Influence of Different Diets on the Degradation of Sulfasalazine by Colon Bacteria Determined Using MimiCol3
Abstract
:1. Introduction
2. Results
2.1. Cultivation and Characterization of Three Different Standard Microbiota
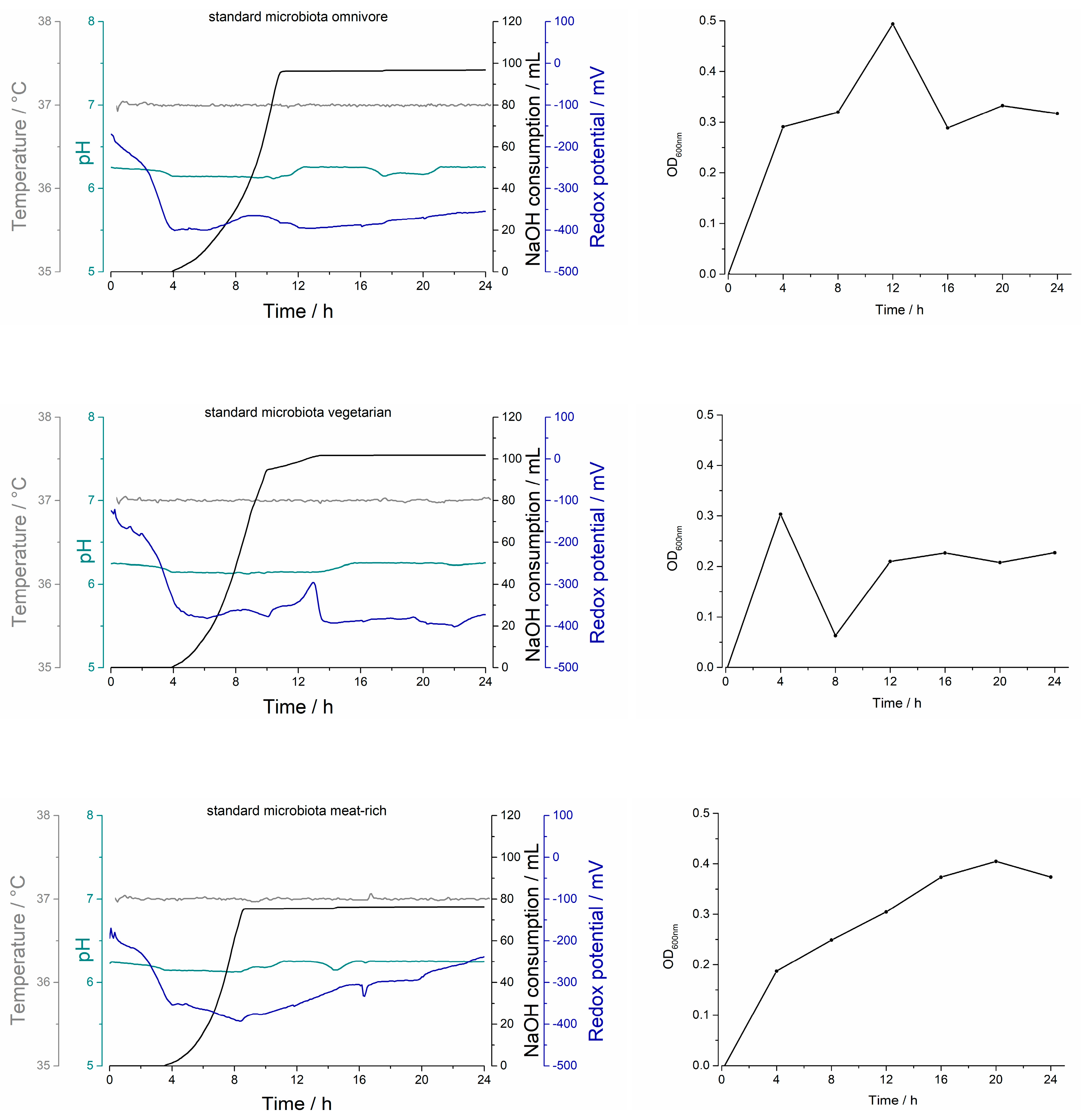
2.1.1. Biostat® A Plus Process Parameters
2.1.2. Optical Density
2.1.3. Characterization of the Three Different Standard Microbiota Using Selective Agar
2.2. Investigation of Sulfasalazine Degradation and Bacterial Growth Using MimiCol3
2.2.1. Process Parameters
2.2.2. Optical Density
2.2.3. Degradation of Sulfasalazine
2.2.4. Determination of the Three Different Standard Microbiota and the Pooled Standard Microbiota Using Selective Agar
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
4.2.1. Preparation of Standard Microbiota
4.2.2. Characterization of Standard Microbiota
4.2.3. Experimental Procedure
4.2.4. Preparation of Calibration Curves
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Hara, A.M.; Shanahan, F. The Gut Flora as a Forgotten Organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doestzada, M.; Vila, A.V.; Zhernakova, A.; Koonen, D.P.Y.; Weersma, R.K.; Touw, D.J.; Kuipers, F.; Wijmenga, C.; Fu, J. Pharmacomicrobiomics: A Novel Route towards Personalized Medicine? Protein Cell 2018, 9, 432–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaut, M. The Intestinal Microbiome: Fundamentals and Influence of Diet. Diabetologe 2016, 12, 386–393. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef]
- Mitrea, L.; Nemeş, S.-A.; Szabo, K.; Teleky, B.-E.; Vodnar, D.-C. Guts Imbalance Imbalances the Brain: A Review of Gut Microbiota Association with Neurological and Psychiatric Disorders. Front. Med. 2022, 9, 813204. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Koontz, J.M.; Dancy, B.C.R.; Horton, C.L.; Stallings, J.D.; DiVito, V.T.; Lewis, J.A. The Role of the Human Microbiome in Chemical Toxicity. Int. J. Toxicol. 2019, 38, 251–264. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut Microbiota: Role in Pathogen Colonization, Immune Responses and Inflammatory Disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- McNeil, N.I. The Contribution of the Large Intestine to Energy Supplies in Man. Am. J. Clin. Nutr. 1984, 39, 338–342. [Google Scholar] [CrossRef]
- Li, Y.; Faden, H.S.; Zhu, L. The Response of the Gut Microbiota to Dietary Changes in the First Two Years of Life. Front. Pharmacol. 2020, 11, 334. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Paslier, D.L.; Batto, J.; Bertalan, M.; Borruel, N.; Casellas, F. Enterotypes of the Human Gut Microbiome. Nature 2013, 473, 174–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmer, J.; Lange, B.; Frick, J.S.; Sauer, H.; Zimmermann, K.; Schwiertz, A.; Rusch, K.; Klosterhalfen, S.; Enck, P. A Vegan or Vegetarian Diet Substantially Alters the Human Colonic Faecal Microbiota. Eur. J. Clin. Nutr. 2012, 66, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The Effects of Vegetarian and Vegan Diets on Gut Microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef] [Green Version]
