The Effects of Marine Fungal Asterripeptides A–C on In Vitro and In Vivo Staphylococcus aureus Skin Infection
Abstract
:1. Introduction
2. Results
2.1. Antibacterial Activity of Asterripeptides A–C
2.2. The Effect on the Viability of S. aureus-Infected HaCaT Keratinocytes
2.3. The Effect of 1–3 on the Proliferation of S. aureus-Infected HaCaT Keratinocytes
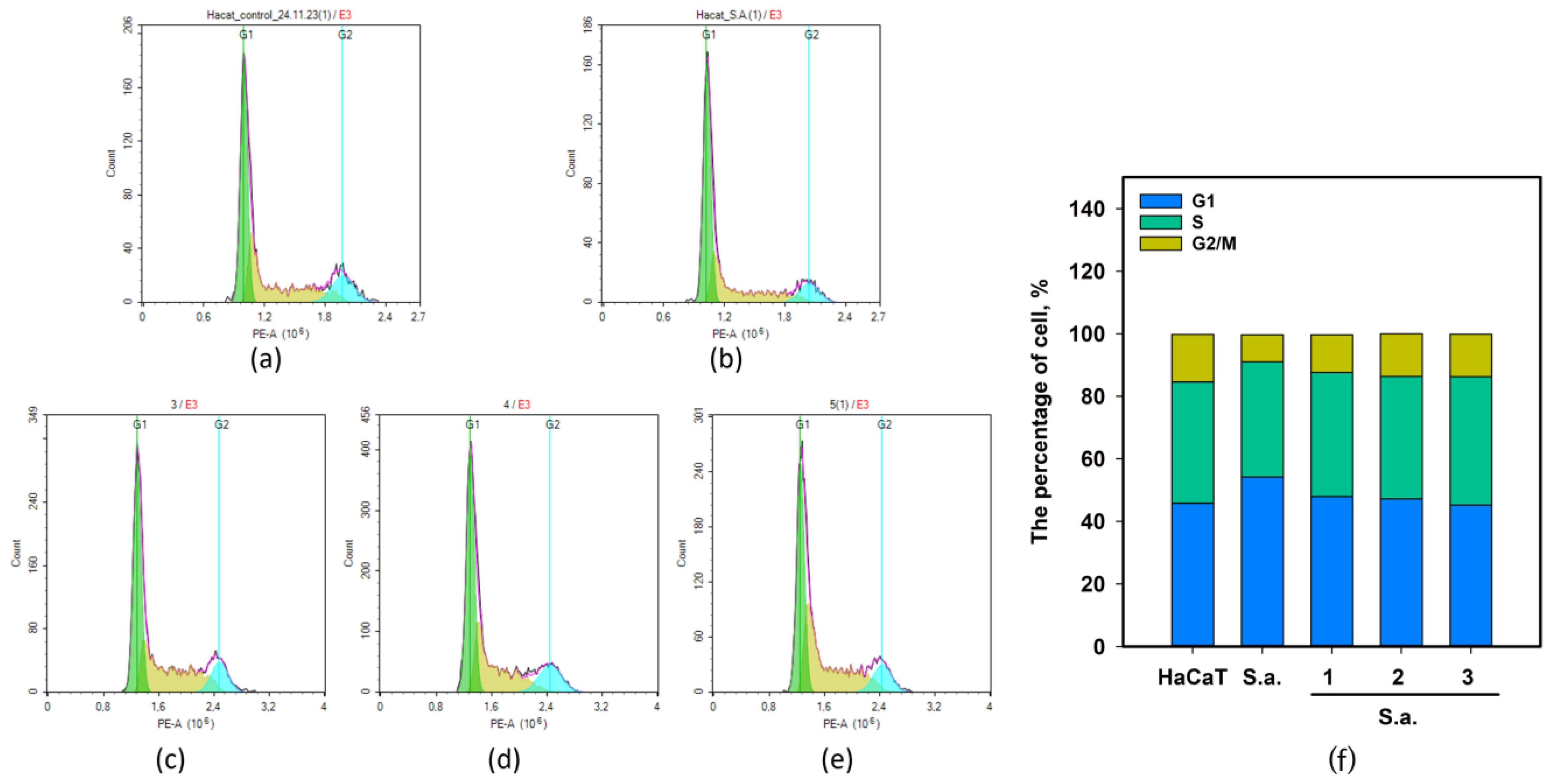
2.4. The Effect of 1–3 on Cell Cycle of S. aureus-Infected HaCaT Keratinocytes
2.5. The Effect of 1–3 on Migration of HaCaT Keratinocytes in In Vitro Model of S. aureus-Infected Skin Wound
