Neuroprotective Properties of Oleanolic Acid—Computational-Driven Molecular Research Combined with In Vitro and In Vivo Experiments
Abstract
:1. Introduction
2. Results
2.1. The Pharmacokinetic In Silico Studies on BBB Permeation
2.2. Anisotropic Membrane-like Systems
2.3. QSAR Analysis for BBB Permeation
n = 40, R2CV = 78.25%, R2pred = 74.02%, S = 0.436
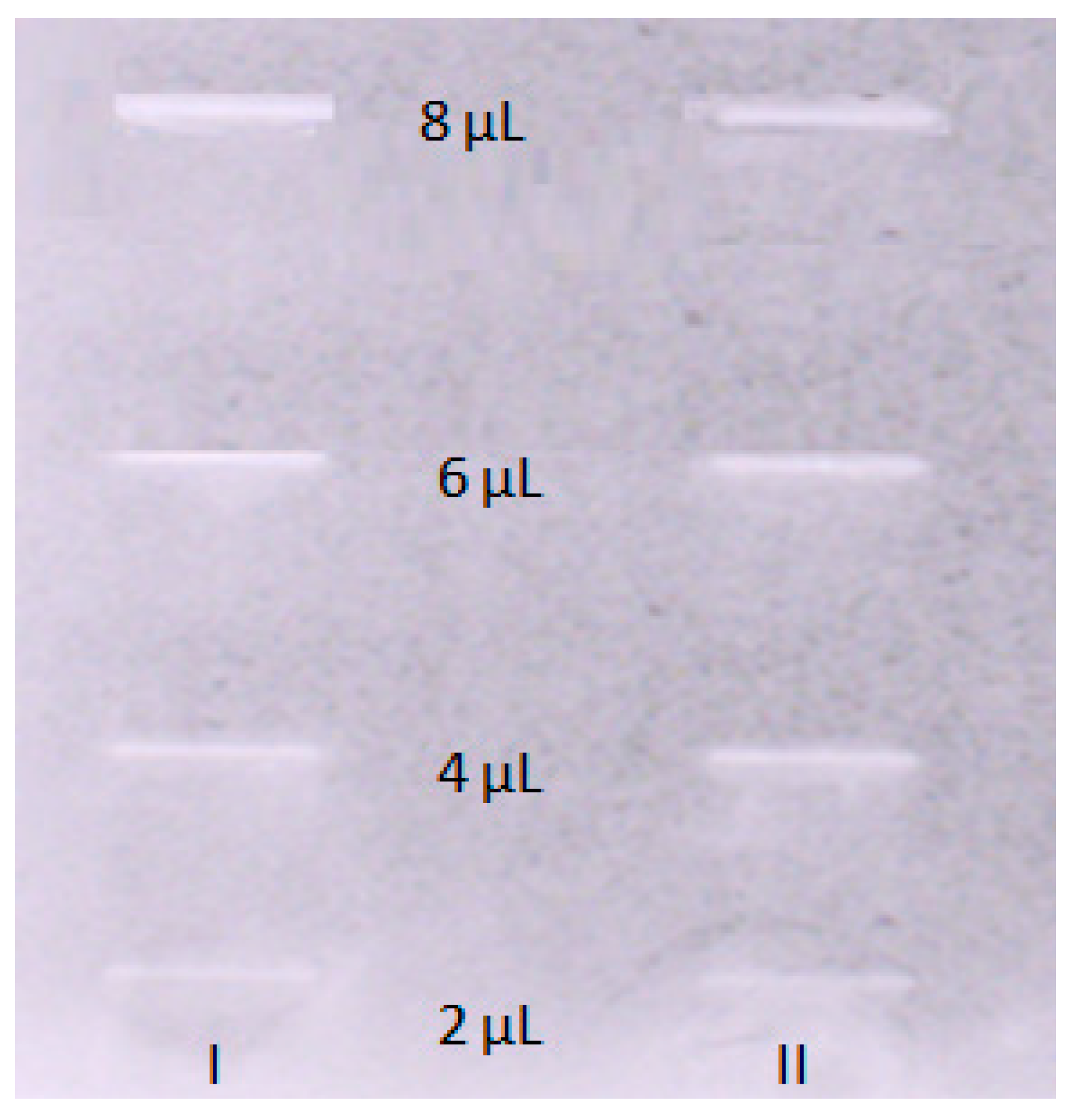
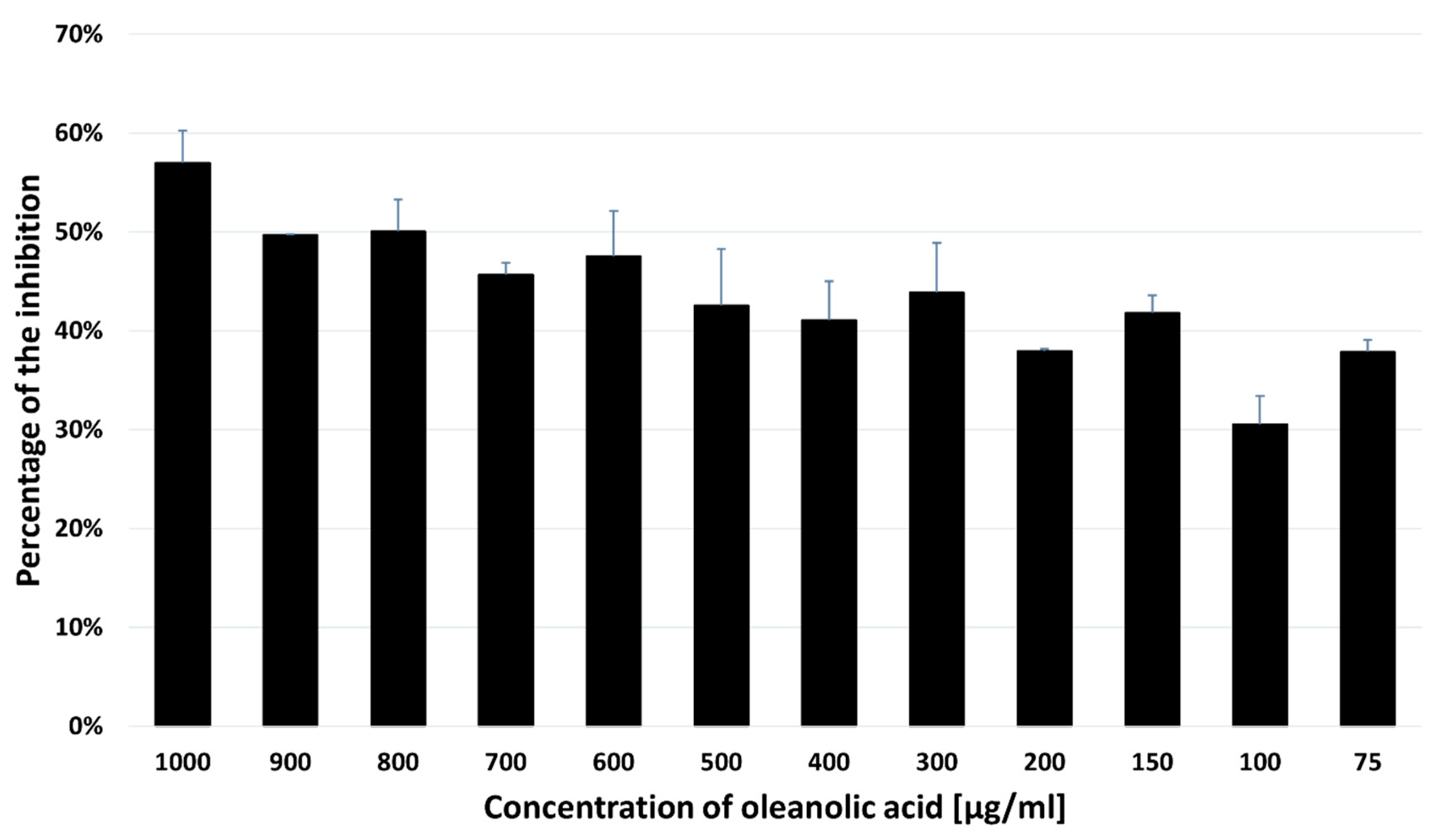
2.4. Acetylcholinesterase Inhibitory Activity of OA
2.5. Free Energy Calculations and Molecular Docking
2.6. Inhibitory Effect of Oleanolic Acid on the SH-SY5Y Cell Viability
2.7. Effect of Oleanolic Acid on the Cell Cycle Analyzed with the Flow Cytometry
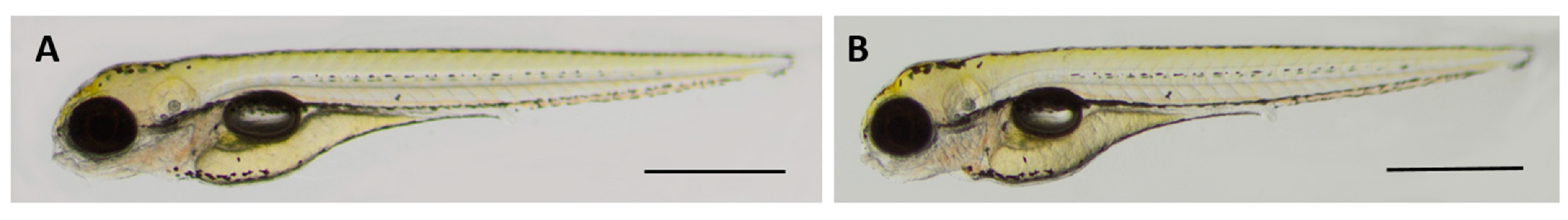
2.8. Oleanolic Acid Is Non-Toxic for Developing Zebrafish
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. In Silico Determination of Blood–Brain Barrier (BBB) Pharmacokinetic Descriptors
4.3. Membrane-like Chromatographic Equipment and Conditions
4.4. Quantitative Structure-Activity Relationship (QSAR) Studies for Estimation of Permeation through the BBB
4.5. TLC-Bioautography Assay toward the Inhibition of Acetylcholinesterase (AChE) Activity
4.6. Molecular Docking
4.7. Free Energy Profiles
4.8. Cytotoxicity Test in Human Neuroblastoma In Vitro
4.8.1. Cell Culture
4.8.2. MTT Assay
4.8.3. Cell Cycle Analysis
4.9. Toxicity Analysis in Zebrafish In Vivo
4.9.1. Animals
4.9.2. Zebrafish Embryo Acute Toxicity Test
4.9.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tarabasz, D.; Szczeblewski, P.; Laskowski, T.; Płaziński, W.; Baranowska-Wójcik, E.; Szwajgier, D.; Kukula-Koch, W.; Meissner, H.O. The Distribution of Glucosinolates in Different Phenotypes of Lepidium peruvianum and Their Role as Acetyl- and Butyrylcholinesterase Inhibitors—In Silico and In Vitro Studies. Int. J. Mol. Sci. 2022, 23, 4858. [Google Scholar] [CrossRef] [PubMed]
- Kukula-Koch, W.; Koch, W.; Czernicka, L.; Głowniak, K.; Asakawa, Y.; Umeyama, A.; Marzec, Z.; Kuzuhara, T. MAO-A Inhibitory Potential of Terpene Constituents from Ginger Rhizomes-A Bioactivity Guided Fractionation. Molecules 2018, 23, 1301. [Google Scholar] [CrossRef] [PubMed]
- Urbain, A.; Simões-Pires, C.A. Thin-Layer Chromatography for the Detection and Analysis of Bioactive Natural Products. In Encyclopedia of Analytical Chemistry; Meyers, R.A., Ed.; Wiley: Hoboken, NJ, USA, 2023. [Google Scholar] [CrossRef]
- Rajmohan, R.; Reddy, P.H. Amyloid-Beta and Phosphorylated Tau Accumulations Cause Abnormalities at Synapses of Alzheimer’s disease Neurons. J. Alzheimer’s Dis. JAD 2017, 57, 975–999. [Google Scholar] [CrossRef] [PubMed]
- Abdykerimova, S.; Sakipova, Z.; Nakonieczna, S.; Koch, W.; Biernasiuk, A.; Grabarska, A.; Malm, A.; Kozhanova, K.; Kukula-Koch, W. Superior Antioxidant Capacity of Berberis iliensis-HPLC-Q-TOF-MS Based Phytochemical Studies and Spectrophotometric Determinations. Antioxidants 2020, 9, 504. [Google Scholar] [CrossRef] [PubMed]
- Smyrska-Wieleba, N.; Mroczek, T. Natural Inhibitors of Cholinesterases: Chemistry, Structure–Activity and Methods of Their Analysis. Int. J. Mol. Sci. 2023, 24, 2722. [Google Scholar] [CrossRef]