- Ferrocino, I.; Di Cagno, R.; De Angelis, M.; Turroni, S.; Vannini, L.; Bancalari, E.; Rantsiou, K.; Cardinali, G.; Neviani, E.; Cocolin, L. Fecal Microbiota in Healthy Subjects Following Omnivore, Vegetarian and Vegan Diets: Culturable Populations and RRNA DGGE Profiling. PLoS ONE 2015, 10, e0128669. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of Diet on the Gut Microbiome and Implications for Human Health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Voreades, N.; Kozil, A.; Weir, T.L. Diet and the Development of the Human Intestinal Microbiome. Front. Microbiol. 2014, 5, 494. [Google Scholar] [CrossRef] [Green Version]
- Palleja, A.; Mikkelsen, K.H.; Forslund, S.K.; Kashani, A.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Liang, S.; Feng, Q.; Zhang, C.; et al. Recovery of Gut Microbiota of Healthy Adults Following Antibiotic Exposure. Nat. Microbiol. 2018, 3, 1255–1265. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between Drugs and the Gut Microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Jourova, L.; Anzenbacher, P.; Anzenbacherova, E. Human Gut Microbiota Plays a Role in the Metabolism of Drugs. Biomed. Pap. 2016, 160, 317–326. [Google Scholar] [CrossRef] [Green Version]
- Chavez-Bravo, E.; Alonso-Calderon, A.I.A.; Sanchez-Calvario, L.; Castaneda-Roldan, E.; Vidal Robles, E.; Salazar-Robles, G. Characterization of the Degradation Products from the Red Dye 40 by Enterobacteria. J. Pure Appl. Microbiol. 2016, 10, 2569–2575. [Google Scholar] [CrossRef]
- Saranraj, P.; Stella, D.; Sivasakthivelan, P. Scholars Research Library Separation, Purification and Characterization of Dye Degrading Enzyme Azoreductase from Bacterial Isolates. Cent. Eur. J. Exp. Biol. 2014, 3, 19–25. [Google Scholar]
- Zahran, S.A.; Ali-Tammam, M.; Hashem, A.M.; Aziz, R.K.; Ali, A.E. Azoreductase Activity of Dye-Decolorizing Bacteria Isolated from the Human Gut Microbiota. Sci. Rep. 2019, 9, 5508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molly, K.; Vande Woestyne, M.; Verstraete, W. Development of a 5-Step Multi-Chamber Reactor as a Simulation of the Human Intestinal Microbial Ecosystem. Appl. Microbiol. Biotechnol. 1993, 39, 254–258. [Google Scholar] [CrossRef]
- O’Farrell, C.; Stamatopoulos, K.; Simmons, M.; Batchelor, H. In Vitro Models to Evaluate Ingestible Devices: Present Status and Current Trends. Adv. Drug Deliv. Rev. 2021, 178, 113924. [Google Scholar] [CrossRef]
- Venema, K. The TNO In Vitro Model of the Colon (TIM-2). In The Impact of Food Bioactives on Health; Springer International Publishing: Cham, Switzerland, 2015; pp. 293–304. ISBN 9783319161044. [Google Scholar]