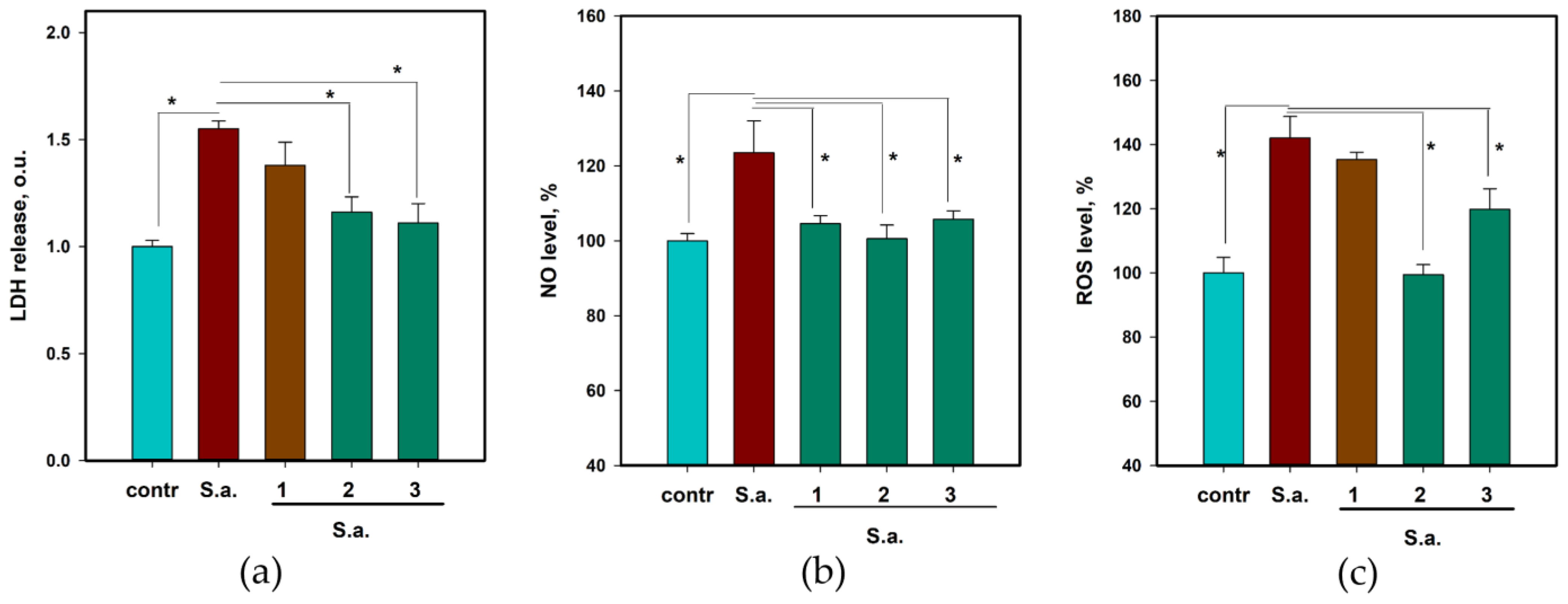
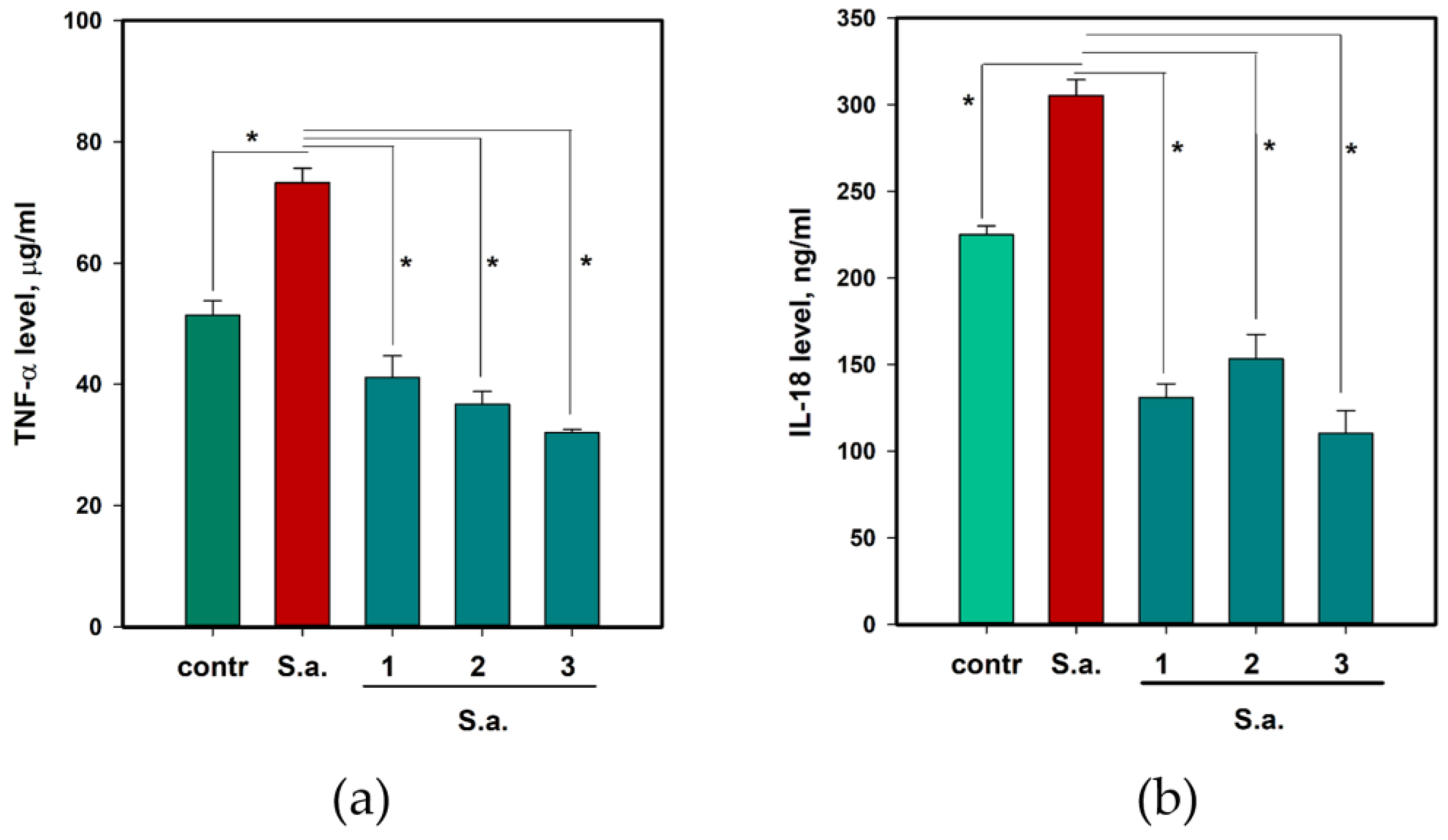
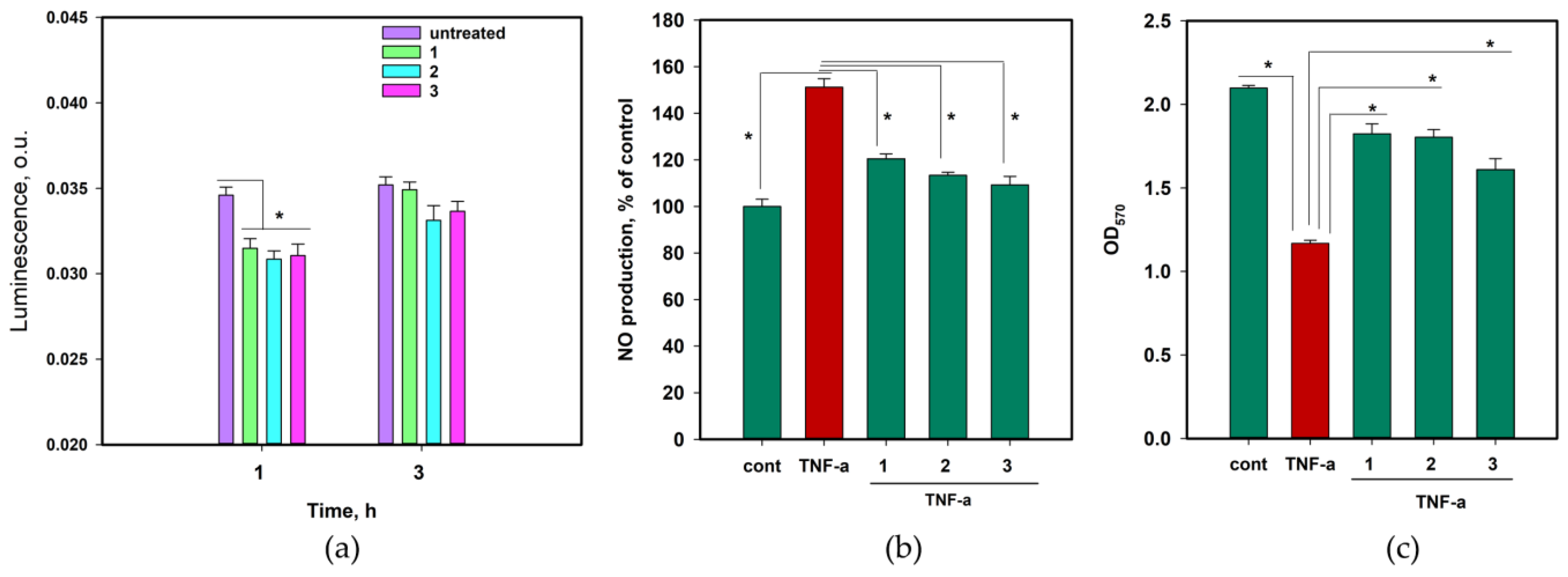
2.6. The Anti-Inflammatory and Antioxidant Activity of 1–3
2.7. The Effect of Asterripeptide C in In Vivo Experiments
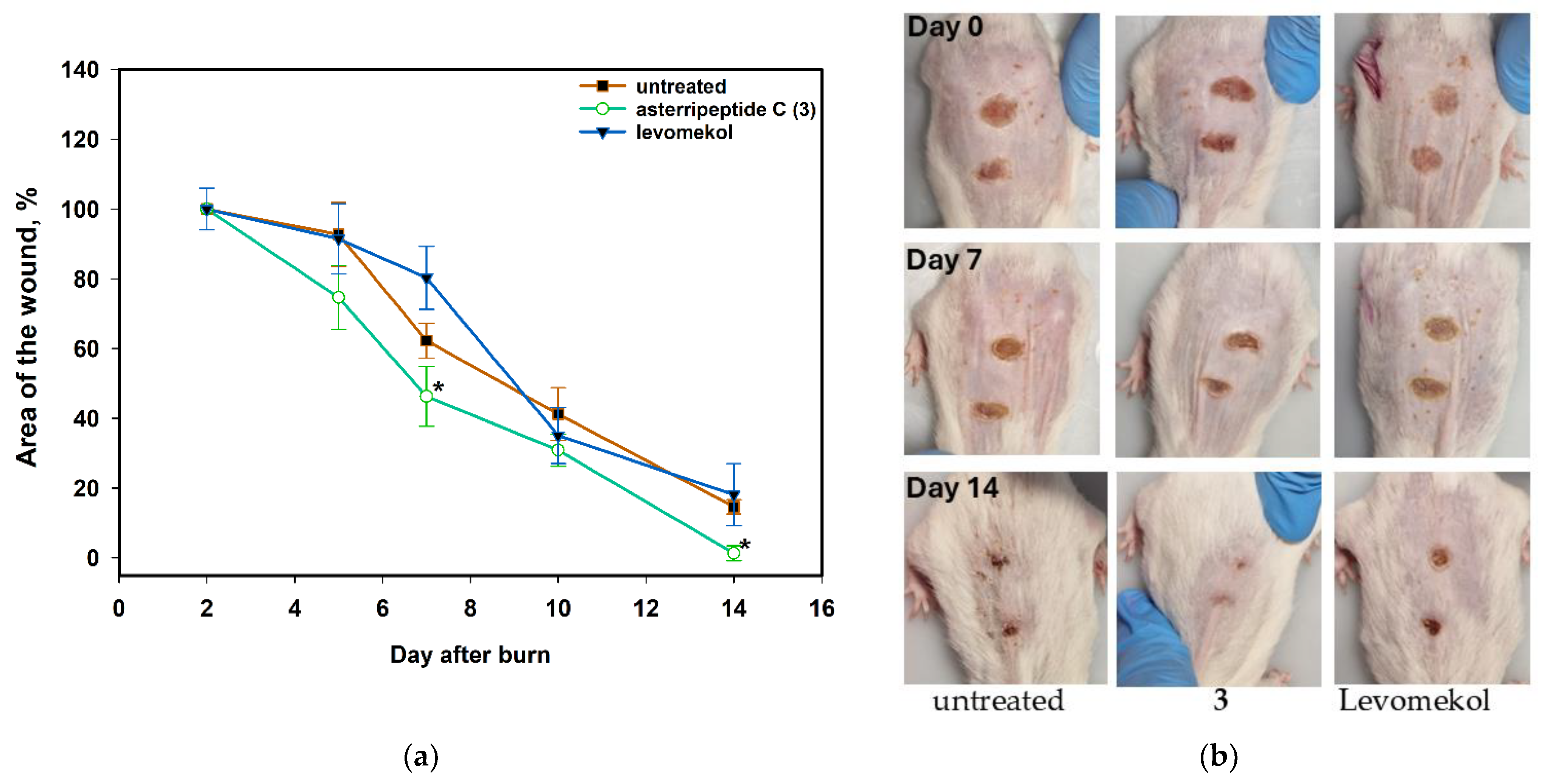
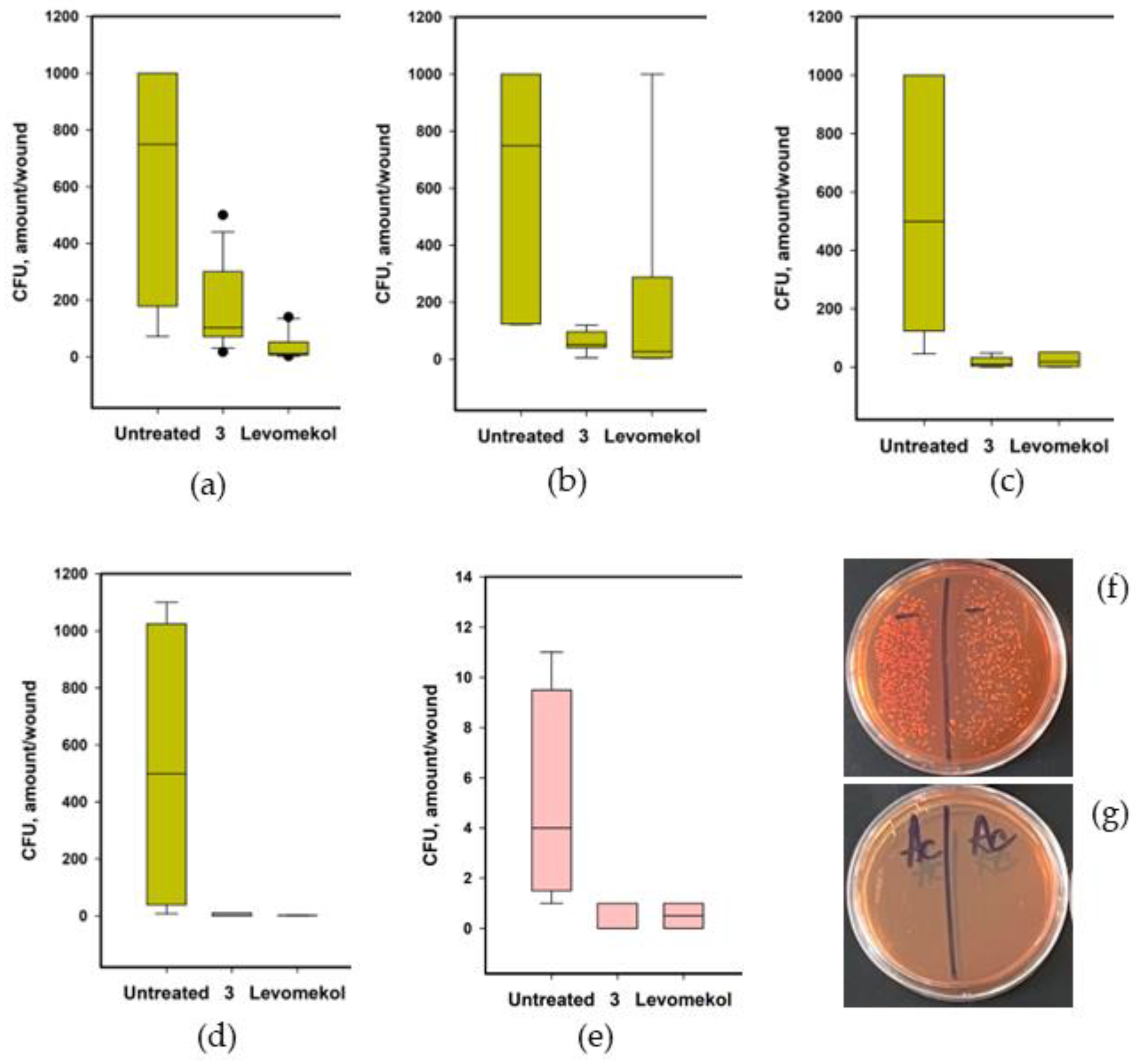
2.7.1. The Effect of Asterripeptide C (3) against Burn Wounds
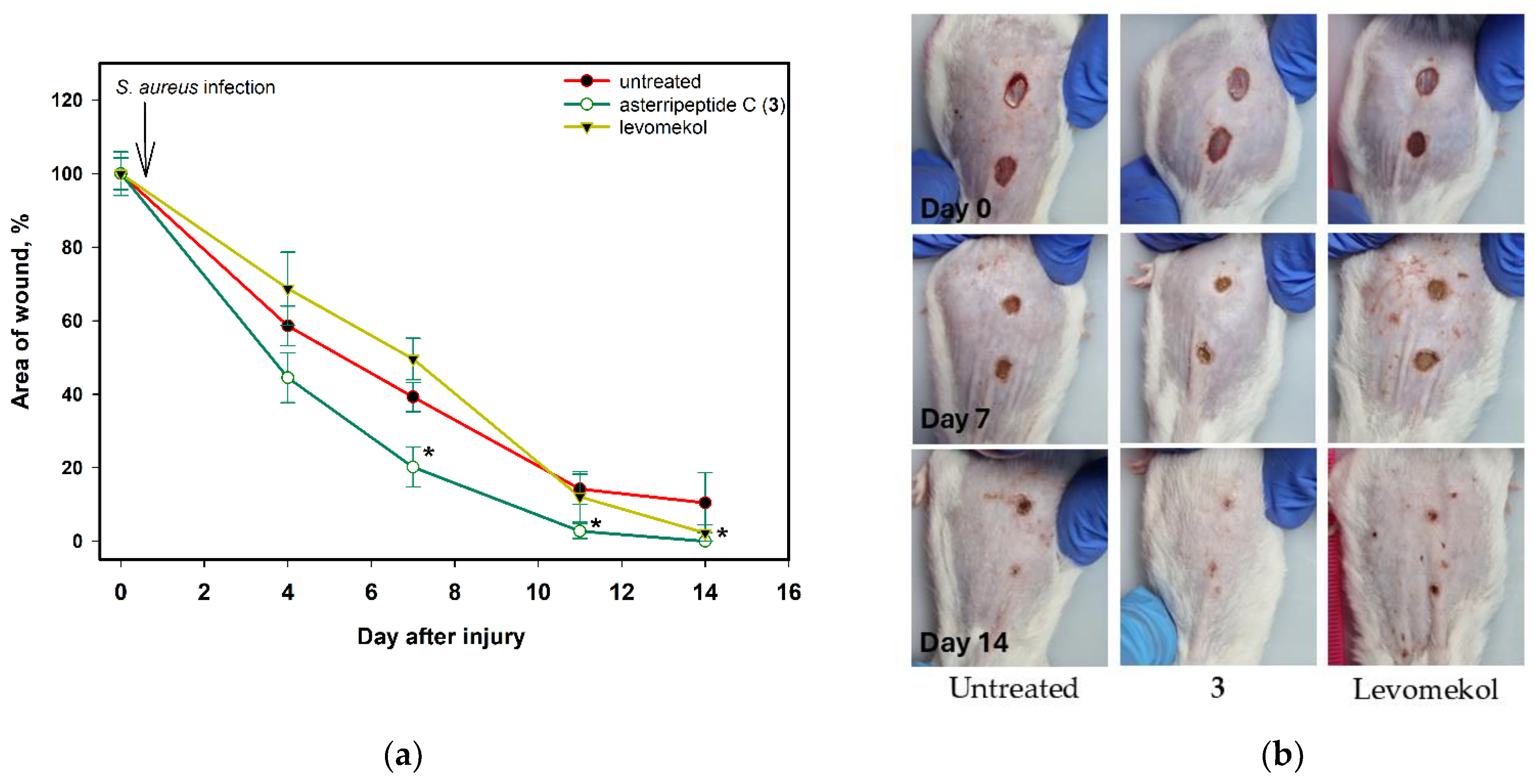
2.7.2. The Effect of Asterripeptide C (3) against S. aureus-Infected Incised Wound
3. Discussion
4. Materials and Methods
4.1. Compounds
4.2. Bacterial Strain and Antimicrobial Assays
4.3. Cell Lines and Culture Conditions
4.4. The Infection of HaCaT Cells with Staphyloccocus aureus
4.5. Cell Viability Assays
4.5.1. Lactate Dehydrogenase (LDH) Release Test
4.5.2. Formazan Production (MTT) Assay
4.6. Cytokines, ROS and NO Level Measure
4.7. TNF-α-Induced Inflammation in HaCaT Cells
4.8. Migration of S. aureus-Infected HaCaT Cells
4.9. Flow Cytometry
4.9.1. Proliferation of S. aureus-Infected HaCaT Cells
4.9.2. Cell Cycle of S. aureus-Infected HaCaT Cells
4.10. Luciferase Measure of the Transcriptional Activity of NF-κB
4.11. Protein Electrophoresis and Immunoblotting
4.12. Molecular Docking
4.13. In Vivo Experiments
4.13.1. Animals and Treatment
4.13.2. Burn Wound
4.13.3. S. aureus-Infected Incised Wound
4.13.4. The Measurements and Peripheral Blood Analysis
4.14. Statistical Data Evaluation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Global Action Plan on Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789241509763 (accessed on 21 June 2024).
