New Pyrazole/Pyrimidine-Based Scaffolds as Inhibitors of Heat Shock Protein 90 Endowed with Apoptotic Anti-Breast Cancer Activity
Abstract
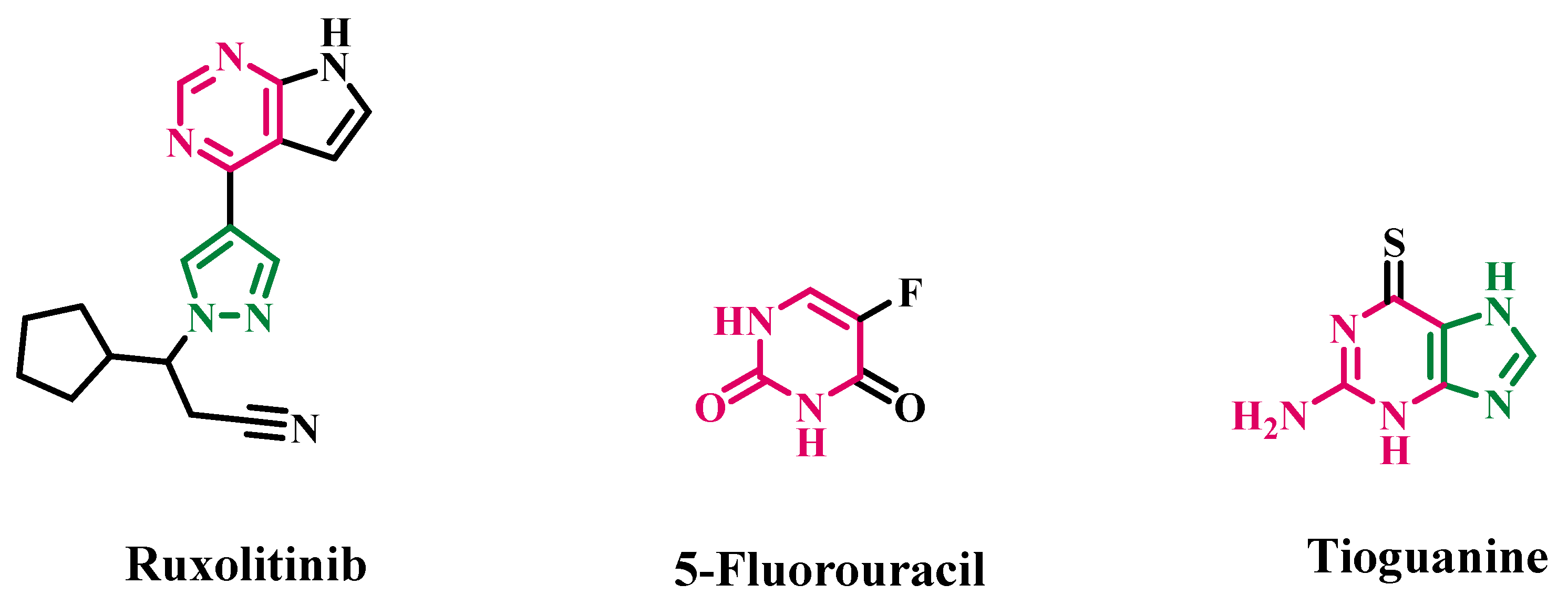
:1. Introduction
2. Results and Discussion
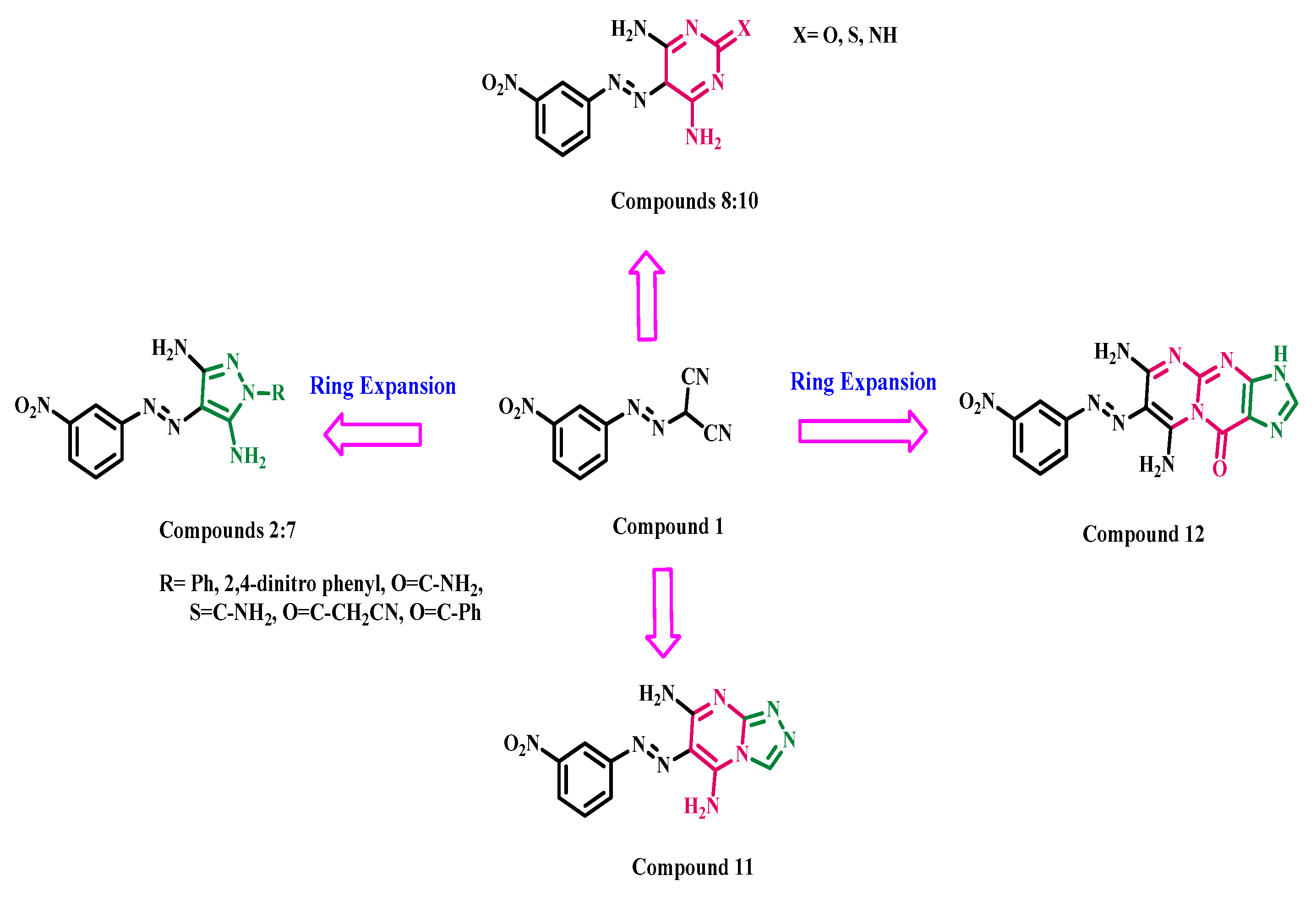
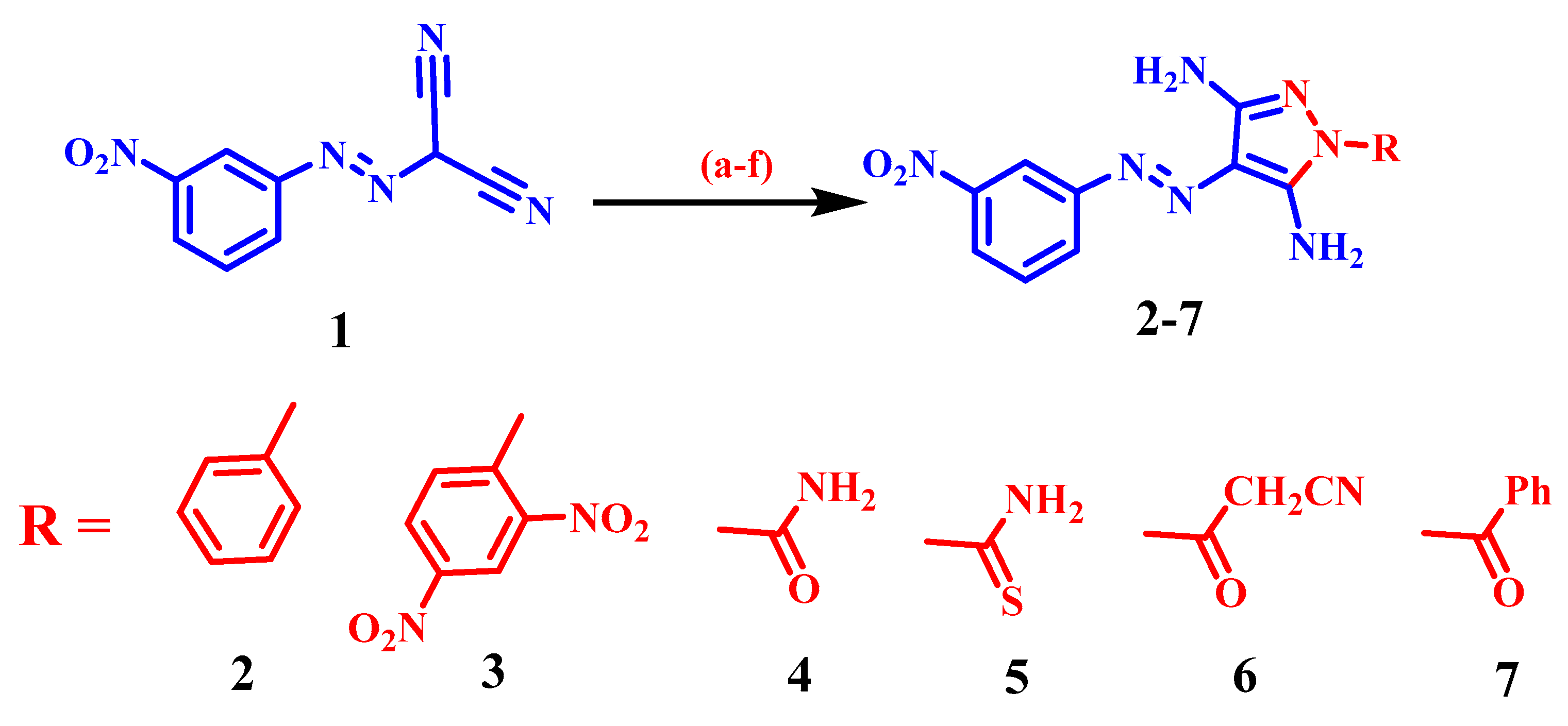
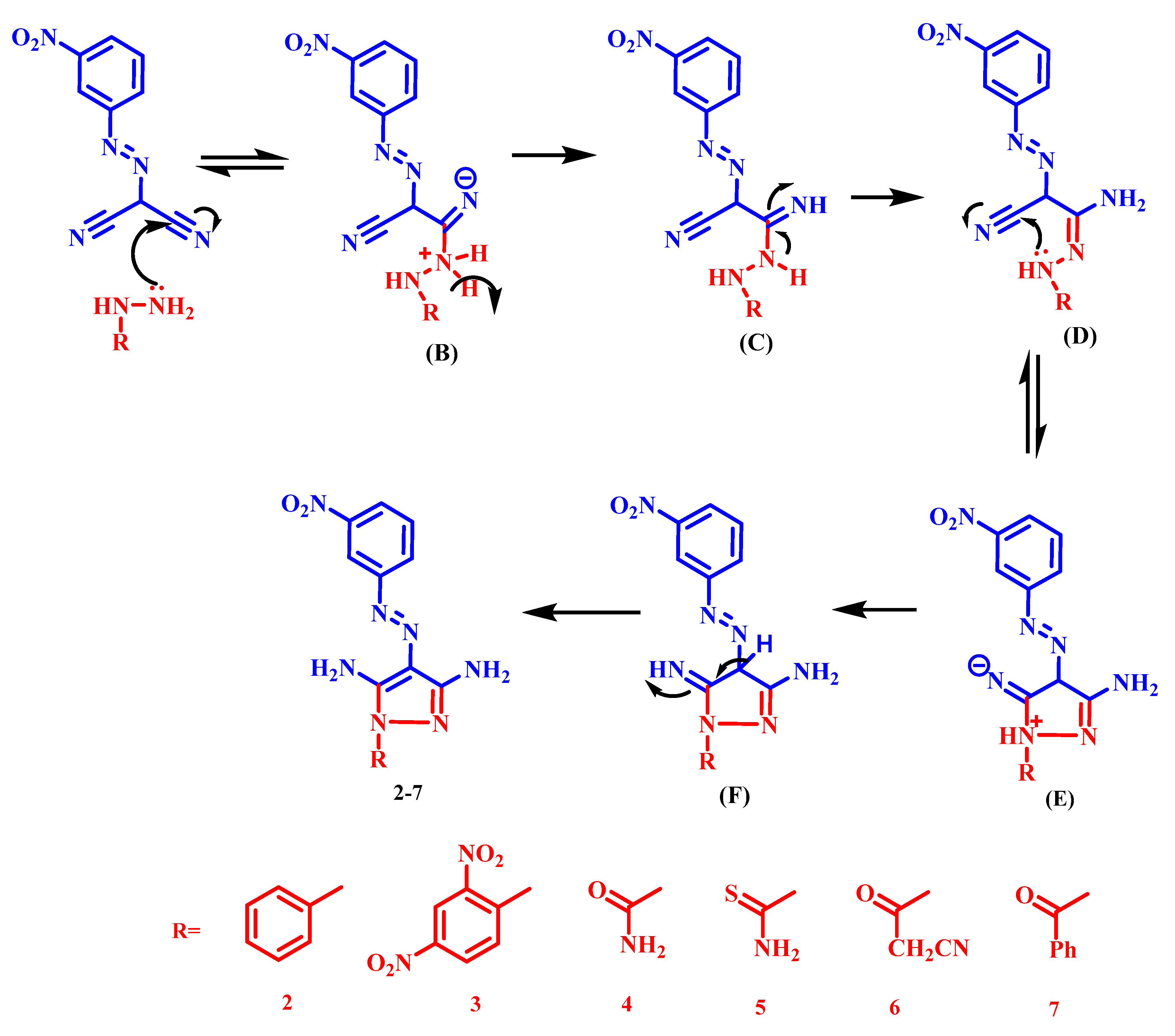
2.1. Chemistry
2.2. Biological Assays
2.2.1. In Vitro Antiproliferative Activity
2.2.2. Effect on Normal Cells
2.2.3. Hsp90 Inhibitory Activity
2.2.4. Activation of Caspases Cascade
2.2.5. BAX and Bcl-2 Assays
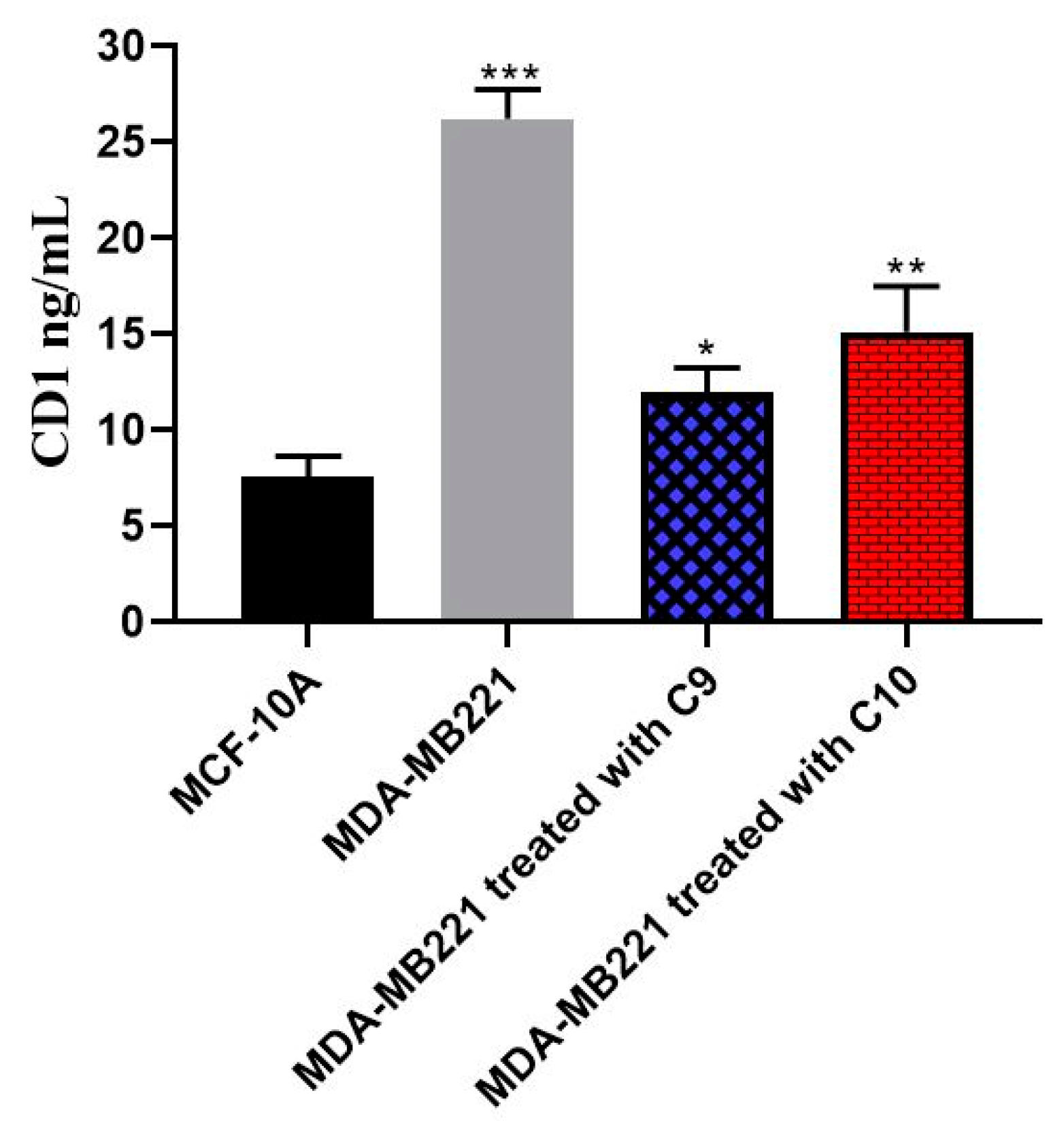
2.2.6. CD1 Inhibition Activity
2.3. Molecular Docking Studies
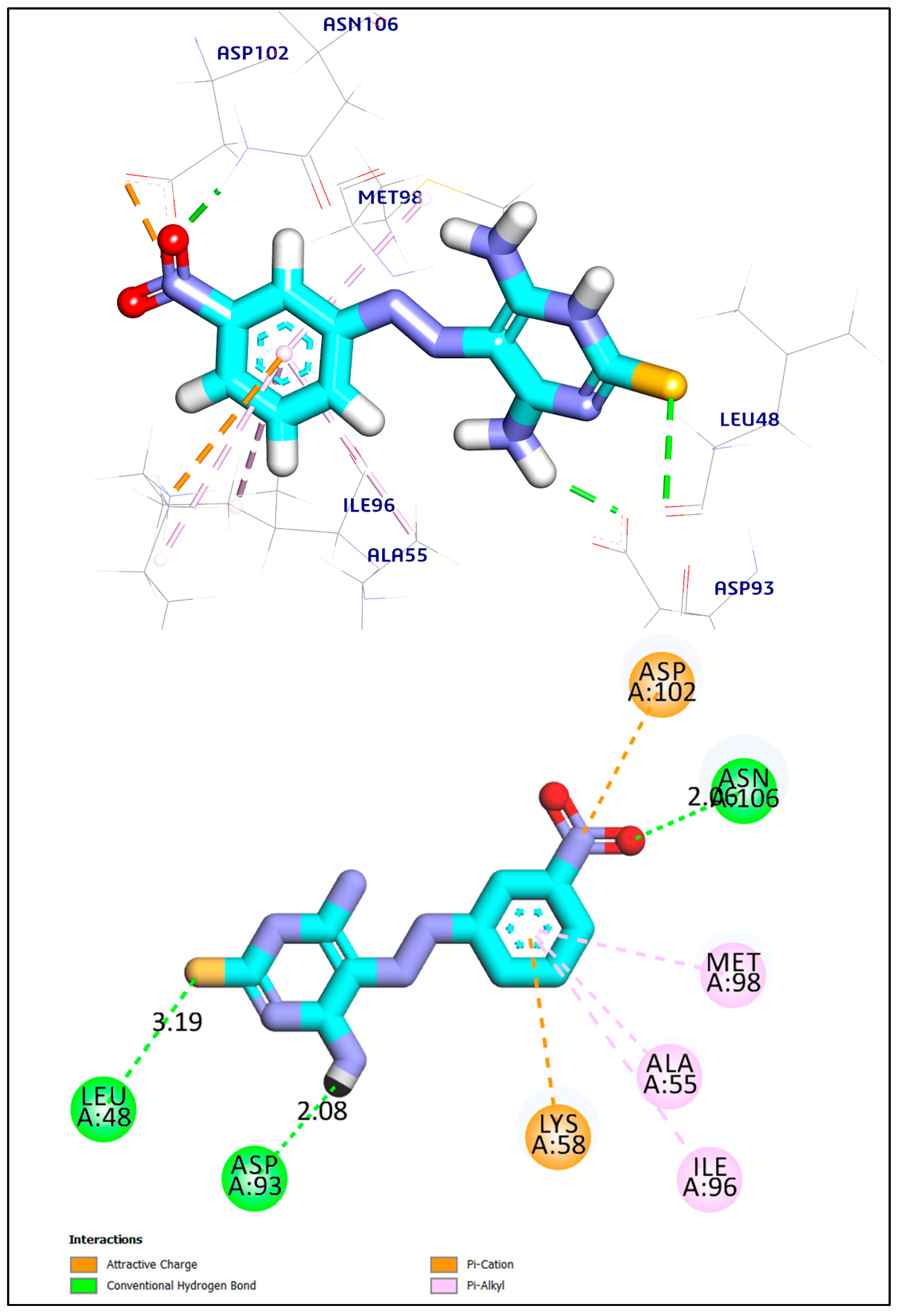
2.3.1. Docking Study
2.3.2. Molecular Dynamics (MD) Simulation Study
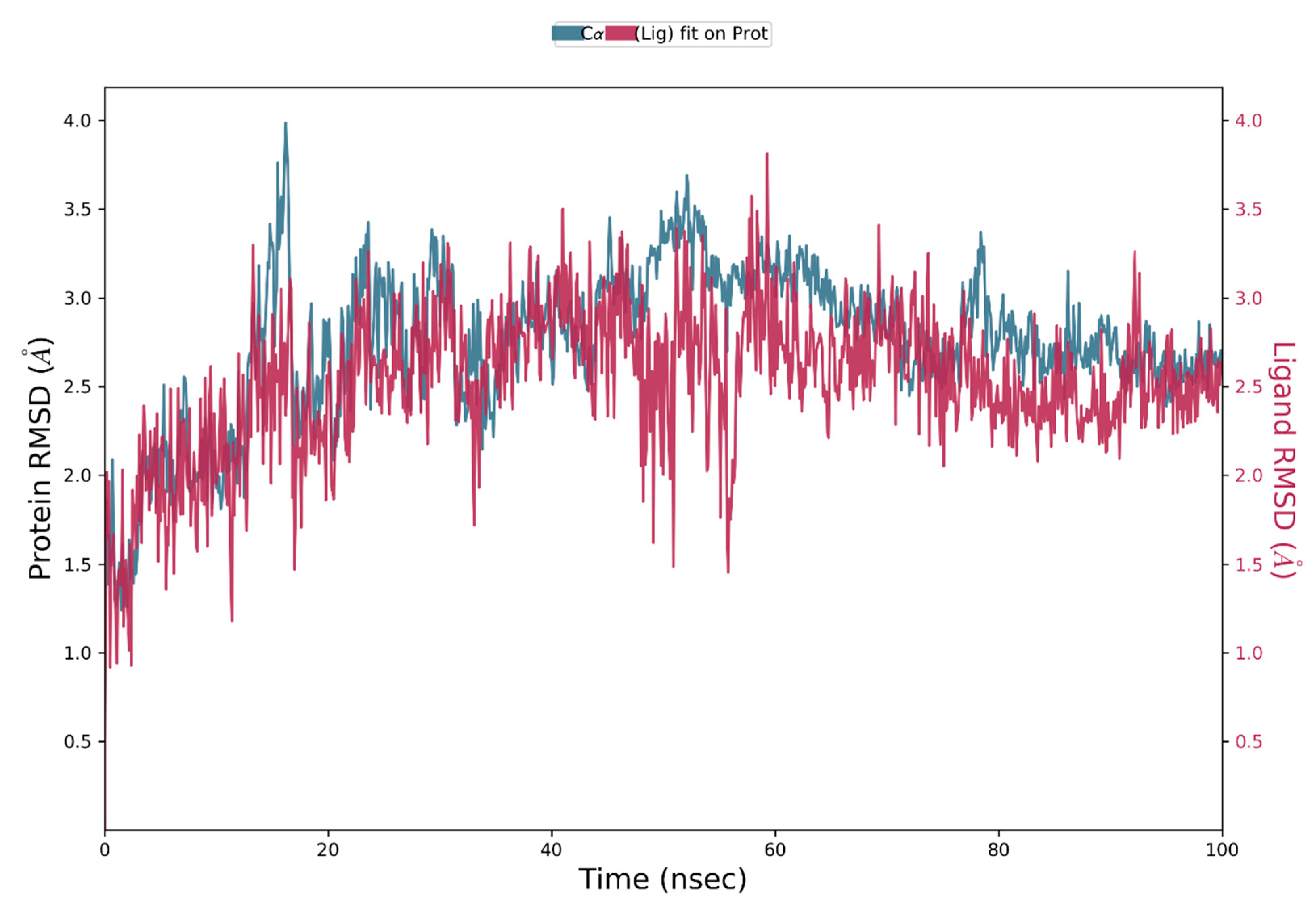
Protein and Ligand RMSD Analysis
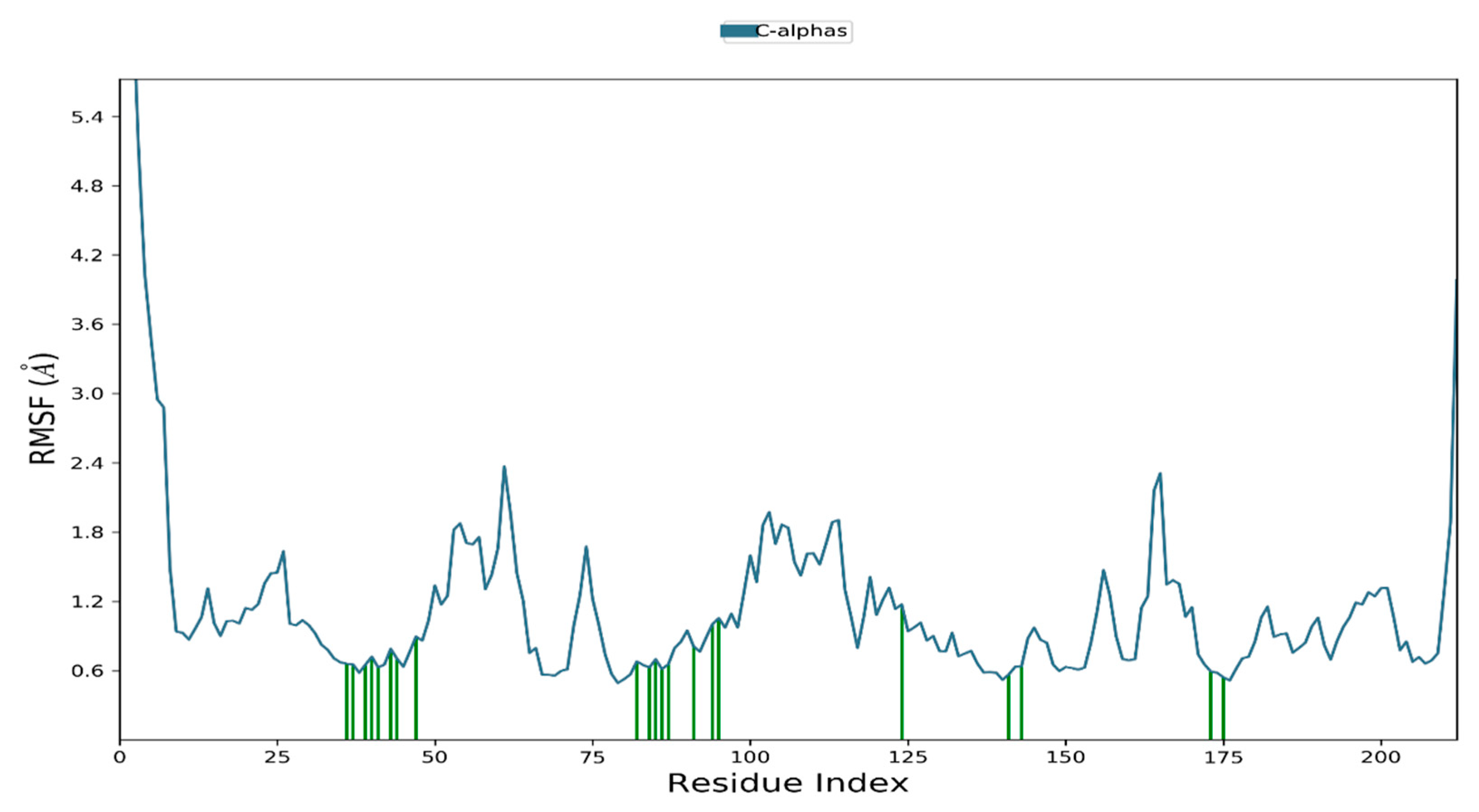
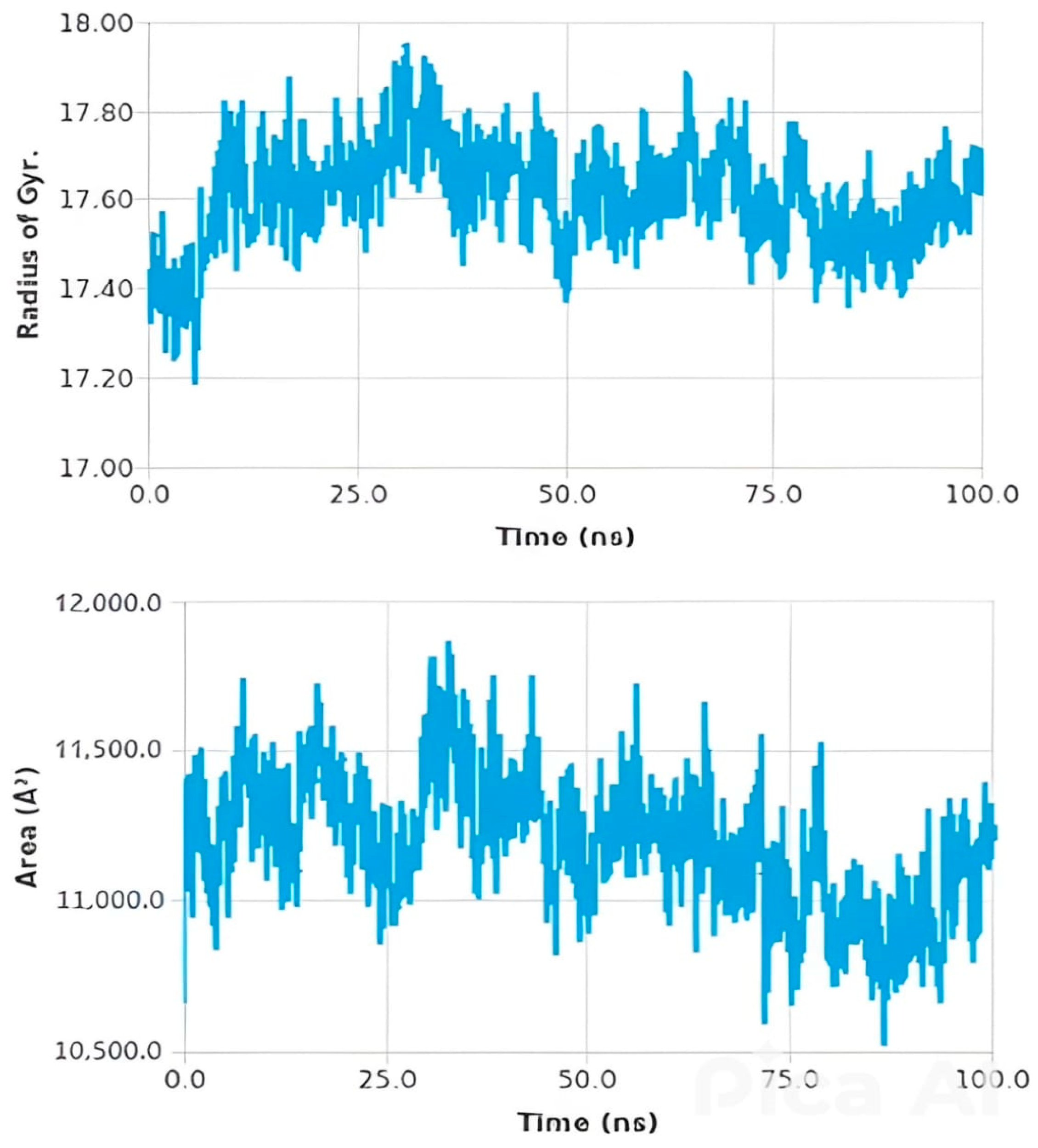
Histogram and Heat Map Analysis
3. Materials and Methods
3.1. Chemistry
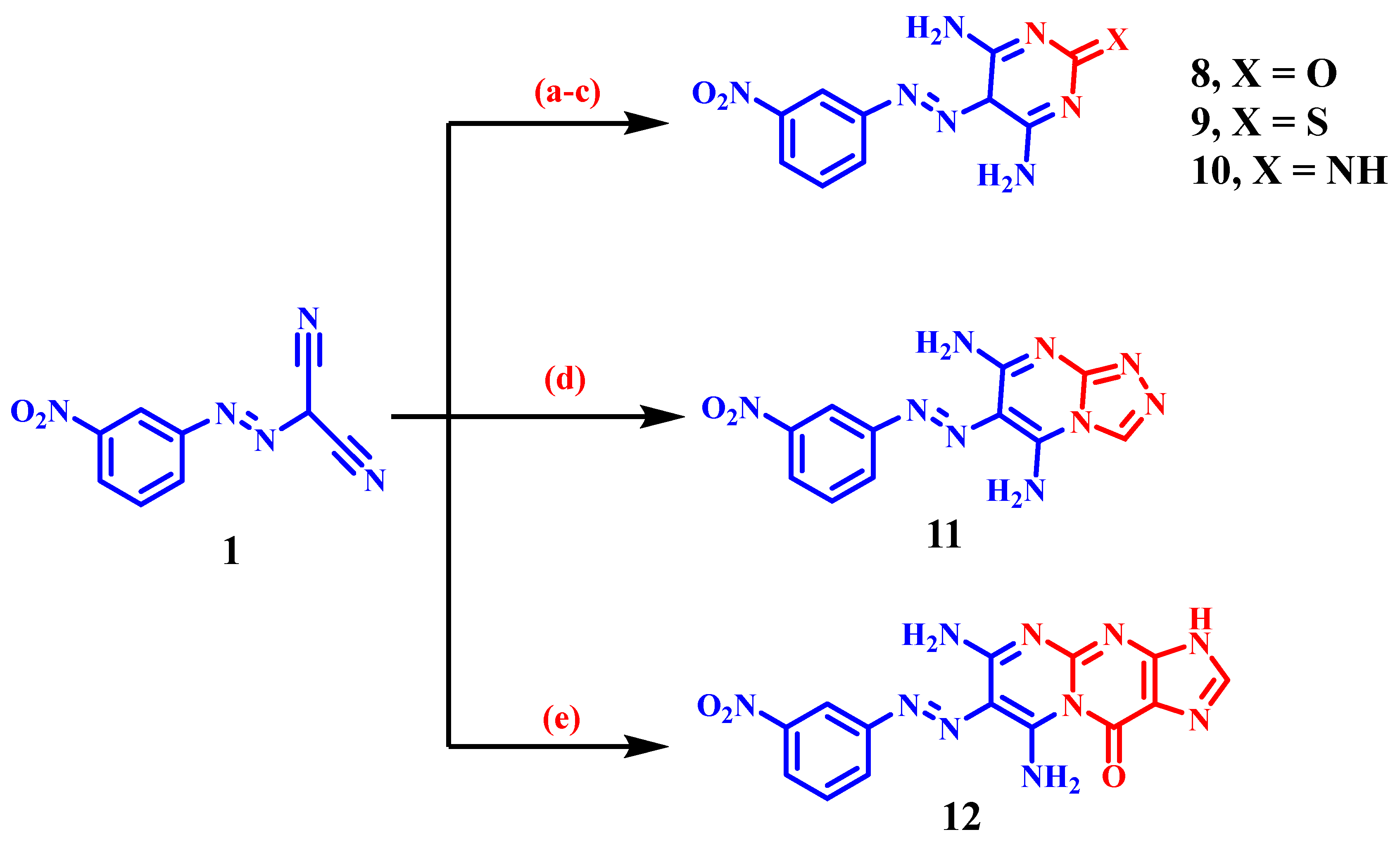
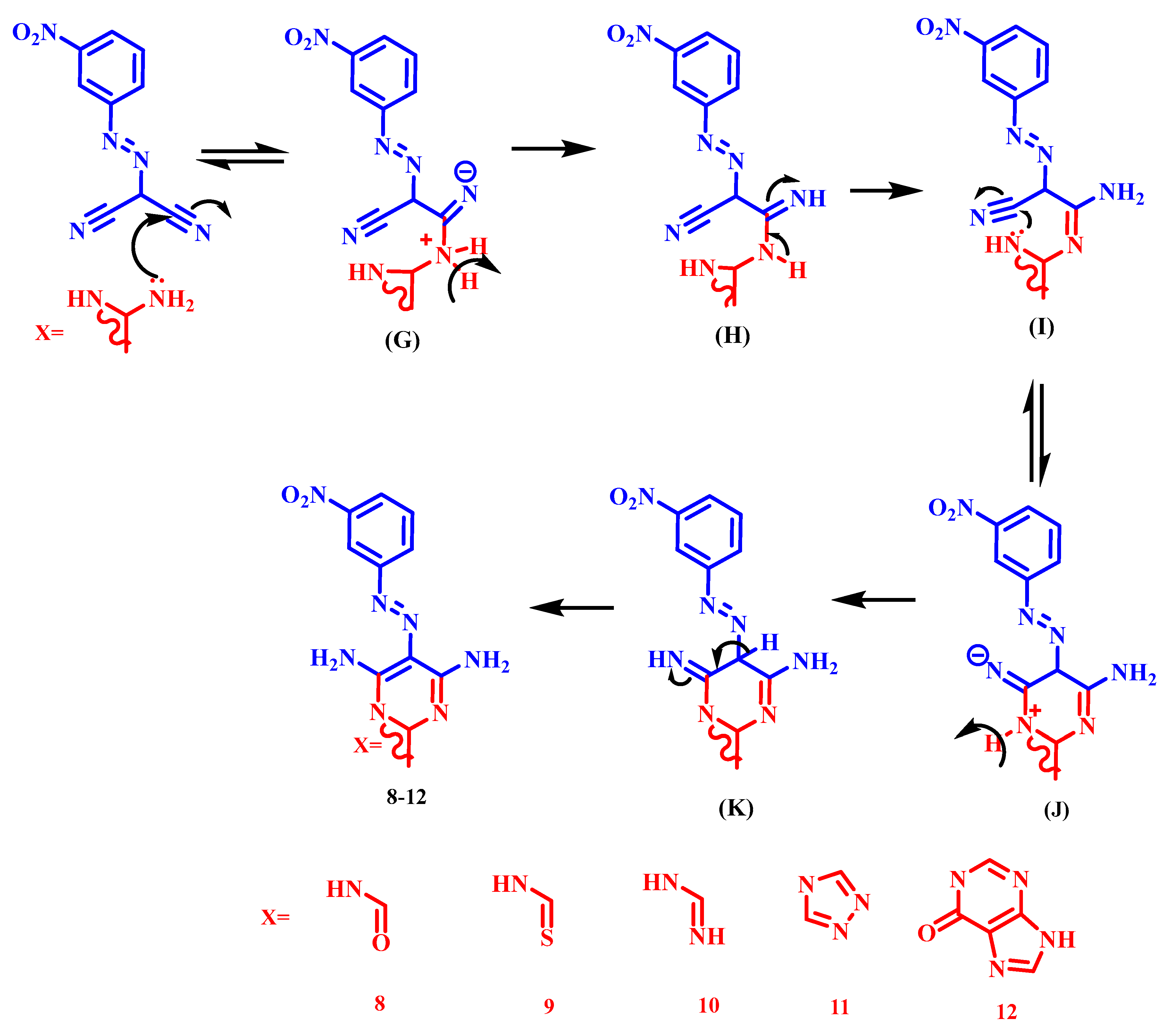
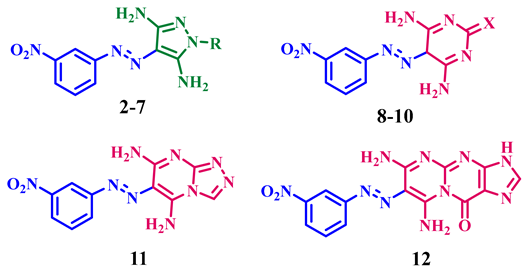
3.1.1. General Synthetic Method of Derivatives (2–12)
4-((3-Nitrophenyl)diazenyl)-1-phenyl-1H-pyrazole-3,5-diamine (2)
1-(2,4-Dinitrophenyl)-4-((3-nitrophenyl)diazenyl)-1H-pyrazole-3,5-diamine (3)
3,5-Diamino-4-((3-nitrophenyl)diazenyl)-1H-pyrazole-1-carboxamide (4)
3,5-Diamino-4-((3-nitrophenyl)diazenyl)-1H-pyrazole-1-carbothioamide (5)
3-(3,5-Diamino-4-((3-nitrophenyl)diazenyl)-1H-pyrazol-1-yl)-3-oxopropanenitrile (6)
(3,5-Diamino-4-((3-nitrophenyl)diazenyl)-1H-pyrazol-1-yl)(phenyl) methanone (7)
4,6-Diamino-5-((3-nitrophenyl)diazenyl)pyrimidin-2(5H)-one (8)
4,6-Diamino-5-((3-nitrophenyl)diazenyl)pyrimidine-2(5H)-thione (9)
2-Imino-5-((3-nitrophenyl)diazenyl)-2,5-dihydropyrimidine-4,6-diamine (10)
6-((3-Nitrophenyl)diazenyl)-[1,2,4]triazolo[4,3-a]pyrimidine-5,7-diamine (11)
6,8-Diamino-7-((3-nitrophenyl)diazenyl)pyrimido[1,2-a]purin-10(3H)-one (12)
3.2. Biological Evaluation
3.2.1. MTT Assay for Cell Viability
3.2.2. Hsp90 Inhibitory Assay
3.2.3. Activation of Caspases
3.2.4. Effects on BAX and Bcl-2 Proteins
3.2.5. Cyclin-D1 Inhibitory Assay
3.2.6. Molecular Modeling
Molecular Docking Study
Molecular Dynamics (MD) Simulation Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Afifi, A.M.; Saad, A.M.; Al-Husseini, M.J.; Elmehrath, A.O.; Northfelt, D.W.; Sonbol, M.B. Causes of death after breast cancer diagnosis: A us population-based analysis. Cancer 2020, 126, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A. (Ed.) Breast cancer statistics: Recent trends. In Breast Cancer Metastasis and Drug Resistance: Challenges and Progress; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–7. [Google Scholar]
