Metformin Therapy Changes Gut Microbiota Alpha-Diversity in COVID-19 Patients with Type 2 Diabetes: The Role of SARS-CoV-2 Variants and Antibiotic Treatment
Abstract
:1. Introduction
2. Results
2.1. Patient Population
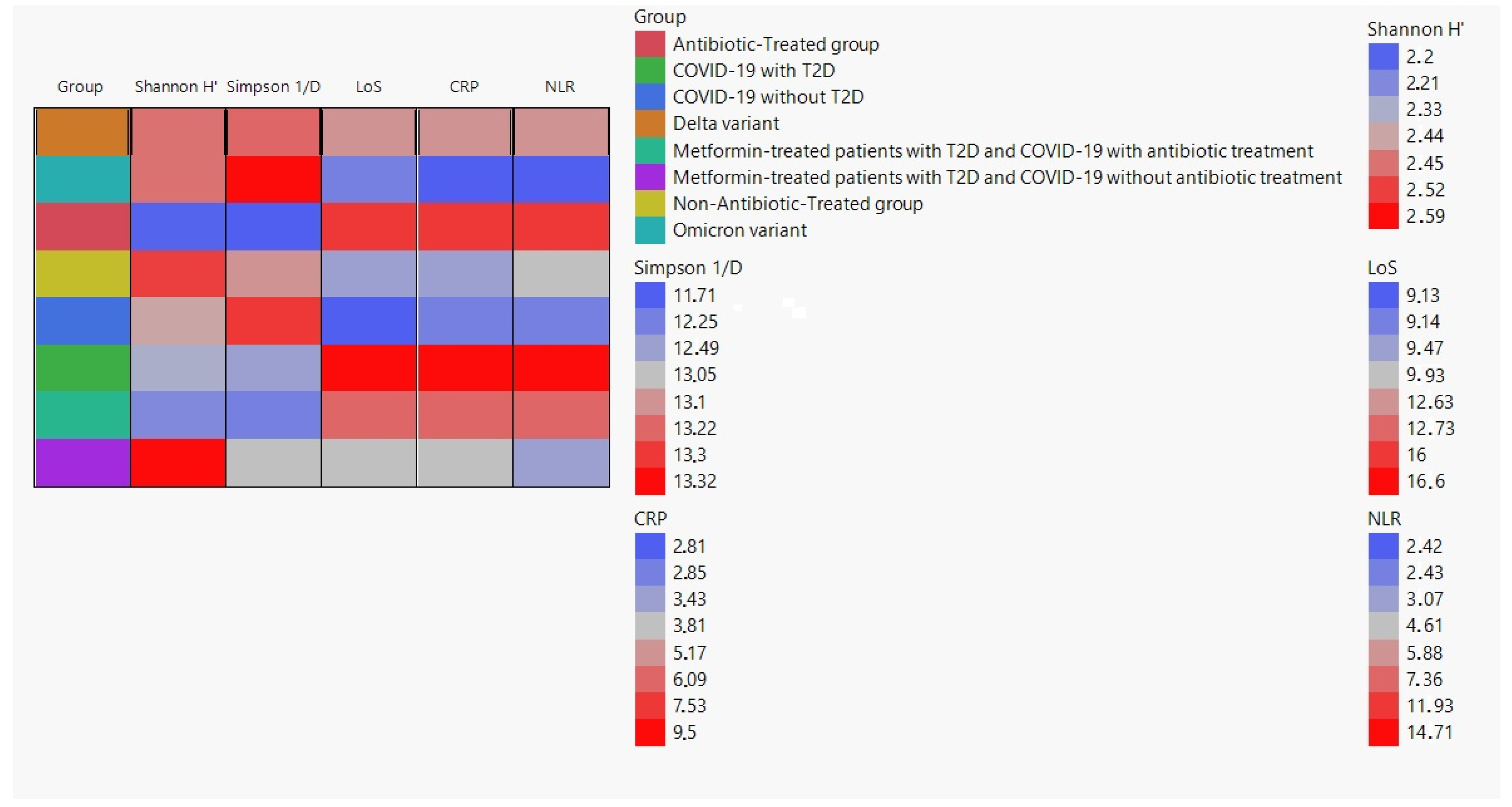
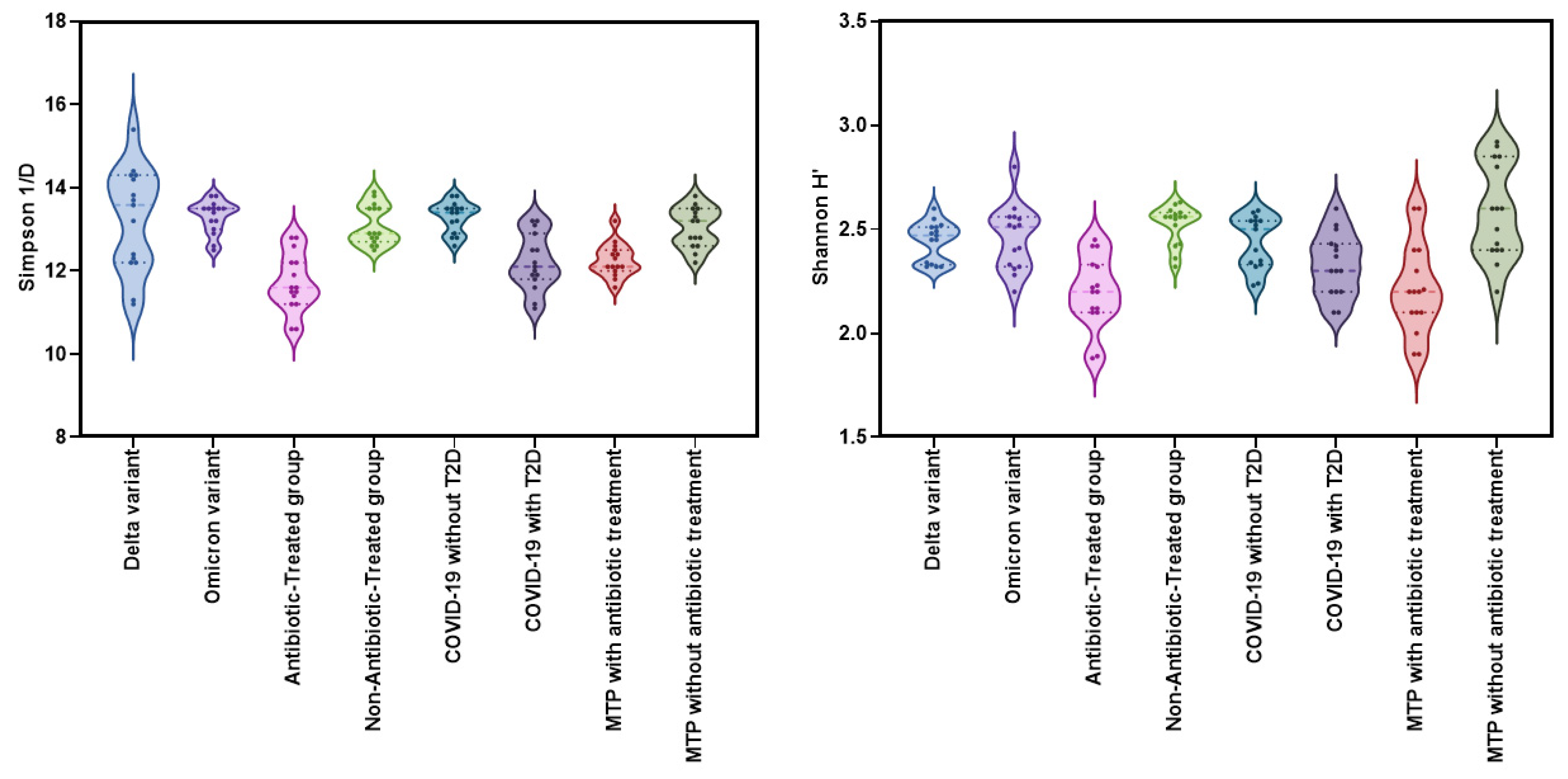
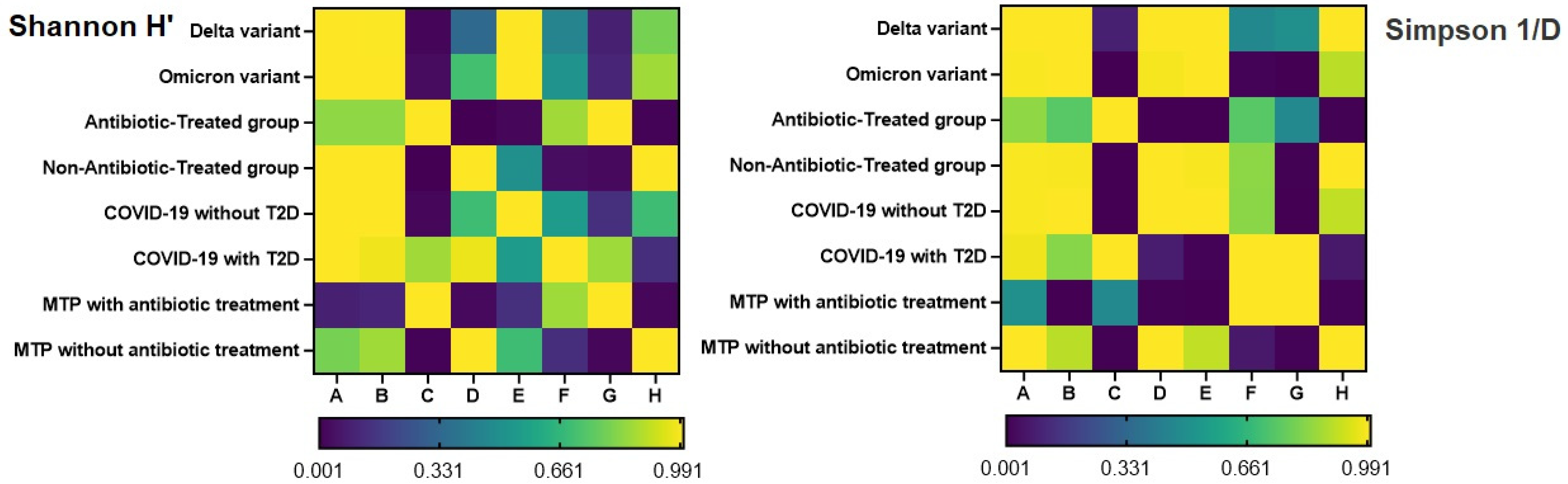
2.2. Alpha-Diversity Analysis
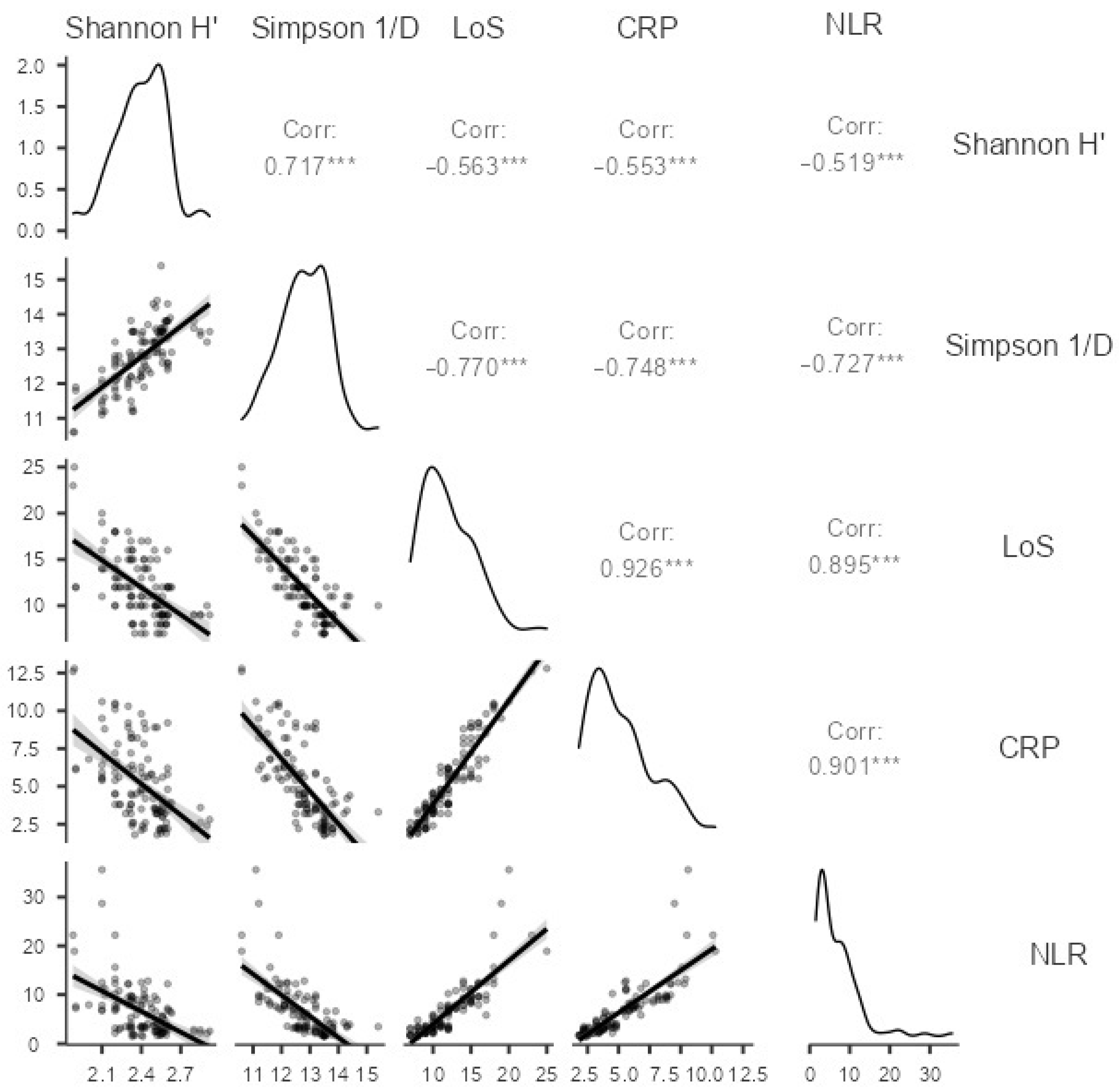
2.3. Correlation between Alpha Diversity Indices and Clinical Parameters
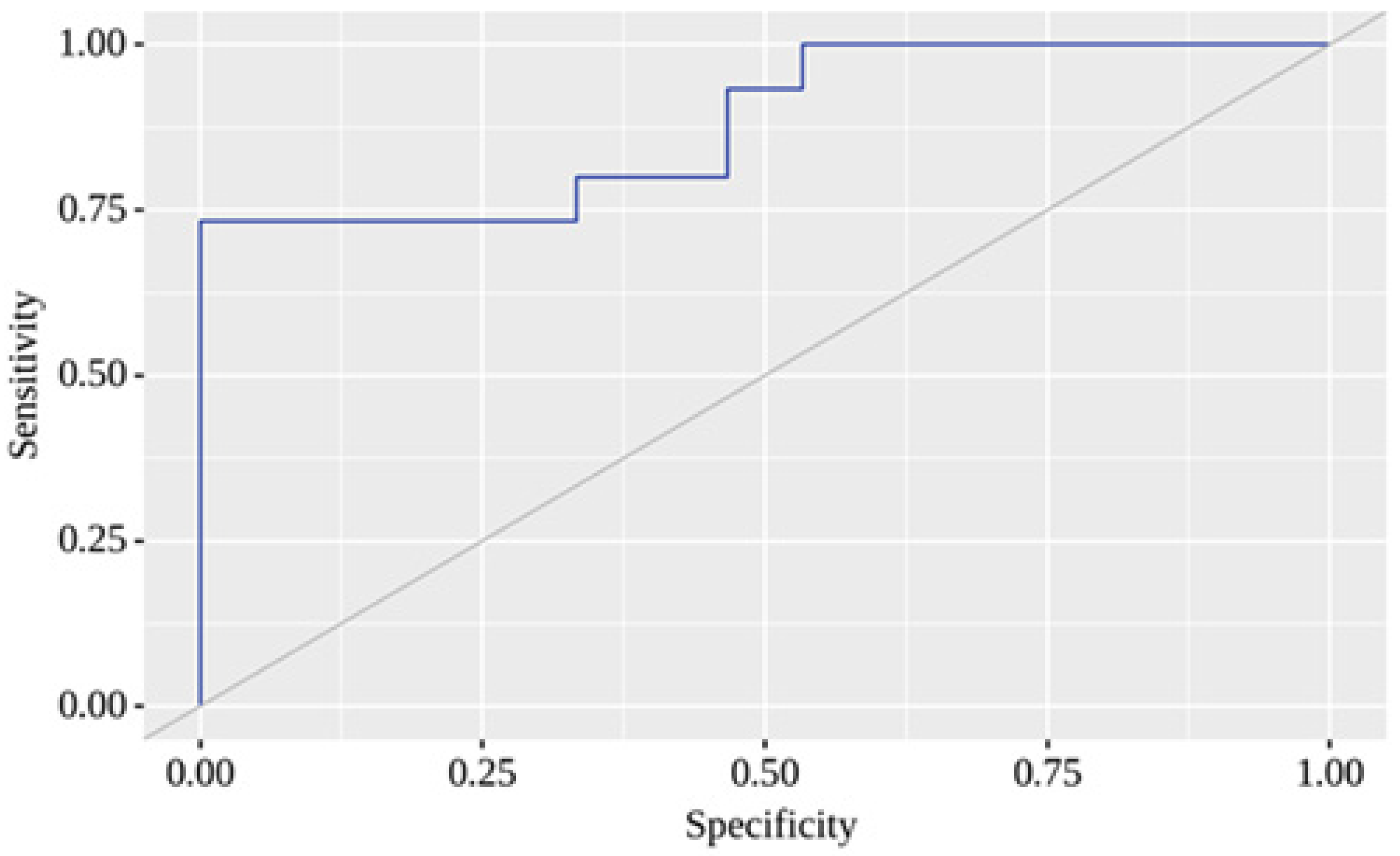
2.4. Binary Logistic Regression
3. Discussion
4. Material and Methods
4.1. Study Design and Sample Collection
4.2. Microbiota Analysis and Calculation of Alpha-Diversity
4.3. Statistical Analysis
5. Limitation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Piermattei, A.; Di Sante, G.; Migliara, G.; Delogu, G.; Ria, F. Immunomodulation by Gut Microbiota: Role of Toll-Like Receptor Expressed by T Cells. J. Immunol. Res. 2014, 2014, 586939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Ejtahed, H.S.; Tito, R.Y.; Siadat, S.D.; Hasani-Ranjbar, S.; Hoseini-Tavassol, Z.; Rymenans, L.; Verbeke, K.; Soroush, A.R.; Raes, J.; Larijani, B. Metformin induces weight loss associated with gut microbiota alteration in non-diabetic obese women: A randomized double-blind clinical trial. Eur. J. Endocrinol. 2019, 180, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, X.; Xu, J.; Lian, F.; Yu, X.; Zhao, Y.; Xu, L.; Zhang, M.; Zhao, X.; Shen, J.; Wu, S.; et al. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: A Multicenter, Randomized, Open Label Clinical Trial. mBio 2018, 9, e02392-17. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, A.L.; Stephens, J.W.; Harris, D.A. Gut microbiota influence in type 2 diabetes mellitus (T2DM). Gut Pathog. 