A Novel Electrochemical Differentiation between Exosomal-RNA of Breast Cancer MCF7 and MCF7/ADR-Resistant Cells
Abstract
:1. Introduction
2. Results and Discussion
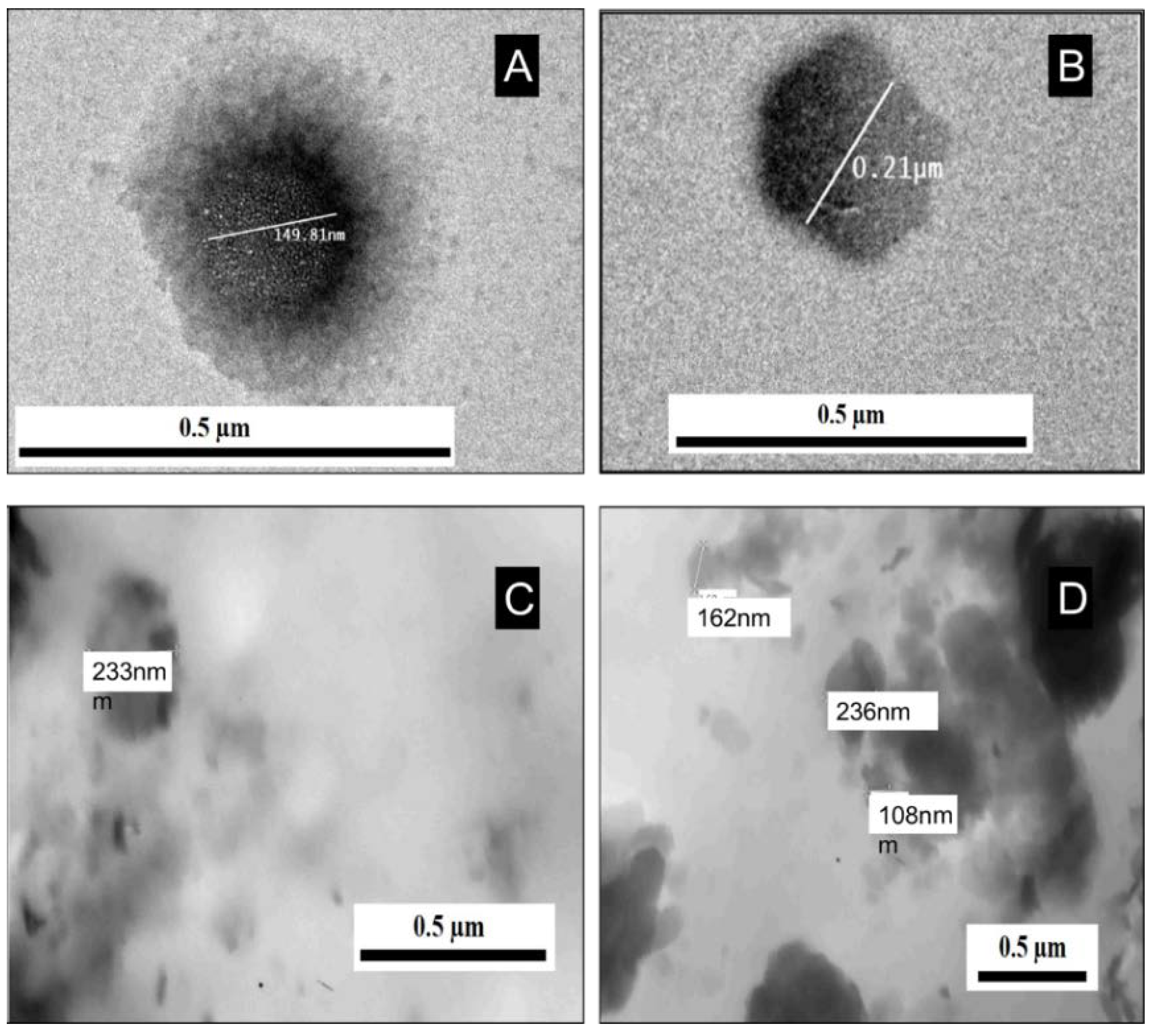
2.1. Physical Characterization of Exosomes and Isolated Exosomal RNAs
2.2. Electrochemical Characterization
2.2.1. Electrochemical Characterization of Exosomal RNA Isolated from MCF7
2.2.2. Electrochemical Characterization of Exosomal RNA Isolated from MCF7/ADR
3. Materials and Methods
3.1. Materials
3.2. Cell Culture
3.3. Exosomes Isolation by Ultracentrifugation and Physical Characterization
3.4. RNA Isolation from Exosomes
3.5. Setup and Conditions for the Electrochemical Detection of Exosomal RNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- WHO. WHO Cancer. Available online: http://www.who.int/cancer/en/ (accessed on 12 May 2019).