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ngumi, M.W. Experiences of Women with Breast Cancer during the COVID-19 Pandemic in Nairobi, Kenya. Ph.D. Thesis, University of Nairobi, Nairobi, Kenya, 2023. [Google Scholar]
- Lakkis, N.A.; Abdallah, R.M.; Musharrafieh, U.M.; Issa, H.G.; Osman, M.H. Epidemiology of breast, corpus uteri, and ovarian cancers in lebanon with emphasis on breast cancer incidence trends and risk factors compared to regional and global rates. Cancer Control 2024, 31, 10732748241236266. [Google Scholar] [CrossRef]
- Zhu, J.W.; Charkhchi, P.; Adekunte, S.; Akbari, M.R. What is known about breast cancer in young women? Cancers 2023, 15, 1917. [Google Scholar] [CrossRef] [PubMed]
- Hoter, A.; El-Sabban, M.E.; Naim, H.Y. The Hsp90 family: Structure, regulation, function, and implications in health and disease. Int. J. Mol. Sci. 2018, 19, 2560. [Google Scholar] [CrossRef]
- Sykut, M.R.H. Heat Shock Protein: Potential Approach for Chemotherapy. Ph.D. Thesis, Brac University, Dhaka, Bangladesh, 2021. [Google Scholar]
- Zhang, M.; Bi, X. Heat shock proteins and breast cancer. Int. J. Mol. Sci. 2024, 25, 876. [Google Scholar] [CrossRef]
- Barrott, J.J.; Haystead, T.A. Hsp90, an unlikely ally in the war on cancer. FEBS J. 2013, 280, 1381–1396. [Google Scholar] [CrossRef] [PubMed]
- Birbo, B.; Madu, E.E.; Madu, C.O.; Jain, A.; Lu, Y. Role of Hsp90 in cancer. Int. J. Mol. Sci. 2021, 22, 10317. [Google Scholar] [CrossRef]
- Maloney, A.; Workman, P. Hsp90 as a new therapeutic target for cancer therapy: The story unfolds. Expert Opin. Biol. Ther. 2002, 2, 3–24. [Google Scholar] [CrossRef] [PubMed]
- Whitesell, L.; Lin, N.U. Hsp90 as a platform for the assembly of more effective cancer chemotherapy. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef] [PubMed]
- Tsygankova, V.; Ya, A.; Shtompel, O.; Romaniuk, O.; Yaikova, M.; Hurenko, A.; Solomyanny, R.; Abdurakhmanova, E.; Klyuchko, S.; Holovchenko, O. Application of synthetic low molecular weight heterocyclic compounds derivatives of pyrimidine, pyrazole and oxazole in agricultural biotechnology as a new plant growth regulating substances. Int. J. Med. Biotechnol. Genet. 2017, 2, 10–32. [Google Scholar]
- Solomyanny, R.; Mrug, G.; Frasinyuk, M.; Shablykin, O.; Brovarets, V. Study of auxin, cytokinin and gibberellin-like activity of heterocyclic compounds derivatives of pyrimidine, pyridine, pyrazole and isoflavones. Eur. J. Biotechnol. Biosci. 2016, 4, 29–44. [Google Scholar]