2021, 13, 50. [Google Scholar] [CrossRef]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Qin, C.; Li, Y.; Wu, Z.; Liu, L. Oat phenolic compounds regulate metabolic syndrome in high fat diet-fed mice via gut microbiota. Food Biosci. 2022, 50, 101946. [Google Scholar] [CrossRef]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [Green Version]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [Green Version]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velásquez-Mejía, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Mannerås-Holm, L.; Ståhlman, M.; Olsson, L.M.; Serino, M.; Planas-Fèlix, M. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, N. Effects of metformin on the gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. 2020, 13, 5003. [Google Scholar] [CrossRef] [PubMed]
- Hussain, I.; Cher, G.L.Y.; Abid, M.A.; Abid, M.B. Role of Gut Microbiome in COVID-19: An Insight Into Pathogenesis and Therapeutic Potential. Front. Immunol. 2021, 12, 765965. [Google Scholar] [CrossRef]
- Petakh, P.; Isevych, V.; Mohammed, I.; Loshak, K.; Poliak, I.; Kamyshnyiy, A. Association between Use of Metformin and Insulin with Hematological Parameters in COVID-19 Patients with Type 2 Diabetes: A Single Center, Cross-Sectional Study. Clin. Diabetol. 2022, 11, 432–433. [Google Scholar] [CrossRef]
- Petakh, P.; Griga, V.; Mohammed, I.B.; Loshak, K.; Poliak, I.; Kamyshnyiy, A. Effects of Metformin, Insulin on Hematological Parameters of COVID-19 Patients with Type 2 Diabetes. Med. Arch. 2022, 76, 329–332. [Google Scholar] [CrossRef]