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kirker, K.R.; Secor, P.R.; James, G.A.; Fleckman, P.; Olerud, J.E.; Stewart, P.S. Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair. Regen. 2009, 17, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Bondareva, N.E.; Soloveva, A.V.; Sheremet, A.B.; Koroleva, E.A.; Kapotina, L.N.; Morgunova, E.Y.; Luyksaar, S.I.; Zayakin, E.S.; Zigangirova, N.A. Preventative treatment with Fluorothiazinon suppressed Acinetobacter baumannii-associated septicemia in mice. J. Antibiot. 2022, 75, 155–163. [Google Scholar] [CrossRef]
- Savitskii, M.V.; Moskaleva, N.E.; Brito, A.; Zigangirova, N.A.; Soloveva, A.V.; Sheremet, A.B.; Bondareva, N.E.; Lubenec, N.L.; Kuznetsov, R.M.; Samoylov, V.M. Pharmacokinetics, quorum-sensing signal molecules and tryptophan-related metabolomics of the novel anti-virulence drug Fluorothiazinon in a Pseudomonas aeruginosa-induced pneumonia murine model. J. Pharm. Biomed. Anal. 2023, 236, 115739. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, S.; Cao, S. Antimicrobial compounds from marine fungi. Phytochem. Rev. 2021, 20, 85–117. [Google Scholar] [CrossRef]
- Deng, Y.; Liu, Y.; Li, J.; Wang, X.; He, S.; Yan, X.; Shi, Y.; Zhang, W.; Ding, L. Marine natural products and their synthetic analogs as promising antibiofilm agents for antibiotics discovery and development. Eur. J. Med. Chem. 2022, 239, 114513. [Google Scholar] [CrossRef]
- Youssef, F.S.; Ashour, M.L.; Singab, A.N.B.; Wink, M. A Comprehensive Review of Bioactive Peptides from Marine Fungi and Their Biological Significance. Mar. Drugs 2019, 17, 559. [Google Scholar] [CrossRef]
- Scopel, M.; Abraham, W.-R.; Henriques, A.T.; Macedo, A.J. Dipeptide cis-cyclo (Leucyl-Tyrosyl) produced by sponge associated Penicillium sp. F37 inhibits biofilm formation of the pathogenic Staphylococcus epidermidis. Bioorg. Med. Chem. Lett. 2013, 23, 624–626. [Google Scholar] [CrossRef]
- Hafez Ghoran, S.; Taktaz, F.; Sousa, E.; Fernandes, C.; Kijjoa, A. Peptides from marine-derived fungi: Chemistry and biological activities. Mar. Drugs 2023, 21, 510. [Google Scholar] [CrossRef]
- Firakova, S.; Proksa, B.; Šturdíková, M. Biosynthesis and biological activity of enniatins. Pharm. Int. J. Pharm. Sci. 2007, 62, 563–568. [Google Scholar]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Kurakado, S.; Matsumoto, Y.; Yoshino, Y.; Sugita, T.; Koyama, K.; Kinoshita, K. Enniatins from a marine-derived fungus Fusarium sp. inhibit biofilm formation by the pathogenic fungus Candida albicans. J. Nat. Med. 2023, 77, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Balkovec, J.M.; Hughes, D.L.; Masurekar, P.S.; Sable, C.A.; Schwartz, R.E.; Singh, S.B. Discovery and development of first in class antifungal caspofungin (CANCIDAS®)—A case study. Nat. Prod. Rep. 2014, 31, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Girich, E.V.; Rasin, A.B.; Popov, R.S.; Yurchenko, E.A.; Chingizova, E.A.; Trinh, P.T.H.; Ngoc, N.T.D.; Pivkin, M.V.; Zhuravleva, O.I.; Yurchenko, A.N. New tripeptide derivatives asperripeptides A-C from Vietnamese mangrove-derived fungus Aspergillus terreus LM.5.2. Mar. Drugs 2022, 20, 77. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Chingizova, E.A.; Oleinikova, G.K.; Starnovskaya, S.S.; Antonov, A.S.; Kirichuk, N.N.; Menshov, A.S.; Popov, R.S.; Kim, N.Y.; Berdyshev, D.V.; et al. Anthraquinone derivatives and other aromatic compounds from marine fungus Asteromyces cruciatus KMM 4696 and their effects against Staphylococcus aureus. Mar. Drugs 2023, 21, 431. [Google Scholar] [CrossRef]
