Up-Regulation of PSMA Expression In Vitro as Potential Application in Prostate Cancer Therapy
Abstract
:1. Introduction
2. Results
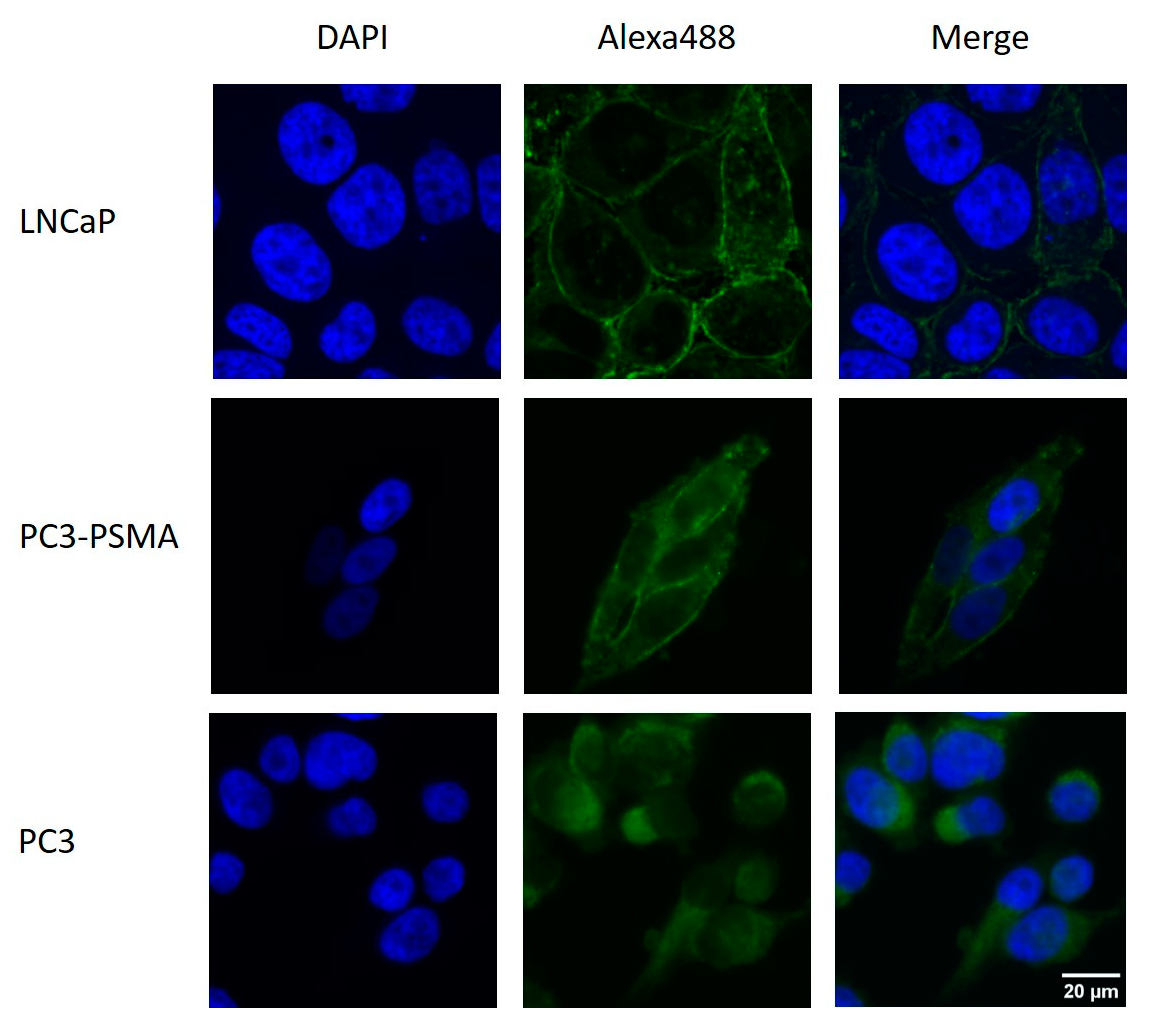
2.1. Immunohistochemical Detection of PSMA Expression
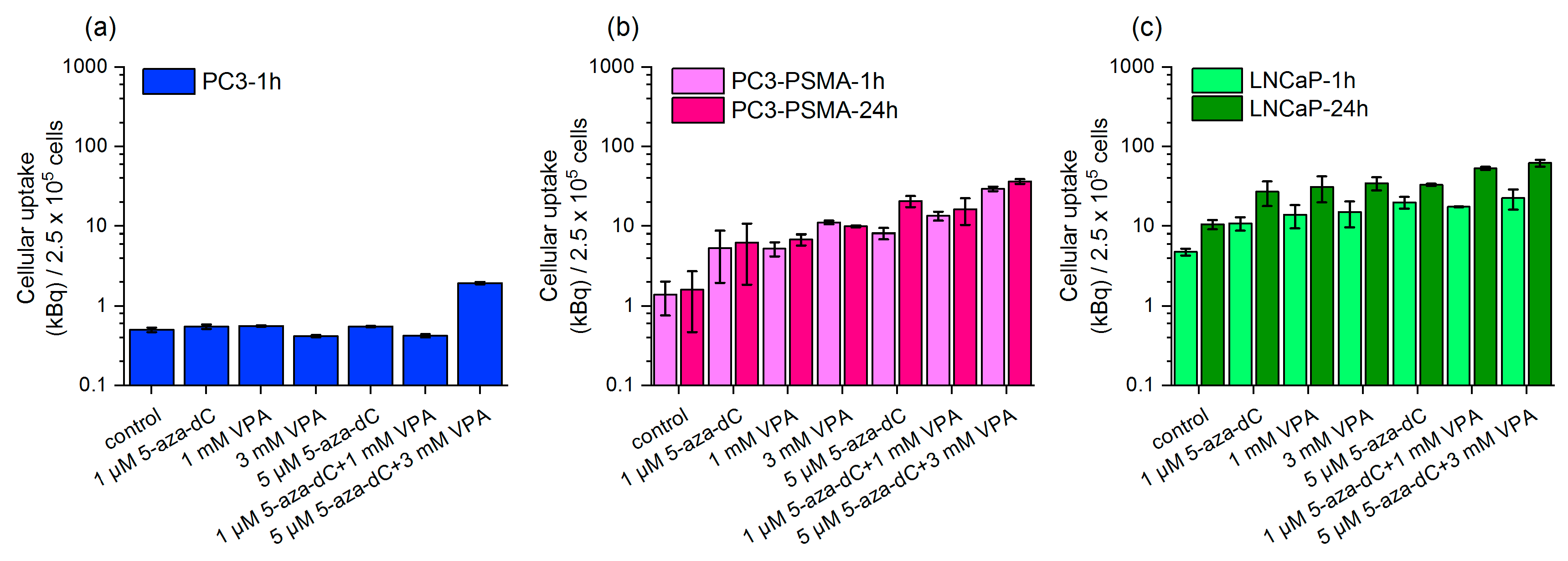
2.2. Uptake of Lu-177–PSMA-617 in Dependency on 5-aza-dC and VPA Stimulation
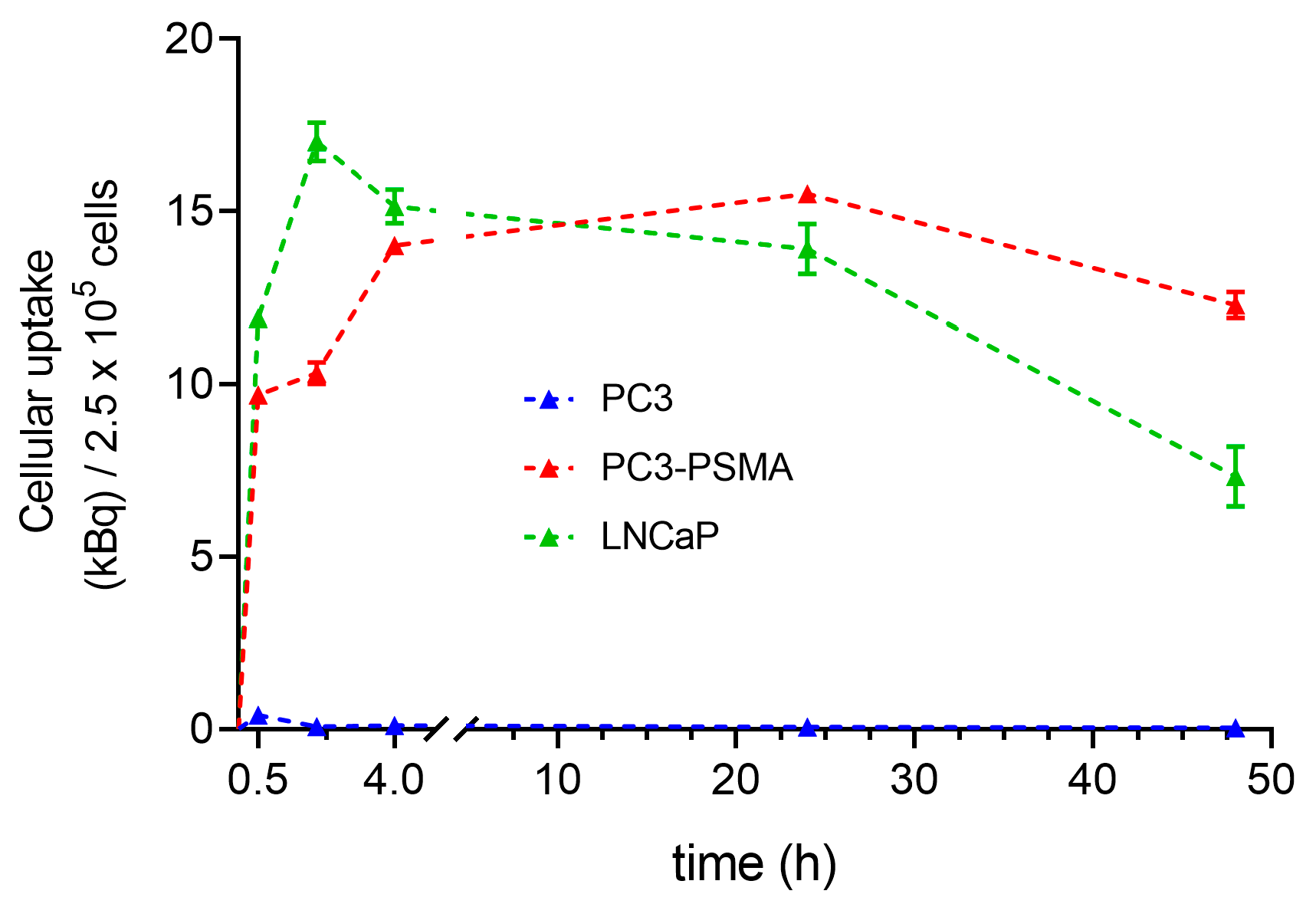
2.3. Time Dependence of Lu-177–PSMA-617 Uptake in PC3, PC3-PSMA, and LNCaP Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunohistochemistry
4.3. Chemical Modulation by 5-aza-dC and VPA
4.4. Radiosynthesis of Lu-177–PSMA-617
4.5. Intracellular Lu-177–PSMA-617 Radionuclide Uptake and Uptake Kinetic
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, X.; Jiang, T.; Gao, C.; Liu, H.; Sun, Y.; Zou, Q.; Tang, R.; Zeng, W. Detection rate of fluorine-18 prostate-specific membrane antigen-1007 PET/CT for prostate cancer in primary staging and biochemical recurrence with different serum PSA levels: A systematic review and meta-analysis. Front. Oncol. 2022, 12, 911146. [Google Scholar] [CrossRef] [PubMed]
- Maurer, T.; Eiber, M.; Schwaiger, M.; Gschwend, J.E. Current use of PSMA-PET in prostate cancer management. Nat. Rev. Urol. 2016, 13, 226–235. [Google Scholar] [CrossRef]
- Perera, M.; Papa, N.; Roberts, M.; Williams, M.; Udovicich, C.; Vela, I.; Christidis, D.; Bolton, D.; Hofman, M.S.; Lawrentschuk, N.; et al. Gallium-68 Prostate-specific Membrane Antigen Positron Emission Tomography in Advanced Prostate Cancer-Updated Diagnostic Utility, Sensitivity, Specificity, and Distribution of Prostate-specific Membrane Antigen-avid Lesions: A Systematic Review and Meta-analysis. Eur. Urol. 2020, 77, 403–417. [Google Scholar] [CrossRef] [PubMed]
- von Eyben, F.E.; Picchio, M.; von Eyben, R.; Rhee, H.; Bauman, G. 68Ga-Labeled Prostate-specific Membrane Antigen Ligand Positron Emission Tomography/Computed Tomography for Prostate Cancer: A Systematic Review and Meta-analysis. Eur. Urol. Focus 2018, 4, 686–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, K.; Bluemel, C.; Weineisen, M.; Schottelius, M.; Wester, H.J.; Czernin, J.; Eberlein, U.; Beykan, S.; Lapa, C.; Riedmiller, H.; et al. Biodistribution and radiation dosimetry for a probe targeting prostate-specific membrane antigen for imaging and therapy. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2015, 56, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Kratochwil, C.; Giesel, F.L.; Stefanova, M.; Benesova, M.; Bronzel, M.; Afshar-Oromieh, A.; Mier, W.; Eder, M.; Kopka, K.; Haberkorn, U. PSMA-Targeted Radionuclide Therapy of Metastatic Castration-Resistant Prostate Cancer with 177Lu-Labeled PSMA-617. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2016, 57, 1170–1176. [Google Scholar] [CrossRef] [Green Version]
- Sartor, O.; de Bono, J.; Chi, K.N.; Fizazi, K.; Herrmann, K.; Rahbar, K.; Tagawa, S.T.; Nordquist, L.T.; Vaishampayan, N.; El-Haddad, G.; et al. Lutetium-177-PSMA-617 for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2021, 385, 1091–1103. [Google Scholar] [CrossRef]
- Berglund, A.; Matta, J.; Encarnacion-Medina, J.; Ortiz-Sanchez, C.; Dutil, J.; Linares, R.; Marcial, J.; Abreu-Takemura, C.; Moreno, N.; Putney, R.; et al. Dysregulation of DNA Methylation and Epigenetic Clocks in Prostate Cancer among Puerto Rican Men. Biomolecules 2021, 12, 2. [Google Scholar] [CrossRef]
- Watt, F.; Martorana, A.; Brookes, D.E.; Ho, T.; Kingsley, E.; O’Keefe, D.S.; Russell, P.J.; Heston, W.D.; Molloy, P.L. A tissue-specific enhancer of the prostate-specific membrane antigen gene, FOLH1. Genomics 2001, 73, 243–254. [Google Scholar] [CrossRef]
- Li, Z.; Li, W.; Xie, J.; Wang, Y.; Tang, A.; Li, X.; Ye, J.; Gui, Y.; Cai, Z. Epigenetic inactivation of PCDH10 in human prostate cancer cell lines. Cell Biol. Int. 2011, 35, 671–676. [Google Scholar] [CrossRef]
- Bates, S.E. Epigenetic Therapies for Cancer. N. Engl. J. Med. 2020, 383, 650–663. [Google Scholar] [CrossRef]
- Dear, A.E. Epigenetic Modulators and the New Immunotherapies. N. Engl. J. Med. 2016, 374, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Noss, K.R.; Wolfe, S.A.; Grimes, S.R. Upregulation of prostate specific membrane antigen/folate hydrolase transcription by an enhancer. Gene 2002, 285, 247–256. [Google Scholar] [CrossRef]