- Petakh, P.; Kamyshna, I.; Nykyforuk, A.; Yao, R.; Imbery, J.F.; Oksenych, V.; Korda, M.; Kamyshnyi, A. Immunoregulatory Intestinal Microbiota and COVID-19 in Patients with Type Two Diabetes: A Double-Edged Sword. J. Viruses 2022, 14, 477. [Google Scholar] [CrossRef]
- Buchynskyi, M.; Kamyshna, I.; Oksenych, V.; Zavidniuk, N.; Kamyshnyi, A. The Intersection of COVID-19 and Metabolic-Associated Fatty Liver Disease: An Overview of the Current Evidence. Viruses 2023, 15, 1072. [Google Scholar] [CrossRef]
- Petakh, P.; Oksenych, V.; Kamyshnyi, A. The F/B ratio as a biomarker for inflammation in COVID-19 and T2D: Impact of metformin. Biomed. Pharmacother. 2023, 163, 114892. [Google Scholar] [CrossRef] [PubMed]
- Gradisteanu Pircalabioru, G.; Grigore, G.A.; Czobor Barbu, I.; Chifiriuc, M.-C.; Savu, O. Impact of COVID-19 on the Microbiome and Inflammatory Status of Type 2 Diabetes Patients. Biomedicines 2023, 11, 179. [Google Scholar] [CrossRef] [PubMed]
- Patangia, D.V.; Anthony Ryan, C.; Dempsey, E.; Paul Ross, R.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiol. Open 2022, 11, e1260. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Raichon, L.; Venzon, M.; Klein, J.; Axelrad, J.E.; Zhang, C.; Sullivan, A.P.; Hussey, G.A.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat. Commun. 2022, 13, 5926. [Google Scholar] [CrossRef]
- Righi, E.; Lambertenghi, L.; Gorska, A.; Sciammarella, C.; Ivaldi, F.; Mirandola, M.; Sartor, A.; Tacconelli, E. Impact of COVID-19 and Antibiotic Treatments on Gut Microbiome: A Role for Enterococcus spp. J. Biomed. 2022, 10, 2786. [Google Scholar] [CrossRef]
- Romani, L.; Del Chierico, F.; Macari, G.; Pane, S.; Ristori, M.V.; Guarrasi, V.; Gardini, S.; Pascucci, G.R.; Cotugno, N.; Perno, C.F.; et al. The Relationship Between Pediatric Gut Microbiota and SARS-CoV-2 Infection. Front. Cell Infect. Microbiol. 2022, 12, 908492. [Google Scholar] [CrossRef]