- Loto, R.; Loto, C.; Popoola, A.; Ranyaoa, M. Pyrimidine derivatives as environmentally-friendly corrosion inhibitors: A review. Int. J. Phys. Sci. 2012, 7, 2136–2144. [Google Scholar]
- Tsygankova, V.; Voloshchuk, I.; Klyuchko, S.; Pilyo, S.; Brovarets, V.; Kovalenko, O. The effect of pyrimidine and pyridine derivatives on the growth and productivity of sorghum. Int. J. Bot. Stud 2022, 7, 19–31. [Google Scholar]
- Choudhary, D.; Garg, S.; Kaur, M.; Sohal, H.S.; Malhi, D.S.; Kaur, L.; Verma, M.; Sharma, A.; Mutreja, V. Advances in the synthesis and bio-applications of pyrazine derivatives: A review. Polycycl. Aromat. Compd. 2023, 43, 4512–4578. [Google Scholar] [CrossRef]
- Kumar, R.; Sharma, R.; Sharma, D.K. Pyrazole; a privileged scaffold of medicinal chemistry: A comprehensive review. Curr. Top. Med. Chem. 2023, 23, 2097–2115. [Google Scholar] [CrossRef] [PubMed]
- Othman, I.M.; Alamshany, Z.M.; Tashkandi, N.Y.; Gad-Elkareem, M.A.; Anwar, M.M.; Nossier, E.S. New pyrimidine and pyrazole-based compounds as potential egfr inhibitors: Synthesis, anticancer, antimicrobial evaluation and computational studies. Bioorg. Chem. 2021, 114, 105078. [Google Scholar] [CrossRef]
- Abdellatif, K.R.; Bakr, R.B. Pyrimidine and fused pyrimidine derivatives as promising protein kinase inhibitors for cancer treatment. Med. Chem. Res. 2021, 30, 31–49. [Google Scholar] [CrossRef]
- Nassar, I.F.; Atta-Allah, S.R.; Elgazwy, A.-S.S.H. A convenient synthesis and molecular modeling study of novel pyrazolo [3, 4-d] pyrimidine and pyrazole derivatives as anti-tumor agents. J. Enzym. Inhib. Med. Chem. 2015, 30, 396–405. [Google Scholar] [CrossRef] [PubMed]
- El-Gamal, M.I.; Zaraei, S.-O.; Madkour, M.M.; Anbar, H.S. Evaluation of substituted pyrazole-based kinase inhibitors in one decade (2011–2020): Current status and future prospects. Molecules 2022, 27, 330. [Google Scholar] [CrossRef]
- Ardevines, S.; Marques-Lopez, E.; Herrera, R.P. Heterocycles in breast cancer treatment: The use of pyrazole derivatives. Curr. Med. Chem. 2023, 30, 1145–1174. [Google Scholar] [CrossRef] [PubMed]
- Amewu, R.K.; Sakyi, P.O.; Osei-Safo, D.; Addae-Mensah, I. Synthetic and naturally occurring heterocyclic anticancer compounds with multiple biological targets. Molecules 2021, 26, 7134. [Google Scholar] [CrossRef]
- Harrison, C.; Kiladjian, J.-J.; Al-Ali, H.K.; Gisslinger, H.; Waltzman, R.; Stalbovskaya, V.; McQuitty, M.; Hunter, D.S.; Levy, R.; Knoops, L. Jak inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med. 2012, 366, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Kverka, M.; Rossmann, P.; Tlaskalova-Hogenova, H.; Klimesova, K.; Jharap, B.; de Boer, N.K.; Vos, R.M.; van Bodegraven, A.A.; Lukas, M.; Mulder, C.J. Safety and efficacy of the immunosuppressive agent 6-tioguanine in murine model of acute and chronic colitis. BMC Gastroenterol. 2011, 11, 47. [Google Scholar] [CrossRef]