- Mroczek, T.; Dymek, A.; Widelski, J.; Wojtanowski, K.K. The Bioassay-Guided Fractionation and Identification of Potent Acetylcholinesterase Inhibitors from Narcissus c.v. ’Hawera’ Using Optimized Vacuum Liquid Chromatography, High Resolution Mass Spectrometry and Bioautography. Metabolites 2020, 10, 395. [Google Scholar] [CrossRef]
- Ouyang, C.; Ma, X.; Zhao, J.; Li, S.; Liu, C.; Tang, Y.; Zhou, J.; Chen, J.; Li, X.; Li, W. Oleanolic acid inhibits mercury chloride induced-liver ferroptosis by regulating ROS/iron overload. Ecotoxicol. Environ. Saf. 2023, 258, 114973. [Google Scholar] [CrossRef]
- Ayeleso, T.B.; Matumba, M.G.; Mukwevho, E. Oleanolic Acid and Its Derivatives: Biological Activities and Therapeutic Potential in Chronic Diseases. Molecules 2017, 22, 1915. [Google Scholar] [CrossRef]
- Castellano, J.M.; Ramos-Romero, S.; Perona, J.S. Oleanolic Acid: Extraction, Characterization and Biological Activity. Nutrients 2022, 14, 623. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, C.; Zhao, F.; Li, D.; Lu, W. Biosynthesis of ursolic acid and oleanolic acid in Saccharomyces cerevisiae. AIChE J. 2018, 64, 3794–3802. [Google Scholar] [CrossRef]
- Soczewiński, E.; Wachtmeister, C.A. The relation between the composition of certain ternary two-phase solvent systems and RM values. J. Chromatogr. 1962, 7, 311–320. [Google Scholar] [CrossRef]
- Janicka, M.; Sztanke, M.; Sztanke, K. Predicting the Blood-Brain Barrier Permeability of New Drug-like Compounds via HPLC with Various Stationary Phases. Molecules 2020, 25, 487. [Google Scholar] [CrossRef] [PubMed]
- Stępnik, K. Biomimetic Chromatographic Studies Combined with the Computational Approach to Investigate the Ability of Triterpenoid Saponins of Plant Origin to Cross the Blood–Brain Barrier. Int. J. Mol. Sci. 2021, 22, 3573. [Google Scholar] [CrossRef]
- Platzer, G.; Mayer, M.; Beier, A.; Brüschweiler, S.; Fuchs, J.E.; Engelhardt, H.; Geist, L.; Bader, G.; Schörghuber, J.; Lichtenecker, R.; et al. PI by NMR: Probing CH–π Interactions in Protein–Ligand Complexes by NMR Spectroscopy. Angew. Chem. Int. Ed. 2020, 59, 14861. [Google Scholar] [CrossRef]
- Ferreira de Freitas, R.; Schapira, M. A systematic analysis of atomic protein–ligand interactions in the PDB. Med. Chem.Commun. 2017, 8, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Nishio, M.; Umezawa, Y.; Fantini, J.; Weiss, M.S.; Chakrabarti, P. CH–π hydrogen bonds in biological macromolecules. Phys.Chem. Chem. Phys. 2014, 16, 12648–12683. [Google Scholar] [CrossRef]
- World Health Organization. Global Action Plan on the Public Health Response to Dementia 2017–2025; WHO: Geneva, Switzerland, 2017; ISBN 978-92-4-151348-7. [Google Scholar]
- Birks, J.; Grimley, E.J. Ginkgo biloba for cognitive impairment and dementia. Cochrane Database Syst. Rev. 2009, 21, CD003120. [Google Scholar] [CrossRef]
- Joo, S.S.; Lee, D.I. Potential effects of microglial activation induced by ginsenoside Rg3 in rat primary culture: Enhancement of type a macrophage scavenger receptor expression. Arch. Pharm. Res. 2005, 28, 1164–1169. [Google Scholar] [CrossRef]
- Chen, F.; Eckman, E.A.; Eckman, C.B. Reductions in levels of the Alzheimer’s amyloid β peptide after oral administration of ginsenosides. FASEB J. 2006, 20, 1269–1271. [Google Scholar] [CrossRef]