- Dyshlovoy, S.A.; Fedorov, S.N.; Shubina, L.K.; Kuzmich, A.S.; Bokemeyer, C.; Keller-von Amsberg, G.; Honecker, F. Aaptamines from the marine sponge Aaptos sp. display anticancer activities in human cancer cell lines and modulate AP-1-, NF-kappaB-, and p53-dependent transcriptional activity in mouse JB6 Cl41 cells. Biomed. Res. Int. 2014, 2014, 469309. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, E.A.; Khmel, O.O.; Nesterenko, L.E.; Aminin, D.L. The Kelch/Nrf2 antioxidant system as a target for some marine fungal metabolites. Oxygen 2023, 3, 374–385. [Google Scholar] [CrossRef]
- Hong, F.; Freeman, M.L.; Liebler, D.C. Identification of Sensor Cysteines in Human Keap1 Modified by the Cancer Chemopreventive Agent Sulforaphane. Chem. Res. Toxicol. 2005, 18, 1917–1926. [Google Scholar] [CrossRef]
- Kanikireddy, V.; Varaprasad, K.; Jayaramudu, T.; Karthikeyan, C.; Sadiku, R. Carboxymethyl cellulose-based materials for infection control and wound healing: A review. Int. J. Biol. Macromol. 2020, 164, 963–975. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Xuan, Y.; Zhang, S. Synthesis and applications of carboxymethyl cellulose hydrogels. Gels 2022, 8, 529. [Google Scholar] [CrossRef]
- Ruwizhi, N.; Aderibigbe, B.A. Cinnamic Acid Derivatives and Their Biological Efficacy. Int. J. Mol. Sci. 2020, 21, 5712. [Google Scholar] [CrossRef] [PubMed]
- Hyun, M.W.; Yun, Y.H.; Kim, J.Y.; Kim, S.H. Fungal and Plant Phenylalanine Ammonia-lyase. Mycobiology 2011, 39, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Li, C.; Luo, D.; Teng, P.; Guo, Z.; Tu, X.; Zou, K.; Gong, D. A new cinnamic acid derivative from plant-derived endophytic fungus Pyronema sp. Nat. Prod. Res. 2017, 31, 2413–2419. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.M.; Gallo, M.A.; Jones, C.A.; LaSitis, P.R.; Rosato, L.M. Role of cinnamate in benzoate production in Penicillium brevicompactum. Phytochemistry 1987, 26, 1413–1415. [Google Scholar] [CrossRef]
- Li, W.; Fan, A.; Wang, L.; Zhang, P.; Liu, Z.; An, Z.; Yin, W.-B. Asperphenamate biosynthesis reveals a novel two-module NRPS system to synthesize amino acid esters in fungi. Chem. Sci. 2018, 9, 2589–2594. [Google Scholar] [CrossRef]
- Guzman, J.D. Natural Cinnamic Acids, Synthetic Derivatives and Hybrids with Antimicrobial Activity. Molecules 2014, 19, 19292–19349. [Google Scholar] [CrossRef]
- Koczurkiewicz-Adamczyk, P.; Klaś, K.; Gunia-Krzyżak, A.; Piska, K.; Andrysiak, K.; Stępniewski, J.; Lasota, S.; Wójcik-Pszczoła, K.; Dulak, J.; Madeja, Z. Cinnamic acid derivatives as cardioprotective agents against oxidative and structural damage induced by doxorubicin. Int. J. Mol. Sci. 2021, 22, 6217. [Google Scholar] [CrossRef]
- Simaan, H.; Lev, S.; Horwitz, B.A. Oxidant-Sensing Pathways in the Responses of Fungal Pathogens to Chemical Stress Signals. Front. Microbiol. 2019, 10, 567. [Google Scholar] [CrossRef]
- Wang, L.; Bi, C.; Cai, H.; Liu, B.; Zhong, X.; Deng, X.; Wang, T.; Xiang, H.; Niu, X.; Wang, D. The therapeutic effect of chlorogenic acid against Staphylococcus aureus infection through sortase A inhibition. Front. Microbiol. 2015, 6, 1031. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.; Luan, Y.; Wang, L.; Gao, Z.; Yang, P.; Jing, S.; Wang, Y.; Xiang, H.; Wang, T.; Wang, D. The combination of salvianolic acid A with latamoxef completely protects mice against lethal pneumonia caused by methicillin-resistant Staphylococcus aureus. Emerg. Microbes Infect. 2020, 9, 169–179. [Google Scholar] [CrossRef]