- Kumaraswamy, A.; Welker Leng, K.R.; Westbrook, T.C.; Yates, J.A.; Zhao, S.G.; Evans, C.P.; Feng, F.Y.; Morgan, T.M.; Alumkal, J.J. Recent Advances in Epigenetic Biomarkers and Epigenetic Targeting in Prostate Cancer. Eur. Urol. 2021, 80, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Kutilin, D. Genetic and epigenetic bases of prostate tumor cell radioresistance. Klin. Onkol. 2021, 34, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Kotzerke, J.; Buesser, D.; Naumann, A.; Runge, R.; Huebinger, L.; Kliewer, A.; Freudenberg, R.; Brogsitter, C. Epigenetic-Like Stimulation of Receptor Expression in SSTR2 Transfected HEK293 Cells as a New Therapeutic Strategy. Cancers 2022, 14, 2513. [Google Scholar] [CrossRef]
- Taelman, V.F.; Radojewski, P.; Marincek, N.; Ben-Shlomo, A.; Grotzky, A.; Olariu, C.I.; Perren, A.; Stettler, C.; Krause, T.; Meier, L.P.; et al. Upregulation of Key Molecules for Targeted Imaging and Therapy. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2016, 57, 1805–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veenstra, M.J.; van Koetsveld, P.M.; Dogan, F.; Farrell, W.E.; Feelders, R.A.; Lamberts, S.W.; de Herder, W.W.; Vitale, G.; Hofland, L.J. Epidrug-induced upregulation of functional somatostatin type 2 receptors in human pancreatic neuroendocrine tumor cells. Oncotarget 2016, 9, 14791–14802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullrich, M.; Richter, S.; Liers, J.; Drukewitz, S.; Friedemann, M.; Kotzerke, J.; Ziegler, C.G.; Nolting, S.; Kopka, K.; Pietzsch, J. Epigenetic drugs in somatostatin type 2 receptor radionuclide theranostics and radiation transcriptomics in mouse pheochromocytoma models. Theranostics 2023, 13, 278–294. [Google Scholar] [CrossRef]
- Pollard, J.H.; Menda, Y.; Zamba, K.D.; Madsen, M.; O’Dorisio, M.S.; O’Dorisio, T.; Bushnell, D. Potential for Increasing Uptake of Radiolabeled (68)Ga-DOTATOC and (123)I-MIBG in Patients with Midgut Neuroendocrine Tumors Using a Histone Deacetylase Inhibitor Vorinostat. Cancer Biother. Radiopharm. 2021, 36, 632–641. [Google Scholar] [CrossRef]