- Li, N.; Liu, B.; Dluzen, D.E.; Jin, Y. Protective effects of ginsenoside Rg2 against glutamate induced neurotoxicity in PC12 cells. J. Ethnopharmacol. 2007, 111, 458–463. [Google Scholar] [CrossRef]
- Joo, S.S.; Yoo, Y.M.; Ahn, B.W.; Nam, S.Y.; Kim, Y.-B.; Hwang, K.W.; Lee, D.I. Prevention of inflammation-mediated neurotoxicity by Rg3 and its role in microglial activation. Biol. Pharm. Bull. 2008, 31, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Shieh, P.C.; Tsao, C.W.; Li, J.S.; Wu, H.T.; Wen, Y.J.; Kou, D.H.; Cheng, J.T. Role of pituitary adenylate cyclase-activating polypeptide (PACAP) in the action of ginsenoside Rh2 against betaamyloid-induced inhibition of rat brain astrocytes. Neurosci. Lett. 2008, 434, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, I.; Yasui, K. Wogonin inhibits inducible prostaglandin E2 production in macrophages. Eur. J. Pharmacol. 2000, 406, 477–481. [Google Scholar] [CrossRef]
- Park, B.K.; Heo, M.Y.; Par, H.; Kim, H.P. Inhibition of TPA-induced cyclooxygenase-2 expression and skin inflammation in mice by wogonin, a plant flavone from Scutellaria radix. Eur. J. Pharmacol. 2001, 425, 153–157. [Google Scholar] [CrossRef]
- Nakamura, N.; Hayasaka, S.; Zhang, X.Y.; Nagaki, Y.; Matsumoto, M.; Hayasaka, Y.; Terasawa, K. Effects of baicalein, baicalin, and wogonin on interleukin-6 and interleukin-8 expression, and nuclear factor-kb binding activities induced by interleukin-1beta in human retinal pigment epithelial cell line. Exp. Eye Res. 2003, 77, 195–202. [Google Scholar] [CrossRef]
- Suk, K.; Lee, H.; Kang, S.S.; Cho, G.J.; Choi, W.S. Flavonoid baicalein attenuates activation induced cell death of brain microglia. J. Pharmacol. Exp. Ther. 2003, 305, 638–645. [Google Scholar] [CrossRef]
- Hosseinzadeh, H.; Ramezani, M.; Shafaei, H.; Taghiabadi, E. Anticonvulsant effect of Berberis integerrima L. root extracts in mice. J. Acupunct. Meridian Stud. 2013, 6, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Ya’u, J.; Yaro, A.H.; Malami, S.; Musa, M.A.; Abubakar, A.; Yahaya, S.M.; Chindo, B.A.; Anuka, J.A.; Hussaini, I.M. Anticonvulsant activity of aqueous fraction of Carissa edulis root bark. Pharm. Biol. 2015, 53, 1329–1338. [Google Scholar] [CrossRef]
- Bhat, J.U.; Nizami, Q.; Asiaf, A.; Parray, S.A.; Ahmad, S.T.; Aslam, M.; Khanam, R.; Mujeeb, M.; Umar, S.; Siddiqi, A. Anticonvulsant activity of methanolic and aqueous extracts of Melissa parviflora in experimentally induced Swiss albino mice. EXCLI J. 2012, 11, 1–6. [Google Scholar]
- Tariq, U.; Butt, M.S.; Pasha, I.; Faisal, M.N. Neuroprotective effects of Olea europaea L. fruit extract against cigarette smoke-induced depressive-like behaviors in Sprague-Dawley rats. J. Food Biochem. 2021, 45, e14014. [Google Scholar] [CrossRef]
- Angeloni, C.; Malaguti, M.; Barbalace, M.C.; Hrelia, S. Bioactivity of Olive Oil Phenols in Neuroprotection. Int. J. Mol. Sci. 2017, 18, 2230. [Google Scholar] [CrossRef] [PubMed]
- dos Santos-Neto, L.L.; Toledo, M.A.d.V.; Medeiros-Souza, P.; de Souza, G.A. The use of herbal medicine in Alzheimer’s disease—A systematic review. Evid. Based Complement. Altern. Med. 2006, 3, 441–445. [Google Scholar] [CrossRef]