- Mohamady, S.; Ismail, M.I.; Mogheith, S.M.; Attia, Y.M.; Taylor, S.D. Discovery of 5-aryl-3-thiophen-2-yl-1h-pyrazoles as a new class of Hsp90 inhibitors in hepatocellular carcinoma. Bioorg. Chem. 2020, 94, 103433. [Google Scholar] [CrossRef]
- Zajec, Ž.; Dernovšek, J.; Distel, M.; Gobec, M.; Tomašič, T. Optimisation of pyrazolo [1,5-a] pyrimidin-7 (4h)-one derivatives as novel Hsp90 c-terminal domain inhibitors against ewing sarcoma. Bioorg. Chem. 2023, 131, 106311. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Abou-Zied, H.A.; Beshr, E.A.; Youssif, B.G.; Hayallah, A.M.; Abdel-Aziz, M. Design, synthesis, antiproliferative actions, and dft studies of new bis–pyrazoline derivatives as dual egfr/brafv600e inhibitors. Int. J. Mol. Sci. 2023, 24, 9104. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Abou-Zied, H.A.; Hisham, M.; Beshr, E.A.; Youssif, B.G.; Bräse, S.; Hayallah, A.M.; Abdel-Aziz, M. Design, synthesis, and biological evaluation of novel 3-cyanopyridone/pyrazoline hybrids as potential apoptotic antiproliferative agents targeting egfr/brafv600e inhibitory pathways. Molecules 2023, 28, 6586. [Google Scholar] [CrossRef] [PubMed]
- Al-Wahaibi, L.H.; Gouda, A.M.; Abou-Ghadir, O.F.; Salem, O.I.; Ali, A.T.; Farghaly, H.S.; Abdelrahman, M.H.; Trembleau, L.; Abdu-Allah, H.H.; Youssif, B.G. Design and synthesis of novel 2,3-dihydropyrazino [1,2-a] indole-1,4-dione derivatives as antiproliferative egfr and brafv600e dual inhibitors. Bioorg. Chem. 2020, 104, 104260. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.; Mahmoud, M.A.; Mostafa, Y.A.; Raslan, A.E.; Youssif, B.G. Novel piperine-carboximidamide hybrids: Design, synthesis, and antiproliferative activity via a multi-targeted inhibitory pathway. J. Enzym. Inhib. Med. Chem. 2023, 38, 376–386. [Google Scholar] [CrossRef]
- Elbastawesy, M.A.; Aly, A.A.; Ramadan, M.; Elshaier, Y.A.; Youssif, B.G.; Brown, A.B.; Abuo-Rahma, G.E.-D.A. Novel pyrazoloquinolin-2-ones: Design, synthesis, docking studies, and biological evaluation as antiproliferative egfr-tk inhibitors. Bioorg. Chem. 2019, 90, 103045. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Youssif, B.G.; Shaykoon, M.S.A.; Abdelrahman, M.H.; Elsadek, B.E.; Aboraia, A.S.; Abuo-Rahma, G.E.-D.A. Utilization of tetrahydrobenzo [4, 5] thieno [2, 3-d] pyrimidinone as a cap moiety in design of novel histone deacetylase inhibitors. Bioorg. Chem. 2019, 91, 103127. [Google Scholar] [CrossRef] [PubMed]