- Beeck, R.; Dols, A.; Weitschies, W.; Schneider, F.; Seradj, D.S.; Krause, J.; Schick, P. An Advanced Bioreactor Simulating Dynamic Physiological Conditions in the Human Ascending Colon: MimiCol3. Pharmaceutics 2022, 14, 1049. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors Affecting the Composition of the Gut Microbiota, and Its Modulation. PeerJ 2019, 2019, e7502. [Google Scholar] [CrossRef] [Green Version]
- Cummings, J.H.; Macfarlane, G.T. The Control and Consequences of Bacterial Fermentation in the Human Colon. J. Appl. Bacteriol. 1991, 70, 443–459. [Google Scholar] [CrossRef]
- Frantz, J.C.; McCallum, R.E. Growth Yields and Fermentation Balance of Bacteroides Fragilis Cultured in Glucose-Enriched Medium. J. Bacteriol. 1979, 137, 1263–1270. [Google Scholar] [CrossRef]
- Rangarajan, A.A.; Chia, H.E.; Azaldegui, C.A.; Olszewski, M.H.; Pereira, G.V.; Koropatkin, N.M.; Biteen, J.S. Ruminococcus Bromii Enables the Growth of Proximal Bacteroides Thetaiotaomicron by Releasing Glucose during Starch Degradation. Microbiol. (United Kingd.) 2022, 168, 001180. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Ding, L.; Huang, Q.; Fu, X.; Liu, F.; Dhital, S.; Zhang, B. In Vitro Colonic Fermentation Profiles and Microbial Responses of Propionylated High-Amylose Maize Starch by Individual Bacteroides-Dominated Enterotype Inocula. Food Res. Int. 2021, 144, 110317. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [Green Version]
- Dr Schoefer, L. Naturheilkunde Metabolische Endotoxinämie Überfordert Die Leber Metabolische Endotoxinämie. Naturheilkd. J. 2017, 19, 2–4. [Google Scholar]
- Arbor, A.; Arbor, A. Culture Enterobacteria. Microbiology 1974, 119, 736–747. [Google Scholar]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007, 5, 1556–1573. [Google Scholar] [CrossRef] [Green Version]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science. 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- Hold, G.L.; Pryde, S.E.; Russell, V.J.; Furrie, E.; Flint, H.J. Assessment of Microbial Diversity in Human Colonic Samples by 16S RDNA Sequence Analysis. FEMS Microbiol. Ecol. 2002, 39, 33–39. [Google Scholar] [CrossRef]
- Christensen, J. Motility of the Colon. In Physiology of the Gastrointestinal Tract, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 1994; pp. 991–1024. [Google Scholar]
- Malbert, C.H. The Ileocolonic Sphincter. Neurogastroenterol. Motil. 2005, 17, 41–49. [Google Scholar] [CrossRef]
- Maier, R.M. Bacterial Growth, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2009; ISBN 9780123705198. [Google Scholar]
- Leibniz-Instiut Kultivierung von Anaerobiern. Available online: https://www.dsmz.de/fileadmin/Bereiche/Microbiology/Dateien/Kultivierungshinweise/Anaerob.pdf (accessed on 21 November 2021).