- Yin, Y.S.; Minacapelli, C.D.; Parmar, V.; Catalano, C.C.; Bhurwal, A.; Gupta, K.; Rustgi, V.K.; Blaser, M.J. Alterations of the fecal microbiota in relation to acute COVID-19 infection and recovery. Mol. Biomed. 2022, 3, 36. [Google Scholar] [CrossRef]
- Rebelo, J.S.; Domingues, C.P.F.; Dionisio, F.; Gomes, M.C.; Botelho, A.; Nogueira, T. COVID-19 Lockdowns May Reduce Resistance Genes Diversity in the Human Microbiome and the Need for Antibiotics. Int. J. Mol. Sci. 2021, 22, 6891. [Google Scholar] [CrossRef]
- Petakh, P.; Kobyliak, N.; Kamyshnyi, A. Gut microbiota in patients with COVID-19 and type 2 diabetes: A culture-based method. Front. Cell. Infect. Microbiol. 2023, 13, 1142578. [Google Scholar] [CrossRef]
- Li, Y.; Peng, Y.; Shen, Y.; Zhang, Y.; Liu, L.; Yang, X. Dietary polyphenols: Regulate the advanced glycation end products-RAGE axis and the microbiota-gut-brain axis to prevent neurodegenerative diseases. Crit. Rev. Food Sci. Nutr. 2022, 19, 1–27. [Google Scholar] [CrossRef]
- Tian, L.; Jin, T. The incretin hormone GLP-1 and mechanisms underlying its secretion. J. Diabetes 2016, 8, 753–765. [Google Scholar] [CrossRef] [Green Version]
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.B.; Gomes, G.F.; Angelim, M.; Souza, G.F.; Muraro, S.P.; Toledo-Teixeira, D.A.; Rattis, B.A.C.; Passos, A.S.; Pral, L.P.; de Rezende Rodovalho, V.; et al. Impact of Microbiota Depletion by Antibiotics on SARS-CoV-2 Infection of K18-hACE2 Mice. Cells 2022, 11, 2572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Trøseid, M.; Holter, J.C.; Holm, K.; Vestad, B.; Sazonova, T.; Granerud, B.K.; Dyrhol-Riise, A.M.; Holten, A.R.; Tonby, K.; Kildal, A.B.; et al. Gut microbiota composition during hospitalization is associated with 60-day mortality after severe COVID-19. Crit. Care 2023, 27, 69. [Google Scholar] [CrossRef]