- Bozin, B.; Mika-Dukic, N.; Samojlik, I.; Jovin, E. Antimicrobial and antioxidant properties of rosemary and sage (Rosmarinus officinalis L. and Salvia officinalis L., Lamiaceae) essential oils. J. Agric. Food. Chem. 2007, 55, 7879–7885. [Google Scholar] [CrossRef] [PubMed]
- Imanshahidi, M.; Hosseinzadeh, H. The pharmacological effects of Salvia species on the central nervous system. Phytother. Res. 2006, 20, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Visioli, F.; Grande, S.; Bogani, P.; Galli, C. The role of antioxidant in the Mediterranean diets: Focus on cancer. Eur. J. Cancer Prev. 2004, 13, 337–343. [Google Scholar] [CrossRef]
- Ninfali, P.; Mea, G.; Giorgini, S.; Rocchi, M.; Bacchiocca, M. Antioxidant capacity of vegetables, spices, and dressings relevant to nutrition. Br. J. Nutr. 2005, 93, 257–266. [Google Scholar] [CrossRef]
- Cheung, S.; Tai, J. Anti-proliferative and antioxidant properties of rosemary Rosmarinus officinalis. Oncol. Rep. 2007, 17, 1525–1531. [Google Scholar] [CrossRef]
- Lin, S.X.; Curtis, M.A.; Sperry, J. Pyridine alkaloids with activity in the central nervous system. Bioorg. Med. Chem. 2020, 28, 115820. [Google Scholar] [CrossRef]
- Arens, H.; Borbe, H.O.; Ulbrich, B.; Stöckigt, J. Detection of pericine, a new CNS-active indole alkaloid from Picralima nitida cell suspension culture by opiate receptor binding studies. Planta Med. 1982, 46, 210–214. [Google Scholar] [CrossRef]
- Paladini, A.C.; Marder, M.; Viola, H.; Wolfman, C.; Wasowski, C.; Medina, J.H. Flavonoids and the central nervous system: From forgotten factors to potent anxiolytic compounds. J. Pharm. Pharmacol. 1999, 51, 519–526. [Google Scholar] [CrossRef]
- Jäger, A.K.; Saaby, L. Flavonoids and the CNS. Molecules 2011, 16, 1471–1485. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Xu, X.; Lin, J.; Cui, X.; Xu, R. Neuroprotection by Saponins. Phytother. Res. 2015, 29, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.S.; Wang, C.Z.; Wicks, S.M.; Qi, L.W. Chemical and pharmacological studies of saponins with a focus on American ginseng. J. Gins. Res. 2010, 34, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Hussain, G.; Huang, J.; Rasul, A.; Anwar, H.; Imran, A.; Maqbool, J.; Razzaq, A.; Aziz, N.; Makhdoom, E.U.H.; Konuk, M.; et al. Putative Roles of Plant-Derived Tannins in Neurodegenerative and Neuropsychiatry Disorders: An Updated Review. Molecules 2019, 24, 2213. [Google Scholar] [CrossRef]
- Takahashi, R.N.; de Lima, T.C.; Morato, G.S. Pharmacological actions of tannic acid; II. Evaluation of CNS activity in animals. Planta Med. 1986, 52, 272–275. [Google Scholar] [CrossRef]
- Awasthi, M.; Upadhyay, A.K.; Singh, S.; Pandey, V.P.; Dwivedi, U.N. Terpenoids as promising therapeutic molecules against Alzheimer’s disease: Amyloid beta- and acetylcholinesterase-directed pharmacokinetic and molecular docking analyses. Mol. Simul. 2018, 44, 1–11. [Google Scholar] [CrossRef]
- Min, S.L.; Liew, S.Y.; Chear, N.J.Y.; Goh, B.H.; Tan, W.N.; Khaw, K.Y. Plant Terpenoids as the Promising Source of Cholinesterase Inhibitors for Anti-AD Therapy. Biology 2022, 11, 307. [Google Scholar] [CrossRef]
- Mony, T.J.; Elahi, F.; Choi, J.W.; Park, S.J. Neuropharmacological Effects of Terpenoids on Preclinical Animal Models of Psychiatric Disorders: A Review. Antioxidants 2022, 11, 1834. [Google Scholar] [CrossRef]