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-Invasive Early Detection of Cancer Four Years before Conventional Diagnosis Using a Blood Test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Boriachek, K.; Islam, M.N.; Lobb, R.; Möller, A.; Hill, M.M.; Hossain, M.S.A.; Nguyen, N.-T.; Shiddiky, M.J.A. An Electrochemical Method for the Detection of Disease-Specific Exosomes. ChemElectroChem 2017, 4, 967–971. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Guo, Z.; Zhao, Y.; Ma, L.; Li, B.; Yang, C. Compound 968 Reverses Adriamycin Resistance in Breast Cancer MCF-7ADR Cells via Inhibiting P-Glycoprotein Function Independently of Glutaminase. Cell. Death Discov. 2021, 7, 204. [Google Scholar] [CrossRef]
- Nasher, F.; Heller, M.; Hathaway, L.J. Streptococcus Pneumoniae Proteins AmiA, AliA, and AliB Bind Peptides Found in Ribosomal Proteins of Other Bacterial Species. Front. Microbiol. 2018, 8, 2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Chen, Z.; Xie, F.; Xie, M.; Liu, N.; Su, Z.; Gu, J.; Zhao, R. (+/−)-Borneol Reverses Mitoxantrone Resistance against P-Glycoprotein. J. Chem. Inf. Model. 2021, 61, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fan, J.; Fan, Z.; Zhang, K. γ-Tocotrienol Reverses Multidrug Resistance of Breast Cancer Cells through the Regulation of the γ-Tocotrienol-NF-ΚB-P-Gp Axis. J. Steroid Biochem. Mol. Biol. 2021, 209, 105835. [Google Scholar] [CrossRef]
- Altun, İ.; Sonkaya, A. The Most Common Side Effects Experienced by Patients Were Receiving First Cycle of Chemotherapy. Iran. J. Public. Health 2018, 47, 1218–1219. [Google Scholar]
- Veziroglu, E.M.; Mias, G.I. Characterizing Extracellular Vesicles and Their Diverse RNA Contents. Front. Genet. 2020, 11, 700. [Google Scholar] [CrossRef]
- Yu, Z.; Lin, S.; Xia, F.; Liu, Y.; Zhang, D.; Wang, F.; Wang, Y.; Li, Q.; Niu, J.; Cao, C.; et al. ExoSD Chips for High-Purity Immunomagnetic Separation and High-Sensitivity Detection of Gastric Cancer Cell-Derived Exosomes. Biosens. Bioelectron. 2021, 194, 113594. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, W.S.; Guo, Y.; Peng, H.; Zhu, M.; Miao, D.; Su, G. Engineering of Exosome-Triggered Enzyme-Powered DNA Motors for Highly Sensitive Fluorescence Detection of Tumor-Derived Exosomes. Biosens. Bioelectron. 2020, 167, 112482. [Google Scholar] [CrossRef]
- Song, F.; Wang, C.; Wang, C.; Gao, J.; Liu, H.; Zhang, Y.; Han, L. Enrichment-Detection Integrated Exosome Profiling Biosensors Promising for Early Diagnosis of Cancer. Anal. Chem. 2021, 93, 4697–4706. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell. Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zhang, B. The Immunomodulation Potential of Exosomes in Tumor Microenvironment. J. Immunol. Res. 2021, 2021, 3710372. [Google Scholar] [CrossRef] [PubMed]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.-J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [Green Version]
- Exosomes. Nat. Biotechnol. 2020, 38, 1150. [CrossRef]
- Pinnell, J.R.; Cui, M.; Tieu, K. Exosomes in Parkinson Disease. J. Neurochem. 2021, 157, 413–428. [Google Scholar] [CrossRef]