- Essa, B.M.; Selim, A.A.; Sayed, G.H.; Anwer, K.E. Conventional and microwave-assisted synthesis, anticancer evaluation, 99mtc-coupling and in-vivo study of some novel pyrazolone derivatives. Bioorg. Chem. 2022, 125, 105846. [Google Scholar] [CrossRef]
- Anwer, K.E.; Sayed, G.H.; Ramadan, R.M. Synthesis, spectroscopic, dft calculations, biological activities and molecular docking studies of new isoxazolone, pyrazolone, triazine, triazole and amide derivatives. J. Mol. Struct. 2022, 1256, 132513. [Google Scholar] [CrossRef]
- Husseiny, E.M.; Abulkhair, H.S.; El-Dydamony, N.M.; Anwer, K.E. Exploring the cytotoxic effect and cdk-9 inhibition potential of novel sulfaguanidine-based azopyrazolidine-3, 5-diones and 3, 5-diaminoazopyrazoles. Bioorg. Chem. 2023, 133, 106397. [Google Scholar] [CrossRef]
- Anwer, K.E.; Hamza, Z.K.; Ramadan, R.M. Synthesis, spectroscopic, dft calculations, biological activity, sar, and molecular docking studies of novel bioactive pyridine derivatives. Sci. Rep. 2023, 13, 15598. [Google Scholar] [CrossRef]
- Naguib, H.; Dauoud, N.; Shaban, S.; Abdelghaffar, N.; Sayed, G.; Anwer, K. Synthesis of pyrazolone derivatives by grinding, microwave, and conventional techniques and their antimicrobial activity. Russ. J. Org. Chem. 2022, 58, 891–904. [Google Scholar] [CrossRef]
- Mahmoud, M.A.; Mohammed, A.F.; Salem, O.I.; Rabea, S.M.; Youssif, B.G. Design, synthesis, and antiproliferative properties of new 1, 2, 3-triazole-carboximidamide derivatives as dual egfr/vegfr-2 inhibitors. J. Mol. Struct. 2023, 1282, 135165. [Google Scholar] [CrossRef]
- El-Sheref, E.M.; Elbastawesy, M.A.; Brown, A.B.; Shawky, A.M.; Gomaa, H.A.; Bräse, S.; Youssif, B.G. Design and synthesis of (2-oxo-1,2-dihydroquinolin-4-yl)-1,2,3-triazole derivatives via click reaction: Potential apoptotic antiproliferative agents. Molecules 2021, 26, 6798. [Google Scholar] [CrossRef] [PubMed]
- Youssif, B.G.; Abdelrahman, M.H.; Abdelazeem, A.H.; Ibrahim, H.M.; Salem, O.I.; Mohamed, M.F.; Treambleau, L.; Bukhari, S.N.A. Design, synthesis, mechanistic and histopathological studies of small-molecules of novel indole-2-carboxamides and pyrazino [1,2-a] indol-1(2h)-ones as potential anticancer agents effecting the reactive oxygen species production. Eur. J. Med. Chem. 2018, 146, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Alosaimy, A.M.; Abouzied, A.S.; Alsaedi, A.M.; Alafnan, A.; Alamri, A.; Alamri, M.A.; Break, M.K.B.; Sabour, R.; Farghaly, T.A. Discovery of novel indene-based hybrids as breast cancer inhibitors targeting Hsp90: Synthesis, bio-evaluation and molecular docking study. Arab. J. Chem. 2023, 16, 104569. [Google Scholar] [CrossRef]