- Fujisaka, S.; Ussar, S.; Clish, C.; Devkota, S.; Dreyfuss, J.M.; Sakaguchi, M.; Soto, M.; Konishi, M.; Softic, S.; Altindis, E.; et al. Antibiotic effects on gut microbiota and metabolism are host dependent. J. Clin. Investig. 2016, 126, 4430–4443. [Google Scholar] [CrossRef]
- Dahiya, D.; Nigam, P.S. Antibiotic-Therapy-Induced Gut Dysbiosis Affecting Gut Microbiota-Brain Axis and Cognition: Restoration by Intake of Probiotics and Synbiotics. Int. J. Mol. Sci. 2023, 24, 3074. [Google Scholar] [CrossRef]
- Martinez, E.; Taminiau, B.; Rodriguez, C.; Daube, G. Gut Microbiota Composition Associated with Clostridioides difficile Colonization and Infection. Pathogens 2022, 11, 781. [Google Scholar] [CrossRef]
- Dong, L.; Li, Y.; Chen, Q.; Liu, Y.; Qiao, Z.; Sang, S.; Zhang, J.; Zhan, S.; Wu, Z.; Liu, L. Research advances of advanced glycation end products in milk and dairy products: Formation, determination, control strategy and immunometabolism via gut microbiota. Food Chem. 2023, 417, 135861. [Google Scholar] [CrossRef]
- Tangvarasittichai, S. Oxidative stress, insulin resistance, dyslipidemia and type 2 diabetes mellitus. World J. Diabetes 2015, 6, 456–480. [Google Scholar] [CrossRef]
- Jansen, T.; Kvandová, M.; Daiber, A.; Stamm, P.; Frenis, K.; Schulz, E.; Münzel, T.; Kröller-Schön, S. The AMP-Activated Protein Kinase Plays a Role in Antioxidant Defense and Regulation of Vascular Inflammation. Antioxidants 2020, 9, 525. [Google Scholar] [CrossRef] [PubMed]