- Taofiq, O.; González-Paramás, A.M.; Barreiro, M.F.; Ferreira, I.C.F.R. Hydroxycinnamic Acids and Their Derivatives: Cosmeceutical Significance, Challenges and Future Perspectives, a Review. Molecules 2017, 22, 281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, J.; Chang, X.; Zhang, C.; Zhou, H.; Liu, M. Ozagrel for acute ischemic stroke: A meta-analysis of data from randomized controlled trials. Neurol. Res. 2012, 34, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Zuo, C.; Yan, F.; Wang, J.; Zhu, Y.; Luo, W.; Liu, Y.; Liang, W.; Yu, W.; Zhang, J.; Peng, D.; et al. Design, synthesis, and evaluation of the novel ozagrel–paeonol codrug with antiplatelet aggregation activities as a potent anti-stroke therapeutic agent. Front. Pharmacol. 2024, 15, 1362857. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Massiah, M.A.; Bozak, R.E.; Hicks, R.J.; Talalay, P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc. Natl. Acad. Sci. USA 2001, 98, 3404–3409. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Wang, H.; Li, S.; Liu, Q.; Sha, H. The Anti-Inflammatory and Anti-Oxidant Mechanisms of the Keap1/Nrf2/ARE Signaling Pathway in Chronic Diseases. Aging Dis. 2019, 10, 637–651. [Google Scholar] [CrossRef]
- Njålsson, R. Glutathione synthetase deficiency. Cell. Mol. Life Sci. CMLS 2005, 62, 1938–1945. [Google Scholar] [CrossRef]
- He, J.; Wu, X.; Huang, S.; Wang, J.; Niu, S.; Chen, M.; Zhang, G.; Cai, S.; Wu, J.; Hong, B. Phenolic Metabolites from a Deep-Sea-Derived Fungus Aspergillus puniceus A2 and Their Nrf2-Dependent Anti-Inflammatory Effects. Mar. Drugs 2022, 20, 575. [Google Scholar] [CrossRef]
- Kwiecinski, J.M.; Horswill, A.R. Staphylococcus aureus bloodstream infections: Pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol. 2020, 53, 51–60. [Google Scholar] [CrossRef]
- Spaan, A.N.; Henry, T.; van Rooijen, W.J.M.; Perret, M.; Badiou, C.; Aerts, P.C.; Kemmink, J.; de Haas, C.J.C.; van Kessel, K.P.M.; Vandenesch, F.; et al. The Staphylococcal Toxin Panton-Valentine Leukocidin Targets Human C5a Receptors. Cell Host Microbe 2013, 13, 584–594. [Google Scholar] [CrossRef]
- Akhmedov, O.R.; Filatova, A.V.; Shomurotov, S.A.; Turaev, A.S. Wound-Healing Activity and Acute and Chronic Toxicity of a Gel Based on Guanidine-Containing Pectin Derivatives. Pharm. Chem. J. 2022, 56, 899–905. [Google Scholar] [CrossRef]
- Campbell, J. High-throughput assessment of bacterial growth inhibition by optical density measurements. Curr. Protoc. Chem. Biol. 2010, 2, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Kifer, D.; Mužinić, V.; Klarić, M.Š. Antimicrobial potency of single and combined mupirocin and monoterpenes, thymol, menthol and 1, 8-cineole against Staphylococcus aureus planktonic and biofilm growth. J. Antibiot. 2016, 69, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Kozlovskiy, S.A.; Pislyagin, E.A.; Menchinskaya, E.S.; Chingizova, E.A.; Sabutski, Y.E.; Polonik, S.G.; Likhatskaya, G.N.; Aminin, D.L. Anti-inflammatory activity of 1,4-naphthoquinones blocking P2X7 purinergic receptors in RAW 264.7 macrophage cells. Toxins 2023, 15, 47. [Google Scholar] [CrossRef]