- Mahmoud, H.K.; Abdelhady, H.A.; Elaasser, M.M.; Hassain, D.Z.H.; Gomha, S.M. Microwave-assisted one-pot three component synthesis of some thiazolyl (hydrazonoethyl)thiazoles as potential anti-breast cancer agents. Polycycl. Aromat. Compd. 2021, 42, 7232–7246. [Google Scholar] [CrossRef]
- Kusuma, B.R.; Khandelwal, A.; Gu, W.; Brown, D.; Liu, W.; Vielhauer, G.; Holzbeierlein, J.; Blagg, B.S.J. Synthesis and biological evaluation of coumarin replacements of novobiocin as Hsp90 inhibitors. Bioorg. Med. Chem. 2014, 22, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.-J.; Han, L.-H.; Cong, R.-S.; Liang, J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. 2005, 37, 719–727. [Google Scholar] [CrossRef]
- Abou-Zied, H.A.; Youssif, B.G.; Mohamed, M.F.; Hayallah, A.M.; Abdel-Aziz, M. Egfr inhibitors and apoptotic inducers: Design, synthesis, anticancer activity and docking studies of novel xanthine derivatives carrying chalcone moiety as hybrid molecules. Bioorg. Chem. 2019, 89, 102997. [Google Scholar] [CrossRef]
- Youssif, B.G.; Mohamed, A.M.; Osman, E.E.A.; Abou-Ghadir, O.F.; Elnaggar, D.H.; Abdelrahman, M.H.; Treamblu, L.; Gomaa, H.A. 5-chlorobenzofuran-2-carboxamides: From allosteric cb1 modulators to potential apoptotic antitumor agents. Eur. J. Med. Chem. 2019, 177, 1–11. [Google Scholar] [CrossRef]
- Cory, S.; Adams, J.M. The bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2002, 2, 647–656. [Google Scholar] [CrossRef]
- Khan, Z.; Bisen, P.S. Oncoapoptotic signaling and deregulated target genes in cancers: Special reference to oral cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2013, 1836, 123–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Regulation of cell cycle progression by growth factor-induced cell signaling. Cells 2021, 10, 3327. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Anders, L. Signaling through cyclin d-dependent kinases. Oncogene 2014, 33, 1890–1903. [Google Scholar] [CrossRef] [PubMed]
- Giacinti, C.; Giordano, A. Rb and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef]
- Gitig, D.M. Molecular Interactions between p27 (Kip1) and p21 (Cip1) and G1 Cyclin/Cdk Complexes. Ph.D. Thesis, Weill Medical College of Cornell University, Ithaca, NY, USA, 2001. [Google Scholar]
- Coqueret, O. Linking cyclins to transcriptional control. Gene 2002, 299, 35–55. [Google Scholar] [CrossRef]
- Mey, A.S.; Juárez-Jiménez, J.; Hennessy, A.; Michel, J. Blinded predictions of binding modes and energies of Hsp90-α ligands for the 2015 d3r grand challenge. Bioorg. Med. Chem. 2016, 24, 4890–4899. [Google Scholar] [CrossRef]