- Meckes, M.; Calzada, F.; Tortoriello, J.; González, J.L.; Martínez, M. Terpenoids Isolated from Psidium guajava Hexane Extract with Depressant Activity on Central Nervous System. Phytother. Res. 1996, 10, 600–603. [Google Scholar] [CrossRef]
- Mohanty, I.R.; Borde, M.; Kumar, S.; Maheshwari, U. Dipeptidyl peptidase IV Inhibitory activity of Terminalia arjuna attributes to its cardioprotective effects in experimental diabetes: In silico, in vitro and in vivo analyses. Phytomedicine 2019, 57, 158–165. [Google Scholar] [CrossRef]
- Pawar, R.S.; Bhutani, K.K. Effect of oleanane triterpenoids from Terminalia arjuna—A cardioprotective drug on the process of respiratory oxyburst. Phytomedicine 2005, 12, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, D.; Vijayvergiya, R.; Dhawan, V. Terminalia arjuna in coronary artery disease: Ethnopharmacology, pre-clinical, clinical & safety evaluation. J. Ethnopharmacol. 2014, 155, 1029–1045. [Google Scholar]
- Sudharsan, P.T.; Mythili, Y.; Selvakumar, E.; Varalakshmi, P. Cardioprotective effect of pentacyclic triterpene, lupeol and its ester on cyclophosphamide-induced oxidative stress. Hum. Exp. Toxicol. 2005, 24, 313–318. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Shafreen, R.B.; Devi, K.P.; Suganthy, N. Assessment of antioxidant, anticholinesterase and antiamyloidogenic effect of Terminalia chebula, Terminalia arjuna and its bioactive constituent 7-Methyl gallic acid—An in vitro and in silico studies. J. Mol. Liq. 2018, 257, 69–81. [Google Scholar] [CrossRef]
- Gupta, D.; Kumar, M. Evaluation of in vitro antimicrobial potential and GC-MS analysis of Camellia sinensis and Terminalia arjuna. Biotechnol. Rep. 2017, 13, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Patra, A.; Samanta, A.; Roy, S.; Mandal, A.; Das Mahapatra, T.; Pradhan, S.; Das, K.; Nandi, D.K. Analysis of phytochemical profile of Terminalia arjuna bark extract with antioxidative and antimicrobial properties. Asian Pac. J. Trop. Biomed. 2013, 3, 960–966. [Google Scholar] [CrossRef]
- Dube, N.; Nimgulkar, C.; Bharatraj, D.K. Validation of therapeutic anti-inflammatory potential of Arjuna KsheeraPaka—A traditional Ayurvedic formulation of Terminalia arjuna. J. Tradit. Complement. Med. 2017, 7, 414–420. [Google Scholar] [CrossRef]
- Ahmad, M.S.; Ahmad, S.; Gautam, B.; Arshad, M.; Afzal, M. Terminalia arjuna, a herbal remedy against environmental carcinogenicity: An in vitro and in vivo study. Egypt. J. Medical Hum. Genet. 2014, 15, 61–67. [Google Scholar] [CrossRef]
- Ruszkowski, P.; Bobkiewicz-Kozlowska, T. Natural Triterpenoids and their Derivatives with Pharmacological Activity Against Neurodegenerative Disorders. Mini-Rev. Org. Chem. 2014, 11, 307–315. [Google Scholar] [CrossRef]
- Bhattacharjee, B.; Pal, P.K.; Ghosh, A.K.; Mishra, S.; Chattopadhyay, A.; Bandyopadhyay, D. Aqueous bark extract of Terminalia arjuna protects against cadmium-induced hepatic and cardiac injuries in male Wistar rats through antioxidative mechanisms. Food Chem. Toxicol. 2019, 124, 249–264. [Google Scholar] [CrossRef]
- Tanaka, H.; Mizojiri, K. Drug-protein binding and blood-brain barrier permeability. J. Pharmacol. Exp. Ther. 1999, 288, 912–918. [Google Scholar] [PubMed]