- Refardt, J.; Klomp, M.J.; van Koetsveld, P.M.; Dogan, F.; Konijnenberg, M.; Brabander, T.; Feelders, R.A.; de Herder, W.W.; Hofland, L.J.; Hofland, J. Effect of epigenetic treatment on SST(2) expression in neuroendocrine tumour patients. Clin. Transl. Med. 2022, 12, e957. [Google Scholar] [CrossRef] [PubMed]
- Liolios, C.; Patsis, C.; Lambrinidis, G.; Tzortzini, E.; Roscher, M.; Bauder-Wust, U.; Kolocouris, A.; Kopka, K. Investigation of Tumor Cells and Receptor-Ligand Simulation Models for the Development of PET Imaging Probes Targeting PSMA and GRPR and a Possible Crosstalk between the Two Receptors. Mol. Pharm. 2022, 19, 2231–2247. [Google Scholar] [CrossRef] [PubMed]
- Dicitore, A.; Bacalini, M.G.; Saronni, D.; Gaudenzi, G.; Cantone, M.C.; Gelmini, G.; Grassi, E.S.; Gentilini, D.; Borghi, M.O.; Di Blasio, A.M.; et al. Role of Epigenetic Therapy in the Modulation of Tumor Growth and Migration in Human Castration-Resistant Prostate Cancer Cells with Neuroendocrine Differentiation. Neuroendocrinology 2022, 112, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.; Choo, N.; Bolton, D.M.; Lawrentschuk, N.; Risbridger, G.P.; Lawrence, M.G.; Taylor, R.A. New approaches to targeting epigenetic regulation in prostate cancer. Curr. Opin. Urol. 2022, 32, 472–480. [Google Scholar] [CrossRef]
- Sayar, E.; Patel, R.A.; Coleman, I.M.; Roudier, M.P.; Zhang, A.; Mustafi, P.; Low, J.Y.; Hanratty, B.; Ang, L.S.; Bhatia, V.; et al. Reversible epigenetic alterations mediate PSMA expression heterogeneity in advanced metastatic prostate cancer. JCI Insight 2023, e162907. [Google Scholar] [CrossRef]
- Meher, N.; Ashley, G.W.; Bidkar, A.P.; Dhrona, S.; Fong, C.; Fontaine, S.D.; Beckford Vera, D.R.; Wilson, D.M.; Seo, Y.; Santi, D.V.; et al. Prostate-Specific Membrane Antigen Targeted Deep Tumor Penetration of Polymer Nanocarriers. ACS Appl. Mater. Interfaces 2022, 14, 50569–50582. [Google Scholar] [CrossRef]
- Luan, X.; Zhou, H.; Chen, Y.; Zhang, X.; Cui, M.; Chen, K.; Xu, X.; Zhang, J.; Xu, B. A Preclinical Study of an (125)I-Labeled PSMA Ligand for Prostate-Cancer Puncture. Pharmaceuticals 2022, 15, 1252. [Google Scholar] [CrossRef]