- Elbastawesy, M.A.; Mohamed, F.A.; Zaki, I.; Alahmdi, M.I.; Alzahrani, S.S.; Alzahrani, H.A.; Gomaa, H.A.; Youssif, B.G. Design, synthesis and antimicrobial activity of novel quinoline-2-one hybrids as promising DNA gyrase and topoisomerase iv inhibitors. J. Mol. Struct. 2023, 1278, 134902. [Google Scholar] [CrossRef]
- Ivanova, L.; Tammiku-Taul, J.; García-Sosa, A.T.; Sidorova, Y.; Saarma, M.; Karelson, M. Molecular dynamics simulations of the interactions between glial cell line-derived neurotrophic factor family receptor gfrα1 and small-molecule ligands. ACS Omega 2018, 3, 11407–11414. [Google Scholar] [CrossRef]
- Frejat, F.O.A.; Zhai, H.; Cao, Y.; Wang, L.; Mostafa, Y.A.; Gomaa, H.A.; Youssif, B.G.; Wu, C. Novel indazole derivatives as potent apoptotic antiproliferative agents by multi-targeted mechanism: Synthesis and biological evaluation. Bioorg. Chem. 2022, 126, 105922. [Google Scholar]







| Cpd. No. | Time “min” | Yield % | YE | RME | OE | AE | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Th. | G. | M.W. | Th. | G. | M.W. | Th. | G. | M.W. | Th. | G. | M.W. | Th. | G. | M.W. | ||
| 2 | 120 | 10 | 1 | 49 | 79 | 92 | 0.4083 | 7.9 | 92.00 | 0.4289 | 0.6915 | 0.8053 | 0.4289 | 0.6915 | 0.8053 | 100 |
| 3 | 300 | 12 | 2 | 52 | 78 | 94 | 0.1733 | 6.5 | 47.00 | 0.4679 | 0.7018 | 0.8458 | 0.4679 | 0.7018 | 0.8458 | 100 |
| 4 | 480 | 13 | 4 | 51 | 79 | 92 | 0.1063 | 6.08 | 23.00 | 0.3976 | 0.6159 | 0.7172 | 0.4469 | 0.6923 | 0.8062 | 88.96 |
| 5 | 600 | 16 | 5 | 48 | 77 | 95 | 0.0800 | 4.81 | 19.00 | 0.4173 | 0.6694 | 0.8256 | 0.4173 | 0.6694 | 0.8256 | 100 |
| 6 | 720 | 14 | 5 | 53 | 77 | 91 | 0.0736 | 5.50 | 18.20 | 0.4623 | 0.6716 | 0.7937 | 0.4623 | 0.6716 | 0.7937 | 100 |
| 7 | 960 | 18 | 5 | 53 | 79 | 95 | 0.0552 | 4.39 | 19.00 | 0.4686 | 0.6984 | 0.8399 | 0.4686 | 0.6984 | 0.8399 | 100 |
| 8 | 240 | 12 | 2 | 51 | 80 | 91 | 0.2125 | 6.67 | 45.50 | 0.4369 | 0.6854 | 0.7796 | 0.4369 | 0.6854 | 0.7796 | 100 |
| 9 | 300 | 15 | 3 | 50 | 78 | 92 | 0.1667 | 5.20 | 30.67 | 0.4318 | 0.6735 | 0.7944 | 0.4318 | 0.6735 | 0.7944 | 100 |
| 10 | 300 | 14 | 3 | 52 | 76 | 94 | 0.1733 | 5.43 | 31.33 | 0.4002 | 0.5849 | 0.7234 | 0.4528 | 0.6617 | 0.8184 | 88.39 |
| 11 | 600 | 16 | 4 | 54 | 76 | 92 | 0.0900 | 4.75 | 23.00 | 0.4680 | 0.6587 | 0.7973 | 0.4680 | 0.6587 | 0.7973 | 100 |
| 12 | 720 | 16 | 4 | 49 | 78 | 90 | 0.0681 | 4.88 | 22.50 | 0.4353 | 0.6929 | 0.7995 | 0.4353 | 0.6929 | 0.7995 | 100 |
| Compound | R | X | Antiproliferative Activity IC50 (µM) | |
|---|---|---|---|---|
| MCF-7 | MDA-MB231 | |||
| 2 | Phenyl | NA | 6.20 ± 0.40 | 46.40 ± 2.40 |
| 3 | 2,4-Dinitro phenyl | NA | 247.30 ± 8.20 | 68.30 ± 3.40 |