- Soares Martins, T.; Trindade, D.; Vaz, M.; Campelo, I.; Almeida, M.; Trigo, G.; da Cruz e Silva, O.A.B.; Henriques, A.G. Diagnostic and Therapeutic Potential of Exosomes in Alzheimer’s Disease. J. Neurochem. 2021, 156, 162–181. [Google Scholar] [CrossRef]
- Zhu, C.; Li, L.; Wang, Z.; Irfan, M.; Qu, F. Recent Advances of Aptasensors for Exosomes Detection. Biosens. Bioelectron. 2020, 160, 112213. [Google Scholar] [CrossRef]
- Chung, I.-M.; Rajakumar, G.; Venkidasamy, B.; Subramanian, U.; Thiruvengadam, M. Exosomes: Current Use and Future Applications. Clin. Chim. Acta 2020, 500, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Zargartalebi, H.; Salahandish, R.; Aburashed, R.; Yong, K.W.; Sanati-Nezhad, A. Emerging Technologies and Commercial Products in Exosome-Based Cancer Diagnosis and Prognosis. Biosens. Bioelectron. 2021, 183, 113176. [Google Scholar] [CrossRef]
- Wang, M.; Pan, Y.; Wu, S.; Sun, Z.; Wang, L.; Yang, J.; Yin, Y.; Li, G. Detection of Colorectal Cancer-Derived Exosomes Based on Covalent Organic Frameworks. Biosens. Bioelectron. 2020, 169, 112638. [Google Scholar] [CrossRef] [PubMed]
- Taller, D.; Richards, K.; Slouka, Z.; Senapati, S.; Hill, R.; Go, D.B.; Chang, H.-C. On-Chip Surface Acoustic Wave Lysis and Ion-Exchange Nanomembrane Detection of Exosomal RNA for Pancreatic Cancer Study and Diagnosis. Lab Chip 2015, 15, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- Exosome RNA. Exosomes—Improved Methods to Characterize Their Morphology, RNA Content, and Surface Protein Biomarkers. Available online: https://exosome-rna.com/exosomes-improved-methods-to-characterize-their-morphology-rna-content-and-surface-protein-biomarkers/ (accessed on 19 January 2022).
- Administrator, E.R. Quality of Extracellular Vesicle Images by Transmission Electron Microscopy Is Operator and Protocol Dependent|Exosome RNA. Available online: https://exosome-rna.com/quality-of-extracellular-vesicle-images-by-transmission-electron-microscopy-is-operator-and-protocol-dependent/ (accessed on 19 January 2022).
- Martins, T.S.; Catita, J.; Martins Rosa, I.; da Cruz e Silva, O.A.B.; Henriques, A.G. Exosome Isolation from Distinct Biofluids Using Precipitation and Column-Based Approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef] [Green Version]
- Beit-Yannai, E.; Tabak, S.; Stamer, W.D. Physical Exosome:Exosome Interactions. J. Cell. Mol. Med. 2018, 22, 2001–2006. [Google Scholar] [CrossRef] [Green Version]
- Prathiba, J.; Malathi, R. Probing RNA–Antibiotic Interactions: A FTIR Study. Mol. Biol. Rep. 2008, 35, 51–57. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, X.; Ji, Z.; Zhu, L.; Ma, N.; Chen, D.; Jia, X.; Tang, J.; Cao, Y. DFT-Calculated IR Spectrum Amide I, II, and III Band Contributions of N-Methylacetamide Fine Components. ACS Omega 2020, 5, 8572–8578. [Google Scholar] [CrossRef] [Green Version]
- IR Spectrum Table. Available online: https://www.sigmaaldrich.com/EG/en/technical-documents/technical-article/analytical-chemistry/photometry-and-reflectometry/ir-spectrum-table (accessed on 24 January 2022).