- Platts, J.A.; Abraham, M.H.; Zhao, Y.H.; Hersey, A.; Ijaz, L.; Butina, D. Correlation and prediction of a large blood-brain distribution data set--an LFER study. Eur. J. Med. Chem. 2001, 36, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, A.; Choi, J.W. Oleanolic Acid Provides Neuroprotection against Ischemic Stroke through the Inhibition of Microglial Activation and NLRP3 Inflammasome Activation. Biomol. Ther. 2022, 30, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ai, Q.; Shi, A.; Wang, N.; Wang, L.; Wei, Y. Oleanolic acid and ursolic acid: Therapeutic potential in neurodegenerative diseases, neuropsychiatric diseases and other brain disorders. Nutr. Neurosci. 2023, 26, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Gudoityte, E.; Arandarcikaite, O.; Mazeikiene, I.; Bendokas, V.; Liobikas, J. Ursolic and Oleanolic Acids: Plant Metabolites with Neuroprotective Potential. Int. J. Mol. Sci. 2021, 22, 4599. [Google Scholar] [CrossRef]
- Fantini, S.; Sassaroli, A.; Tga’valekos, K.T.; Kornbluth, J. Cerebral blood flow and autoregulation: Current measurement techniques and prospects for noninvasive optical methods. Neurophotonics 2016, 3, 031411. [Google Scholar] [CrossRef]
- Liu, X.; Smith, B.J.; Chen, C.; Callegari, E.; Becker, S.L.; Chen, X.; Cianfrogna, J.; Doran, A.C.; Doran, S.D.; Gibbs, J.P.; et al. Use of a Physiologically Based Pharmacokinetic Model to Study the Time to Reach Brain Equilibrium: An Experimental Analysis of the Role of Blood-Brain Barrier Permeability, Plasma Protein Binding, and Brain Tissue Binding. J. Pharmacol. Exp. Ther. 2005, 313, 1254–1262. [Google Scholar] [CrossRef]
- Talevi, A.; Bellera, C.L. Free Drug Theory. In The ADME Encyclopedia; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Hammarlund-Udenaes, M.; Fridén, M.; Syvänen, S.; Gupta, A. On the rate and extent of drug delivery to the brain. Pharm. Res. 2008, 25, 1737–1750. [Google Scholar] [CrossRef]
- Kalvass, J.C.; Maurer, T.S.; Pollack, G.M. Use of plasma and brain unbound fractions to assess the extent of brain distribution of 34 drugs: Comparison of unbound concentration ratios to in vivo p-glycoprotein efflux ratios. Drug Metab. Dispos. 2007, 35, 660–666. [Google Scholar] [CrossRef]
- Banks, W.A.; Kastin, A.J. Peptides and the blood-brain barrier: Lipophilicity as a predictor of permeability. Brain Res. Bull. 1985, 15, 287–292. [Google Scholar] [CrossRef]
- Chikhale, E.G.; Ng, K.-Y.; Burton, P.S.; Borchardt, R.T. Hydrogen bonding potential as a determinant of the in vitro and in situ blood-brain barrier permeability of peptides. Pharm. Res. 1994, 11, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Abbott, N.J. Prediction of blood–brain barrier permeation in drug discovery from in vivo, in vitro and in silico models. Drug Discov. Today Technol. 2004, 1, 407–416. [Google Scholar] [CrossRef]
- Clark, D.E. In silico prediction of blood–brain barrier permeation. Drug Discov. Today 2003, 8, 927–933. [Google Scholar] [CrossRef]
- Stéen, E.J.L.; Vugts, D.J.; Windhorst, A.D. The Application of in silico Methods for Prediction of Blood-Brain Barrier Permeability of Small Molecule PET Tracers. Front. Nucl. Med. 2022, 2, 853475. [Google Scholar] [CrossRef]