- Petakh, P.; Loshak, K.; Kamyshnyi, A. Hematological features of patients with type 2 diabetes depending on the variant of SARS-CoV-2. Fiziolohichnyĭ Zhurnal 2023, 69, 35–42. [Google Scholar] [CrossRef]
- Usman, A.; Bliden, K.P.; Cho, A.; Walia, N.; Jerjian, C.; Singh, A.; Kundan, P.; Duhan, S.; Tantry, U.S.; Gurbel, P.A. Metformin use in patients hospitalized with COVID-19: Lower inflammation, oxidative stress, and thrombotic risk markers and better clinical outcomes. J. Thromb. Thrombolysis 2022, 53, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Kamyshnyi, O.; Matskevych, V.; Lenchuk, T.; Strilbytska, O.; Storey, K.; Lushchak, O. Metformin to decrease COVID-19 severity and mortality: Molecular mechanisms and therapeutic potential. Biomed. Pharmacother. 2021, 144, 112230. [Google Scholar] [CrossRef]
- Buchynskyi, M.; Kamyshna, I.; Lyubomirskaya, K.; Moshynets, O.; Kobyliak, N.; Oksenych, V.; Kamyshnyi, A. Efficacy of interferon alpha for the treatment of hospitalized patients with COVID-19: A meta-analysis. Front. Immunol. 2023, 14, 1069894. [Google Scholar] [CrossRef]
- Kamyshnyi, A.; Koval, H.; Kobevko, O.; Buchynskyi, M.; Oksenych, V.; Kainov, D.; Lyubomirskaya, K.; Kamyshna, I.; Potters, G.; Moshynets, O. Therapeutic Effectiveness of Interferon-α2b against COVID-19 with Community-Acquired Pneumonia: The Ukrainian Experience. Int. J. Mol. Sci. 2023, 24, 6887. [Google Scholar] [CrossRef]
- Ianevski, A.; Ahmad, S.; Anunnitipat, K.; Oksenych, V.; Zusinaite, E.; Tenson, T.; Bjørås, M.; Kainov, D.E. Seven classes of antiviral agents. Cell. Mol. Life Sci. 2022, 79, 605. [Google Scholar] [CrossRef]
- Ianevski, A.; Simonsen, R.M.; Myhre, V.; Tenson, T.; Oksenych, V.; Bjørås, M.; Kainov, D.E. DrugVirus.info 2.0: An integrative data portal for broad-spectrum antivirals (BSA) and BSA-containing drug combinations (BCCs). Nucleic Acids Res. 2022, 50, W272–W275. [Google Scholar] [CrossRef]