| 4 | CO-NH2 | NA | 236.40 ± 6.90 | 14.50 ± 1.10 |
| 5 | CS-NH2 | NA | 85.90 ± 3.80 | 19.90 ± 1.30 |
| 6 | CO-CH2CN | NA | 234.50 ± 6.50 | 35.90 ± 2.10 |
| 7 | CO-phenyl | NA | 221.40 ± 6.40 | 72.50 ± 3.30 |
| 8 | NA | O | 33.60 ± 2.00 | 39.80 ± 2.50 |
| 9 | NA | S | 26.10 ± 1.30 | 4.70 ± 0.20 |
| 10 | NA | NH | 7.70 ± 0.50 | 10.70 ± 0.40 |
| 11 | NA | NA | 237.20 ± 7.00 | 25.80 ± 1.80 |
| 12 | NA | NA | 244.40 ± 8.00 | 57.30 ± 2.90 |
| Doxorubicin [47] | -- | -- | 33.20 ± 3.50 | 3.20 ± 0.10 |
| Cisplatin [48] | -- | -- | 3.70 ± 0.35 | -- |
| MCF-10A IC50 (µM) | Compound 9 | Compound 10 |
| 30.70 ± 0.94 | 37.30 ± 1.40 |
| Compound | Hsp90 (IC50 = µM) |
|---|---|
| 9 | 2.44 ± 0.08 |
| 10 | 7.30 ± 0.24 |
| Novobiocin | 1.14 ± 0.04 |
| Normal Cell Line MCF-10A | Untreated MDA-MB231 Cell Line | Compound 9 | Compound 10 | |
|---|---|---|---|---|
| Caspase-3 ng/mL | 5.65 ± 0.95 | 2.40 ± 0.60 | 9.30 ± 0.40 | 8.20 ± 0.30 |
| Caspase-8 ng/mL | 5.30 ± 0.50 | 2.00 ± 0.33 | 7.60 ± 0.20 | 8.50 ± 0.20 |
| Normal MCF-10A Cell Line | Untreated MDA-MB231 Cell Line | Compound 9 | Compound 10 | |
|---|---|---|---|---|
| Bcl2 ng/mL | 5.50 ± 0.50 | 28.60 ± 0.40 | 21.46 ± 0.60 | 15.10 ± 0.85 |
| BAX ng/mL | 8.95 ± 0.30 | 2.10 ± 0.20 | 7.50 ± 0.40 | 6.30 ± 0.90 |
| MCF-10A | MDA-MB231 | Compound 9 | Compound 10 | |
|---|---|---|---|---|
| CD1 ng/mL | 7.571 ± 1.10 | 26.30 ± 1.50 | 11.95 ± 1.30 | 15.15 ± 2.40 |
| Ligand | RMSD Value (Å) | Docking Score (kcal/mol) | Interactions | |
|---|---|---|---|---|
| H.B | Pi-Interactions | |||
| Compound 9 | 1.41 | −6.45 | 3 | 5 |
| Co-crystalized ligand | 1.02 | −6.75 | 2 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Wahaibi, L.H.; Elbastawesy, M.A.I.; Abodya, N.E.; Youssif, B.G.M.; Bräse, S.; Shabaan, S.N.; Sayed, G.H.; Anwer, K.E. New Pyrazole/Pyrimidine-Based Scaffolds as Inhibitors of Heat Shock Protein 90 Endowed with Apoptotic Anti-Breast Cancer Activity. Pharmaceuticals 2024, 17, 1284. https://doi.org/10.3390/ph17101284
Al-Wahaibi LH, Elbastawesy MAI, Abodya NE, Youssif BGM, Bräse S, Shabaan SN, Sayed GH, Anwer KE. New Pyrazole/Pyrimidine-Based Scaffolds as Inhibitors of Heat Shock Protein 90 Endowed with Apoptotic Anti-Breast Cancer Activity. Pharmaceuticals. 2024; 17(10):1284. https://doi.org/10.3390/ph17101284
Chicago/Turabian StyleAl-Wahaibi, Lamya H., Mohammed A. I. Elbastawesy, Nader E. Abodya, Bahaa G. M. Youssif, Stefan Bräse, Sara N. Shabaan, Galal H. Sayed, and Kurls E. Anwer. 2024. "New Pyrazole/Pyrimidine-Based Scaffolds as Inhibitors of Heat Shock Protein 90 Endowed with Apoptotic Anti-Breast Cancer Activity" Pharmaceuticals 17, no. 10: 1284. https://doi.org/10.3390/ph17101284