- Radan, M.; Djikic, T.; Obradovic, D.; Nikolic, K. Application of in vitro PAMPA technique and in silico computational methods for blood-brain barrier permeability prediction of novel CNS drug candidates. Eur. J. Pharms. Sci. 2022, 168, 106056. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wei, T.; Goldberg, H.; Wang, W.; Cullion, K.; Kohane, D.S. Getting Drugs Across Biological Barriers. Adv. Mater. 2017, 29, 1606596. [Google Scholar] [CrossRef]
- Oldendorf, W.H. Lipid solubility and drug penetration of the blood brain barrier. Proc. Soc. Exp. Biol. Med. 1974, 147, 813–815. [Google Scholar] [CrossRef]
- Aryal, M.; Arvanitis, C.D.; Alexander, P.M.; McDannold, N. Ultrasound-mediated blood-brain barrier disruption for targeted drug delivery in the central nervous system. Adv. Drug Deliv. Rev. 2014, 72, 94–109. [Google Scholar] [CrossRef]
- Feher, M.; Sourial, E.; Schmidt, J.M. A simple model for the prediction of blood-brain partitioning. Int J. Pharm. 2000, 201, 239–247. [Google Scholar] [CrossRef]
- Liu, R.; Sun, H.; So, S.-S. Development of quantitative structure − property relationship models for early ADME evaluation in drug discovery. 2. Blood-brain barrier penetration. J. Chem. Inf. Comput. Sci. 2001, 41, 1623–1632. [Google Scholar] [CrossRef]
- Prasanna, S.; Doerksen, R.J. Topological polar surface area: A useful descriptor in 2D-QSAR. Curr. Med. Chem. 2009, 16, 21–41. [Google Scholar] [CrossRef]
- Van de Waterbeemd, H. In silico models to predict oral absorption. In Comprehensive Medicinal Chemistry II; Taylor, J.B., Triggle, D.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 669–697. [Google Scholar]
- Barbato, F. The Use of Immobilised Artificial Membrane (IAM) Chromatography for Determination of Lipophilicity. Curr. Comput. Drug Des. 2006, 2, 341–352. [Google Scholar] [CrossRef]
- Ogden, P.B.; Coym, J.W. Retention mechanism of a cholesterol-coated C18 stationary phase: Van’t Hoff and Linear Solvation Energy Relationships (LSER) approaches. J. Chromatogr. A 2011, 1218, 2936–2943. [Google Scholar] [CrossRef]
- Stavrakov, G.; Philipova, I.; Lukarski, A.; Atanasova, M.; Zheleva, D.; Zhivkova, Z.D.; Ivanov, S.; Atanasova, T.; Konstantinov, S.; Doytchinova, I. Galantamine-curcumin hybrids as dual-site binding acetylcholinesterase inhibitors. Molecules 2020, 25, 3341. [Google Scholar] [CrossRef] [PubMed]
- Stępnik, K.; Kukula-Koch, W.; Plazinski, W.; Gawel, K.; Gaweł-Bęben, K.; Khurelbat, D.; Boguszewska-Czubara, A. Significance of Astragaloside IV from the Roots of Astragalus mongholicus as an Acetylcholinesterase Inhibitor—From the Computational and Biomimetic Analyses to the In Vitro and In Vivo Studies of Safety. Int. J. Mol. Sci. 2023, 24, 9152. [Google Scholar] [CrossRef]
- Mente, S.; Lombardo, F. A recursive-partitioning model for blood–brain barrier permeation. J. Comput. Aided Mol. Des. 2005, 19, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Kukula-Koch, W.; Mroczek, T. Application of hydrostatic CCC-TLC-HPLC-ESI-TOF-MS for the bioguided fractionation of anticholinesterase alkaloids from Argemonemexicana L. roots. Anal. Bioanal. Chem. 2015, 407, 2581–2589. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://cactus.nci.nih.gov/translate (accessed on 10 June 2023).