- Ayranci, R.; Torlak, Y.; Ak, M. Non-Enzymatic Electrochemical Detection of Glucose by Mixed-Valence Cobalt Containing Keggin Polyoxometalate/Multi-Walled Carbon Nanotube Composite. J. Electrochem. Soc. 2019, 166, B205. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, X.; Zhang, Y.; Li, W.; Hao, X.; Liu, T.; Liang, X.; Liu, F.; Liu, F.; Yan, X.; et al. High-Performance Electrochemical Sensor Based on Mn1-XZnxFe2O4 Nanoparticle/Nafion-Modified Glassy Carbon Electrode for Pb2+ Detection. J. Electrochem. Soc. 2019, 166, B341. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y.; Zhang, J.; Zhou, J. Electrochemical Sensing Platform Based on Three-Dimensional Holey Graphene for Highly Selective and Ultra-Sensitive Detection of Ascorbic Acid, Uric Acid, and Nitrite. J. Electrochem. Soc. 2019, 166, B787. [Google Scholar] [CrossRef]
- Boriachek, K.; Umer, M.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.-T.; Shiddiky, M.J.A. An Amplification-Free Electrochemical Detection of Exosomal MiRNA-21 in Serum Samples. Analyst 2018, 143, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Kaewnu, K.; Promsuwan, K.; Kanatharana, P.; Thavarungkul, P.; Limbut, W. A Simple and Sensitive Electrochemical Sensor for Chloramphenicol Detection in Pharmaceutical Samples. J. Electrochem. Soc. 2020, 167, 087506. [Google Scholar] [CrossRef]
- Bachurski, D.; Schuldner, M.; Nguyen, P.-H.; Malz, A.; Reiners, K.S.; Grenzi, P.C.; Babatz, F.; Schauss, A.C.; Hansen, H.P.; Hallek, M.; et al. Extracellular Vesicle Measurements with Nanoparticle Tracking Analysis—An Accuracy and Repeatability Comparison between NanoSight NS300 and ZetaView. J. Extracell. Vesicles 2019, 8, 1596016. [Google Scholar] [CrossRef] [PubMed]










| Cell | Exosomes Size Based on Zetasizer | Zeta Potential |
|---|---|---|
| MCF7 | 173.7 nm | −11.3 mV |
| MCF7/ADR resistant | 158.7 nm | −11.6 mV |
| SH-SY5Y | 154.6 nm | −51.0 mV |
| HEPG2 | 174.1 nm | −73.8 mV |
| Parameter | Electrochemical Technique | ||||
|---|---|---|---|---|---|
| CV | SWV | DPV | NPV | SWV (Peak Current) | |
| R2 | 0.96 | 0.95 | 0.84 | 0.84 | 0.78 |
| LOD | 9.1 | 7.4 | 42.2 | 7.2 | 41.5 |
| Parameter | Electrochemical Technique | |||
|---|---|---|---|---|
| CV | SWV | DPV | NPV | |
| R2 | 0.93 | 0.97 | 0.95 | 0.98 |
| LOD | 16.9 | 4.8 | 7.9 | 14.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelaziz, M.H.; El Sawy, E.N.; Abdelnaser, A. A Novel Electrochemical Differentiation between Exosomal-RNA of Breast Cancer MCF7 and MCF7/ADR-Resistant Cells. Pharmaceuticals 2023, 16, 540. https://doi.org/10.3390/ph16040540
Abdelaziz MH, El Sawy EN, Abdelnaser A. A Novel Electrochemical Differentiation between Exosomal-RNA of Breast Cancer MCF7 and MCF7/ADR-Resistant Cells. Pharmaceuticals. 2023; 16(4):540. https://doi.org/10.3390/ph16040540
Chicago/Turabian StyleAbdelaziz, Mohammed H., Ehab N. El Sawy, and Anwar Abdelnaser. 2023. "A Novel Electrochemical Differentiation between Exosomal-RNA of Breast Cancer MCF7 and MCF7/ADR-Resistant Cells" Pharmaceuticals 16, no. 4: 540. https://doi.org/10.3390/ph16040540
APA StyleAbdelaziz, M. H., El Sawy, E. N., & Abdelnaser, A. (2023). A Novel Electrochemical Differentiation between Exosomal-RNA of Breast Cancer MCF7 and MCF7/ADR-Resistant Cells. Pharmaceuticals, 16(4), 540. https://doi.org/10.3390/ph16040540