- Ianevski, A.; Yao, R.; Simonsen, R.M.; Myhre, V.; Ravlo, E.; Kaynova, G.D.; Zusinaite, E.; White, J.M.; Polyak, S.J.; Oksenych, V.; et al. Mono- and combinational drug therapies for global viral pandemic preparedness. iScience 2022, 25, 104112. [Google Scholar] [CrossRef]
- American Society for Microbiology. Biochemical Tests for the Identification of Aerobic Bacteria. In Clinical Microbiology Procedures Handbook; Wiley: Hoboken, NJ, USA, 2016; pp. 3.17.11.11–13.17.48.13. [Google Scholar] [CrossRef]


| Group | Age (Mean ± SD) | Male (%) | Female (%) |
|---|---|---|---|
| Delta variant | 55.1 ± 11.4 | 53.4 | 46.6 |
| Omicron variant | 56.7 ± 4.9 | 46.6 | 53.4 |
| Antibiotic-treated group | 55.3 ± 11.8 | 46.6 | 53.4 |
| Non-antibiotic-treated group | 43.6 ± 9.7 | 40.0 | 60.0 |
| COVID-19 without T2D | 55.4 ± 5.0 | 53.4 | 46.6 |
| COVID-19 with T2D | 61.5 ± 8.0 | 53.4 | 46.6 |
| Metformin-treated patients with T2D and COVID-19 with antibiotic treatment | 55.2 ± 5.4 | 53.4 | 46.6 |
| Metformin-treated patients with T2D and COVID-19 without antibiotic treatment | 50.1 ± 6.6 | 40.0 | 60.0 |
| Group | Simpson 1/D (Mean ± SD) | Shannon H′ (Mean ± SD) | LoS (Mean ± SD) | CRP (Mean ± SD) | NLR (Mean ± SD) |
|---|---|---|---|---|---|
| Delta variant | 13.2 ± 1.19 | 2.4 ± 0.09 | 12.6 ± 2.47 | 5.1 ± 1.54 | 5.8 ± 2.47 |
| Omicron variant | 13.3 ± 0.40 | 2.4 ± 0.16 | 9.1 ± 1.56 | 2.8 ± 0.72 | 2.4 ± 0.66 |
| Antibiotic-treated group | 11.7 ± 0.70 | 2.2 ± 0.17 | 16.0 ± 3.54 | 7.5 ± 2.47 | 11.9 ± 3.79 |
| Non-antibiotic-treated group | 13.1 ± 0.47 | 2.5 ± 0.09 | 9.4 ± 1.76 | 3.4 ± 0.97 | 4.6 ± 1.26 |