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 13, 17. [Google Scholar] [CrossRef]
- Agarwal, R.; Smith, J.C. Speed vs Accuracy: Effect on Ligand Pose Accuracy of Varying Box Size and Exhaustiveness in AutoDock Vina. Mol. Inform. 2023, 42, 2200188. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 30, 455–461. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindalhl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction models for water in relation to protein hydration. In Intermolecular Forces; Pullman, B., Ed.; The Jerusalem Symposia on Quantum Chemistry and Biochemistry; Springer: Dordrecht, The Netherlands, 1981; Volume 14. [Google Scholar] [CrossRef]
- Kukol, A. Lipid Models for United-Atom Molecular Dynamics Simulations of Proteins. J. Chem. Theor. Comput. 2009, 5, 615–626. [Google Scholar] [CrossRef]
- Stroet, M.; Caron, B.; Visscher, K.M.; Geerke, D.P.; Malde, A.K.; Mark, A.E. Automated Topology Builder Version 3.0: Prediction of Solvation Free Enthalpies in Water and Hexane. J. Chem. Theor. Comput. 2018, 14, 5834–5845. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Hub, J.S.; de Groot, B.L.; van der Spoel, D. g_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J. Chem. Theor. Comput. 2010, 12, 3713–3720. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theor. Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Available online: https://www.oecd-ilibrary.org/environment/test-no-236-fish-embryo-acute-toxicity-fet-test_9789264203709-en (accessed on 13 June 2023).
- Nakonieczna, S.; Grabarska, A.; Gawel, K.; Wróblewska-Łuczka, P.; Czerwonka, A.; Stepulak, A.; Kukula-Koch, W. Isoquinoline Alkaloids from Coptis chinensis Franch: Focus on Coptisine as a Potential Therapeutic Candidate against Gastric Cancer Cells. Int. J. Mol. Sci. 2022, 23, 10330. [Google Scholar] [CrossRef] [PubMed]
- Gawel, K.; Turski, W.A.; van der Ent, W.; Mathai, B.J.; Kirstein-Smardzewska, K.J.; Simonsen, A.; Esguerra, C.V. Phenotypic Characterization of Larval Zebrafish (Danio rerio) with Partial Knockdown of the cacna1a Gene. Mol. Neurobiol. 2020, 57, 1904–1916. [Google Scholar] [CrossRef] [PubMed]




| logBB in silico | logPS | logPSFb | Fu | Fb | logPow | TPSA [Å2] | MW [g/mol] |
|---|---|---|---|---|---|---|---|
| −0.45 | −4.3 | −6.1 | 0.0055 | 0.02 | 11.108 | 57.53 | 456.7 |
| Membrane-like System | logkw | s | R2 |
|---|---|---|---|
| IAM | 1.656 | 1.515 | 0.991 |
| CHOL | 2.361 | 2.037 | 0.993 |
| ISRP | 0.637 | 1.853 | 0.983 |
| Membrane-like System | Logkw | logPcw | ΔlogP | E |
|---|---|---|---|---|
| ISRP | 0.637 | 8.995 | 8.358 | 1.46 |
| IAM | 1.656 | 7.339 | ||
| CHOL | 2.361 | 6.634 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stępnik, K.; Kukula-Koch, W.; Plazinski, W.; Rybicka, M.; Gawel, K. Neuroprotective Properties of Oleanolic Acid—Computational-Driven Molecular Research Combined with In Vitro and In Vivo Experiments. Pharmaceuticals 2023, 16, 1234. https://doi.org/10.3390/ph16091234
Stępnik K, Kukula-Koch W, Plazinski W, Rybicka M, Gawel K. Neuroprotective Properties of Oleanolic Acid—Computational-Driven Molecular Research Combined with In Vitro and In Vivo Experiments. Pharmaceuticals. 2023; 16(9):1234. https://doi.org/10.3390/ph16091234
Chicago/Turabian StyleStępnik, Katarzyna, Wirginia Kukula-Koch, Wojciech Plazinski, Magda Rybicka, and Kinga Gawel. 2023. "Neuroprotective Properties of Oleanolic Acid—Computational-Driven Molecular Research Combined with In Vitro and In Vivo Experiments" Pharmaceuticals 16, no. 9: 1234. https://doi.org/10.3390/ph16091234
APA StyleStępnik, K., Kukula-Koch, W., Plazinski, W., Rybicka, M., & Gawel, K. (2023). Neuroprotective Properties of Oleanolic Acid—Computational-Driven Molecular Research Combined with In Vitro and In Vivo Experiments. Pharmaceuticals, 16(9), 1234. https://doi.org/10.3390/ph16091234