- Weimin, S.; Abula, A.; Qianghong, D.; Wenguang, W. Chimeric cytokine receptor enhancing PSMA-CAR-T cell-mediated prostate cancer regression. Cancer Biol. Ther. 2020, 21, 570–580. [Google Scholar] [CrossRef]
- Motawi, T.K.; Darwish, H.A.; Diab, I.; Helmy, M.W.; Noureldin, M.H. Combinatorial strategy of epigenetic and hormonal therapies: A novel promising approach for treating advanced prostate cancer. Life Sci. 2018, 198, 71–78. [Google Scholar] [CrossRef]
- Meller, B.; Bremmer, F.; Sahlmann, C.O.; Hijazi, S.; Bouter, C.; Trojan, L.; Meller, J.; Thelen, P. Alterations in androgen deprivation enhanced prostate-specific membrane antigen (PSMA) expression in prostate cancer cells as a target for diagnostics and therapy. EJNMMI Res. 2015, 5, 66. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Xu, W.; Xiao, Y.T.; Huang, H.; Gu, D.; Ren, S. Targeting signaling pathways in prostate cancer: Mechanisms and clinical trials. Signal. Transduct. Target. Ther. 2022, 7, 198. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Kim, S.C.; Kim, S.H.; Yoon, J.H.; Moon, K.H.; Cheon, S.H.; Kwon, T.; Kim, Y.M.; Park, J.W.; Lee, S.H.; et al. Docetaxel Enhances Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand-Mediated Apoptosis in Prostate Cancer Cells via Epigenetic Gene Regulation by Enhancer of Zeste Homolog 2. World J. Mens. Health 2023, 41, e2. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, M.; Wang, Y.; Luo, J. Curcumin sensitizes prostate cancer cells to radiation partly via epigenetic activation of miR-143 and miR-143 mediated autophagy inhibition. J. Drug Target. 2017, 25, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Van de Wiele, C.; Sathekge, M.; de Spiegeleer, B.; De Jonghe, P.J.; Debruyne, P.R.; Borms, M.; Beels, L.; Maes, A. PSMA expression on neovasculature of solid tumors. Histol. Histopathol. 2020, 35, 919–927. [Google Scholar] [CrossRef]
- Uijen, M.J.M.; Derks, Y.H.W.; Merkx, R.I.J.; Schilham, M.G.M.; Roosen, J.; Prive, B.M.; van Lith, S.A.M.; van Herpen, C.M.L.; Gotthardt, M.; Heskamp, S.; et al. PSMA radioligand therapy for solid tumors other than prostate cancer: Background, opportunities, challenges, and first clinical reports. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4350–4368. [Google Scholar] [CrossRef]