| COVID-19 without T2D | 13.2 ± 0.37 | 2.4 ± 0.12 | 9.1 ± 1.59 | 2.8 ± 0.71 | 2.4 ± 0.64 |
| COVID-19 with T2D | 12.5 ± 0.62 | 2.3 ± 0.15 | 16.6 ± 1.84 | 9.5 ± 0.73 | 14.7 ± 7.95 |
| Metformin-treated patients with T2D and COVID-19 with antibiotic treatment | 12.2 ± 0.39 | 2.2 ± 0.21 | 12.7 ± 1.33 | 6.0 ± 0.38 | 7.3 ± 0.45 |
| Metformin-treated patients with T2D and COVID-19 without antibiotic treatment | 13.0 ± 0.48 | 2.6 ± 0.23 | 9.9 ± 1.09 | 3.8 ± 0.89 | 3.07 ± 0.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petakh, P.; Kamyshna, I.; Oksenych, V.; Kainov, D.; Kamyshnyi, A. Metformin Therapy Changes Gut Microbiota Alpha-Diversity in COVID-19 Patients with Type 2 Diabetes: The Role of SARS-CoV-2 Variants and Antibiotic Treatment. Pharmaceuticals 2023, 16, 904. https://doi.org/10.3390/ph16060904
Petakh P, Kamyshna I, Oksenych V, Kainov D, Kamyshnyi A. Metformin Therapy Changes Gut Microbiota Alpha-Diversity in COVID-19 Patients with Type 2 Diabetes: The Role of SARS-CoV-2 Variants and Antibiotic Treatment. Pharmaceuticals. 2023; 16(6):904. https://doi.org/10.3390/ph16060904
Chicago/Turabian StylePetakh, Pavlo, Iryna Kamyshna, Valentyn Oksenych, Denis Kainov, and Aleksandr Kamyshnyi. 2023. "Metformin Therapy Changes Gut Microbiota Alpha-Diversity in COVID-19 Patients with Type 2 Diabetes: The Role of SARS-CoV-2 Variants and Antibiotic Treatment" Pharmaceuticals 16, no. 6: 904. https://doi.org/10.3390/ph16060904
APA StylePetakh, P., Kamyshna, I., Oksenych, V., Kainov, D., & Kamyshnyi, A. (2023). Metformin Therapy Changes Gut Microbiota Alpha-Diversity in COVID-19 Patients with Type 2 Diabetes: The Role of SARS-CoV-2 Variants and Antibiotic Treatment. Pharmaceuticals, 16(6), 904. https://doi.org/10.3390/ph16060904










