that allosterically inhibits factor XIa and reduces thrombosis in vivo without enhanced risk of bleeding. J. Thromb. Haemost. 2019, 17, 2110–2122. [Google Scholar] [CrossRef] [PubMed]
- Kiyan, Y.; Tkachuk, S.; Kurselis, K.; Shushakova, N.; Stahl, K.; Dawodu, D.; Kiyan, R.; Chichkov, B.; Haller, H. Heparanase-2 protects from LPS-mediated endothelial injury by inhibiting TLR4 signalling. Sci. Rep. 2019, 9, 13591. [Google Scholar] [CrossRef] [Green Version]
- Shamdani, S.; Chantepie, S.; Flageollet, C.; Henni-Chebra, N.; Jouan, Y.; Eymard, F.; Hay, E.; Cohen-Solal, M.; Papy-Garcia, D.; Chevalier, X.; et al. Heparan sulfate functions are altered in the osteoarthritic cartilage. Arthritis Res. Ther. 2020, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Meyer, G.; Besse, B.; Doubre, H.; Charles-Nelson, A.; Aquilanti, S.; Izadifar, A.; Azarian, R.; Monnet, I.; Lamour, C.; Descourt, R.; et al. Anti-tumour effect of low molecular weight heparin in localised lung cancer: A phase III clinical trial. Eur. Respir. J. 2018, 52, 1801220. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Teng, L.; Zhou, J.; Yan, Y.; Zhang, Y.; Lv, G.; Chen, J. Design and synthesis of a native heparin disaccharide grafted poly-2-aminoethyl methacrylate glycopolymer for inhibition of melanoma cell metastasis. Int. J. Biol. Macromol. 2019, 126, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Yin, S.; Xu, S.; Ran, G.; Deng, M.; Mei, L.; Tang, X.; Rao, J.; Li, M.; Zhang, Z.; et al. Low molecular weight heparin-coated and dendrimer-based core-shell nanoplatform with enhanced immune activation and multiple anti-metastatic effects for melanoma treatment. Theranostics 2019, 9, 337–354. [Google Scholar] [CrossRef]
- Feeley, B.T.; Liu, N.Q.; Conduah, A.H.; Krenek, L.; Roth, K.; Dougall, W.C.; Huard, J.; Dubinett, S.; Lieberman, J.R. Mixed metastatic lung cancer lesions in bone are inhibited by noggin overexpression and rank: Fc administration. J. Bone Miner. Res. 2006, 21, 1571–1580. [Google Scholar] [CrossRef]
- Nagata, K.; Kumasaka, K.; Browne, K.D.; Li, S.; St-Pierre, J.; Cognetti, J.; Marks, J.; Johnson, V.E.; Smith, D.H.; Pascual, J.L. Unfractionated heparin after tbi reduces in vivo cerebrovascular inflammation, brain edema and accelerates cognitive recovery. J. Trauma Acute Care Surg. 2016, 81, 1088–1094. [Google Scholar] [CrossRef] [Green Version]
- Al Faruque, H.; Kang, J.H.; Hwang, S.R.; Sung, S.; Alam, M.M.; Sa, K.H.; Nam, E.J.; Byun, Y.R.; Kang, Y.M. Stepwise inhibition of T cell recruitment at post-capillary venules by orally active desulfated heparins in inflammatory arthritis. PLoS ONE 2017, 12, e0176110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murali, S.; Rai, B.; Dombrowski, C.; Lee, J.L.J.; Lim, Z.X.H.; Bramono, D.S.; Ling, L.; Bell, T.; Hinkley, S.; Nathan, S.S.; et al. Affinity-selected heparan sulfate for bone repair. Biomaterials 2013, 34, 5594–5605. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heide, F.; Koch, M.; Stetefeld, J. Heparin Mimetics and Their Impact on Extracellular Matrix Protein Assemblies. Pharmaceuticals 2023, 16, 471. https://doi.org/10.3390/ph16030471
Heide F, Koch M, Stetefeld J. Heparin Mimetics and Their Impact on Extracellular Matrix Protein Assemblies. Pharmaceuticals. 2023; 16(3):471. https://doi.org/10.3390/ph16030471
Chicago/Turabian StyleHeide, Fabian, Manuel Koch, and Jörg Stetefeld. 2023. "Heparin Mimetics and Their Impact on Extracellular Matrix Protein Assemblies" Pharmaceuticals 16, no. 3: 471. https://doi.org/10.3390/ph16030471
APA StyleHeide, F., Koch, M., & Stetefeld, J. (2023). Heparin Mimetics and Their Impact on Extracellular Matrix Protein Assemblies. Pharmaceuticals, 16(3), 471. https://doi.org/10.3390/ph16030471