- Feldmann, A.; Arndt, C.; Bergmann, R.; Loff, S.; Cartellieri, M.; Bachmann, D.; Aliperta, R.; Hetzenecker, M.; Ludwig, F.; Albert, S.; et al. Retargeting of T lymphocytes to PSCA- or PSMA positive prostate cancer cells using the novel modular chimeric antigen receptor platform technology “UniCAR”. Oncotarget 2017, 8, 31368–31385. [Google Scholar] [CrossRef] [Green Version]
- Capozza, M.; Stefania, R.; Dinatale, V.; Bitonto, V.; Conti, L.; Grange, C.; Skovronova, R.; Terreno, E. A Novel PSMA-Targeted Probe for NIRF-Guided Surgery and Photodynamic Therapy: Synthesis and Preclinical Validation. Int. J. Mol. Sci. 2022, 23, 12878. [Google Scholar] [CrossRef]
- Feldmann, A.; Stamova, S.; Bippes, C.C.; Bartsch, H.; Wehner, R.; Schmitz, M.; Temme, A.; Cartellieri, M.; Bachmann, M. Retargeting of T cells to prostate stem cell antigen expressing tumor cells: Comparison of different antibody formats. Prostate 2011, 71, 998–1011. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Runge, R.; Naumann, A.; Miederer, M.; Kotzerke, J.; Brogsitter, C. Up-Regulation of PSMA Expression In Vitro as Potential Application in Prostate Cancer Therapy. Pharmaceuticals 2023, 16, 538. https://doi.org/10.3390/ph16040538
Runge R, Naumann A, Miederer M, Kotzerke J, Brogsitter C. Up-Regulation of PSMA Expression In Vitro as Potential Application in Prostate Cancer Therapy. Pharmaceuticals. 2023; 16(4):538. https://doi.org/10.3390/ph16040538
Chicago/Turabian StyleRunge, Roswitha, Anne Naumann, Matthias Miederer, Joerg Kotzerke, and Claudia Brogsitter. 2023. "Up-Regulation of PSMA Expression In Vitro as Potential Application in Prostate Cancer Therapy" Pharmaceuticals 16, no. 4: 538. https://doi.org/10.3390/ph16040538
APA StyleRunge, R., Naumann, A., Miederer, M., Kotzerke, J., & Brogsitter, C. (2023). Up-Regulation of PSMA Expression In Vitro as Potential Application in Prostate Cancer Therapy. Pharmaceuticals, 16(4), 538. https://doi.org/10.3390/ph16040538