- Croucher, S.C.; Houston, A.P.; Bayliss, C.E.; Turner, R.J. Bacterial Populations Associated with Different Regions of the Human Colon Wall. Appl. Environ. Microbiol. 1983, 45, 1025–1033. [Google Scholar] [CrossRef]
- Beeck, R.; Glöckl, G.; Krause, J.; Schick, P.; Weitschies, W. Mimicking the Dynamic Colonic Microbiota in Vitro to Gain a Better Understanding on the in Vivo Metabolism of Xenobiotics: Degradation of Sulfasalazine. Int. J. Pharm. 2021, 603, 120704. [Google Scholar] [CrossRef] [PubMed]
- Sandborn, W.J.; Hanauer, S.B. Systematic Review: The Pharmacokinetic Profiles of Oral Mesalazine Formulations and Mesalazine pro-Drugs Used in the Management of Ulcerative Colitis. Aliment. Pharmacol. Ther. 2003, 17, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Degnan, B.A.; Macfarlane, G.T. Transport and Metabolism of Glucose and Arabinose in Bifidobacterium Breve. Arch. Microbiol. 1993, 160, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Rafii, F.; Franklin, W.; Cerniglia, C.E. Azoreductase Activity of Anaerobic Bacteria Isolated from Human Intestinal Microflora. Appl. Environ. Microbiol. 1990, 56, 2146–2151. [Google Scholar] [CrossRef] [PubMed]
- Kellow, J.E.; Borody, T.J.; Phillips, S.F.; Haddad, A.C.; Brown, M.L. Sulfapyridine Appearance in Plasma after Salicylazosulfapyridine. Another Simple Measure of Intestinal Transit. Gastroenterology 1986, 91, 396–400. [Google Scholar] [CrossRef]
- Minekus, M.; Smeets-Peeters, M.; Bernalier, A.; Marol-Bonnin, S.; Havenaar, R.; Marteau, P.; Alric, M.; Fonty, G.; Huis In’t Veld, J.H. A Computer-Controlled System to Simulate Conditions of the Large Intestine with Peristaltic Mixing, Water Absorption and Absorption of Fermentation Products. Appl. Microbiol. Biotechnol. 1999, 53, 108–114. [Google Scholar] [CrossRef]








| Solution | Compound | Concentration g/L |
|---|---|---|
| 1 | Dipotassium hydrogen phosphate | 2.50 |
| Sodium chloride | 4.50 | |
| Iron(II)sulfate heptahydrate | 0.005 | |
| 2 | Calcium chloride dihydrate | 0.45 |
| Magnesium sulfate heptahydrate | 0.50 | |
| 3 | Pectin (apple) | 0.60 |
| Xylan (beech) | 0.60 | |
| Arabinogalactan | 0.60 | |
| Amylopectin (corn) | 0.60 | |
| Starch (potato) | 5.00 | |
| 4 | L-cysteine | 0.40 |
| 5 | Meat peptones | 3.00 |
| Casein (cow’s milk) | 3.00 | |
| Ox bile | 0.05 | |
| 6 | Polysorbate 80 | 2.00 |
| 7 | Hemin | 0.01 |
| Agar Media | Bacteria Species |
|---|---|
| Tryptic soy agar (TSA/CASO) | Total aerobic |
| MacConkey agar | Enterobacteriaceae |
| Man, Rogosa and Sharpe (MRS) agar | Lactobacilli |
| Modified reinforced clostridial medium | Clostridia |
| Bifidus Selective Medium (BSM) agar | Bifidobacteria |
| Modified Schaedler agar | Bacteroides |
| Schaedler agar | Total anaerobes |
| Experiment | Vessel 1 | Vessel 2 | Vessel 3 |
|---|---|---|---|
| 1 | SMomni | SMveget | SMmeat |
| 2 | SMmeat | SMomni | SMveget |
| 3 | SMveget | SMmeat | SMomni |
| 4 | SMpooled | SMpooled | SMpooled |
| 5 | SMomni | SMveget | SMmeat |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seradj, D.-S.; Beeck, R.; Haase, A.; Krause, J.; Schick, P.; Weitschies, W. Influence of Different Diets on the Degradation of Sulfasalazine by Colon Bacteria Determined Using MimiCol3. Pharmaceuticals 2023, 16, 1128. https://doi.org/10.3390/ph16081128
Seradj D-S, Beeck R, Haase A, Krause J, Schick P, Weitschies W. Influence of Different Diets on the Degradation of Sulfasalazine by Colon Bacteria Determined Using MimiCol3. Pharmaceuticals. 2023; 16(8):1128. https://doi.org/10.3390/ph16081128
Chicago/Turabian StyleSeradj, Dariah-Sohreh, Regine Beeck, Annika Haase, Julius Krause, Philipp Schick, and Werner Weitschies. 2023. "Influence of Different Diets on the Degradation of Sulfasalazine by Colon Bacteria Determined Using MimiCol3" Pharmaceuticals 16, no. 8: 1128. https://doi.org/10.3390/ph16081128