- Girich, E.V.; Trinh, P.T.; Nesterenko, L.E.; Popov, R.S.; Kim, N.Y.; Rasin, A.B.; Menchinskaya, E.S.; Kuzmich, A.S.; Chingizova, E.A.; Minin, A.S.; et al. Absolute stereochemistry and cytotoxic effects of vismione E from marine sponge-derived fungus Aspergillus sp. 1901NT-1.2.2. Int. J. Mol. Sci. 2023, 24, 8150. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks Iii, C.L.; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Haberthür, U.; Caflisch, A. FACTS: Fast analytical continuum treatment of solvation. J. Comput. Chem. 2008, 29, 701–715. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef]

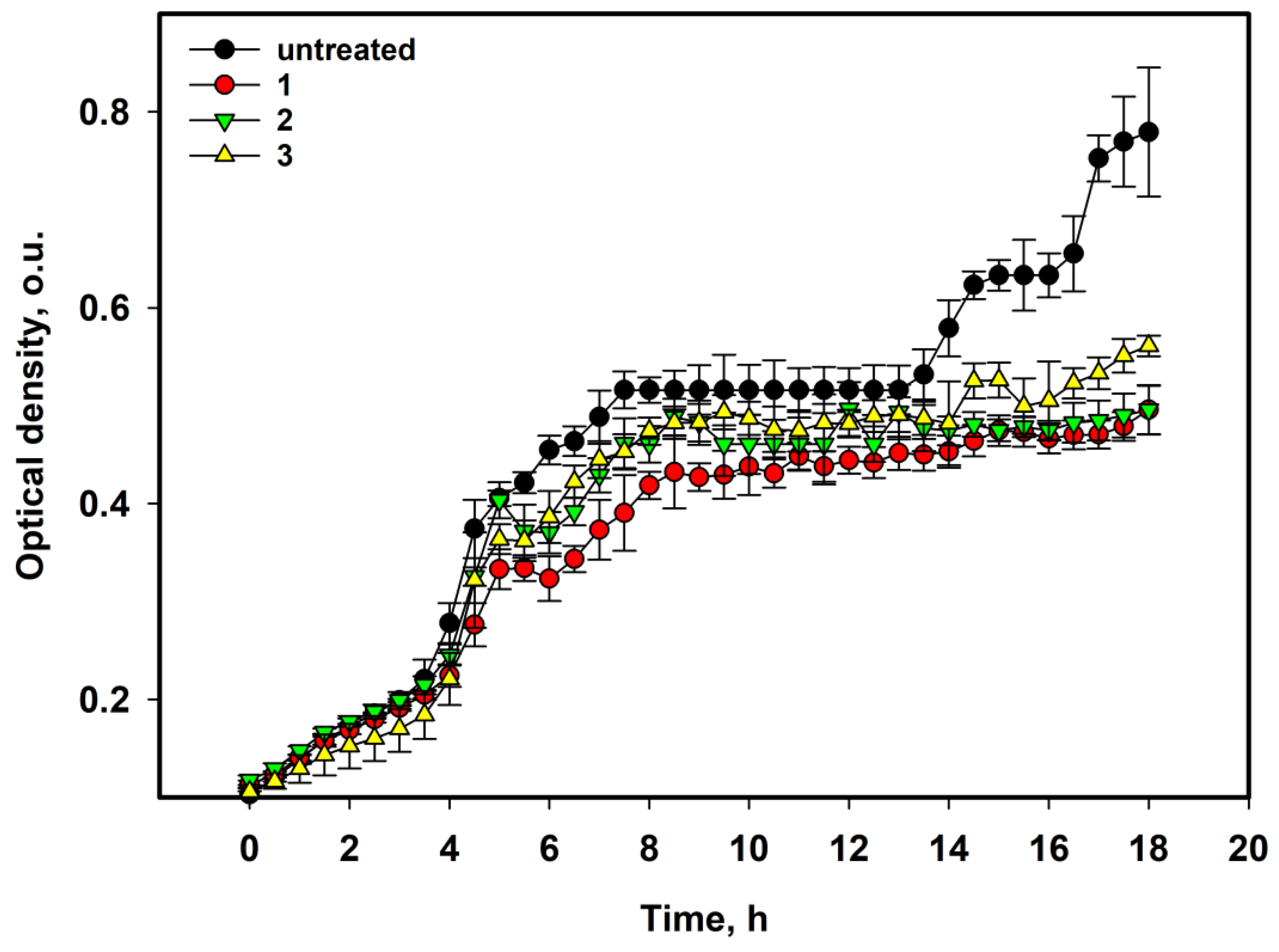
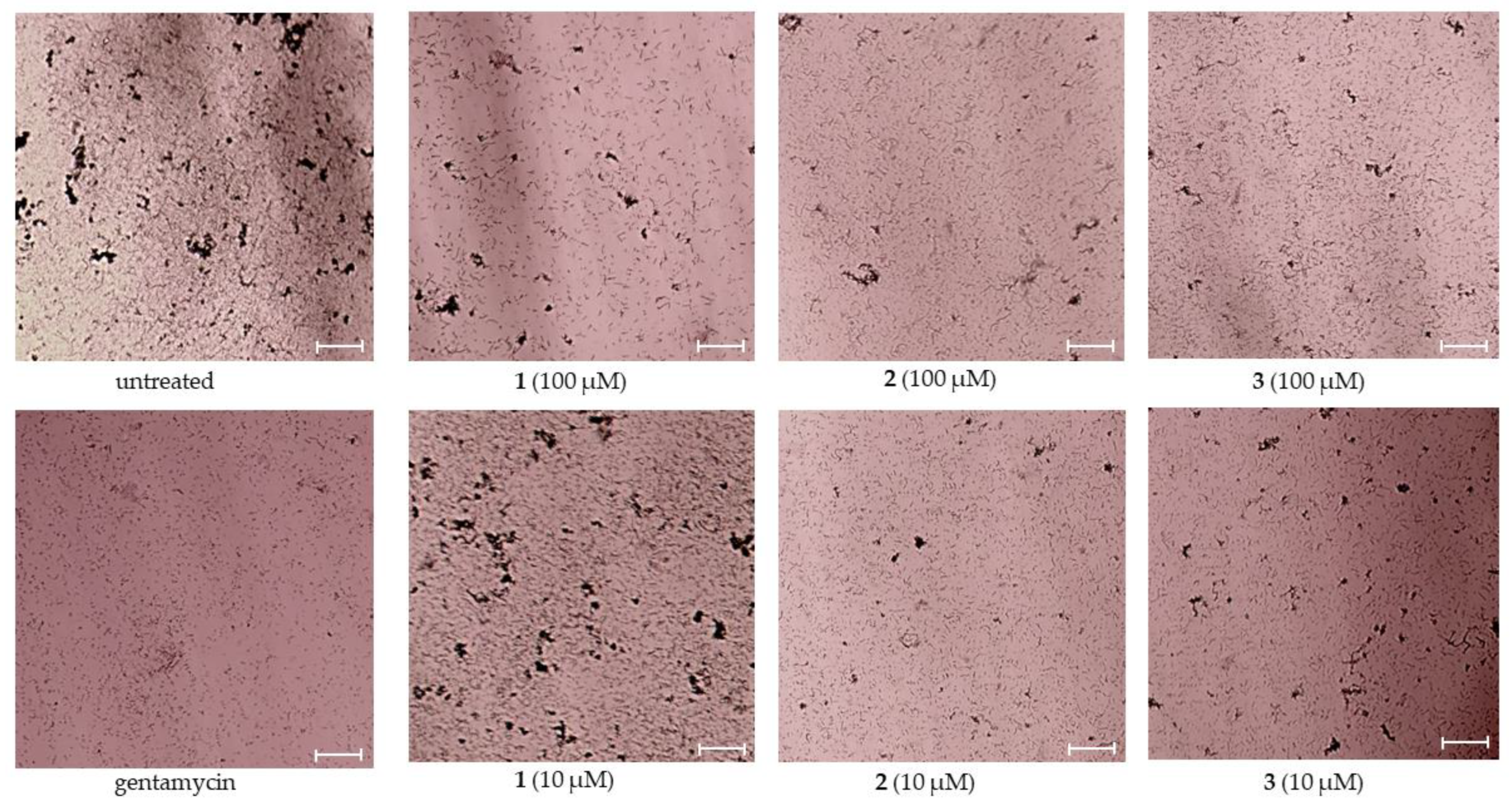





| Compound | Growth Inhibition, % | Biofilm Formation Inhibition, % | |||
|---|---|---|---|---|---|
| 100 µM | 50 µM | 25 µM | 100 µM | 10 µM | |
| 1 | 57.33 ± 1.12 | 52.85 ± 2.01 | - | 52.33 ± 2.06 | 62.79 ± 1.49 |
| 2 | 49.79 ± 0.78 | 28.96 ± 1.22 | - | 54.04 ± 4.99 | 52.63 ± 3.99 |
| 3 | 35.30 ± 1.15 | 34.85 ± 3.40 | 13.56 ± 1.08 | 53.79 ± 6.99 | 61.92 ± 5.79 |
| Compound | ΔG, kcal/mol | FF Score, kcal/mol | H-Binding Interactions, Å | Hydrophobic Interactions |
|---|---|---|---|---|
| 1 | −10.0638 | −1084.234 | Gly605 … O, 3.709 Gly367 … O, 1.933 | Ala556, Ile416, Gly603, Leu365, Ala366, Ile559, Val512, Cys513, Val514, Val561, Thr560, Ala607, Cys368, Val420 |
| 2 | −9.657 | −1045.076 | Ile559 … O, 2.352 | Ala556, Gly364, Gly603, Leu365, Ala336, Leu557, Gly558, Ile559, Val512, Cys513, Thr560, Val561, Val514, Val420, Gly419, Ser486 |
| 3 | −9.429 | −1071.740 | Ile559 … O, 2.208 | Ala556, Gly364, Val604, Gly509, Leu365, Ala510, Leu557, Val465, Ala366, Val418, Val606, Ile559, Gly367, Cys368, Val369, Thr560, Ala466, Cys513, Val514 |
| Indicator | Intact | Untreated | 3 | Base | Levomekol |
|---|---|---|---|---|---|
| Day 18 | |||||
| WBC, 109/L | 6.447 ± 1.526 | 6.813 ± 1.985 | 4.750 ± 1.208 | 4.983 ± 0.818 | 6.397 ± 1.970 |
| Neu, 109/L | 1.703 ± 0.418 | 3.615 ± 0.526 | 1.377 ± 0.471 | 1.483 ± 0.301 | 2.167 ± 0.669 |
| Lym, 109/L | 4.167 ± 0.708 | 3.597 ± 0.230 | 3.073 ± 0.681 | 3.150 ± 0.630 | 3.847 ± 1.217 |
| Mon, 109/L | 0.347 ± 0.121 | 0.197 ± 0.087 | 0.147 ± 0.047 | 0.150 ± 0.044 | 0.197 ± 0.072 |
| Eos, 109/L | 0.230 ± 0.070 | 0.190 ± 0.075 | 0.150 ± 0.026 | 0.197 ± 0.072 | 0.187 ± 0.042 |
| Bas, 109/L | 0.000 ± 0.000 | 0.013 ± 0.006 | 0.003 ± 0.006 | 0.003 ± 0.006 | 0.000 ± 0.000 |
| RBC, 1012/L | 9.307 ± 0.341 | 8.653 ± 1.286 | 9.620 ± 0.310 | 9.620 ± 0.495 | 9.253 ± 0.146 |
| HGB, g/L | 148.00 ± 9.54 | 134.00 ± 28.00 | 152.33 ± 4.04 | 152.00 ± 7.00 | 140.00 ± 2.00 |
| PLT, 109/L | 376.00 ± 69.60 | 549.33 ± 73.71 | 328.00 ± 16.97 | 524.00 ± 73.54 | 468.00 ± 167.10 |
| Indicator | Intact | Untreated | 3 | Base | Levomekol |
|---|---|---|---|---|---|
| Day 4 | |||||
| WBC, 109/L | 4.463 ± 1.042 | 5.820 ± 2.260 | 4.820 ± 1.000 | 5.680 ± 0.612 | 4.557 ± 1.330 |
| Neu, 109/L | 1.290 ± 0.365 | 2.798 ± 0.391 | 1.953 ± 0.534 | 2.057 ± 0.283 | 1.870 ± 0.678 |
| Lym, 109/L | 2.773 ± 0.391 | 2.647 ± 0.615 | 2.610 ± 0.548 | 3.243 ± 0.344 | 2.447 ± 0.621 |
| Mon, 109/L | 0.197 ± 0.115 | 0.277 ± 0.096 | 0.133 ± 0.029 | 0.227 ± 0.091 | 0.130 ± 0.010 |
| Eos, 109/L | 0.200 ± 0.176 | 0.117 ± 0.012 | 0.120 ± 0.036 | 0.153 ± 0.012 | 0.110 ± 0.044 |
| Bas, 109/L | 0.003 ± 0.006 | 0.000 ± 0.000 | 0.003 ± 0.006 | 0.000 ± 0.000 | 0.000 ± 0.000 |
| RBC, 1012/L | 9.363 ± 0.317 | 10.143 ± 0.151 | 9.857 ± 0.446 | 9.787 ± 0.147 | 9.363 ± 0.887 |
| HGB, g/L | 145.333 ± 7.506 | 152.667 ± 3.215 | 155.000 ± 6.083 | 155.667 ± 1.528 | 156.000 ± 10.440 |
| PLT, 109/L | 377.333 ± 47.606 | 587.667 ± 34.686 | 495.333 ± 58.398 | 554.667 ± 71.699 | 563.000 ± 136.451 |
| Day 12 | |||||
| WBC, 109/L | 6.177 ± 1.236 | 2.520 ± 1.942 | 4.535 ± 0.932 | 4.497 ± 0.391 | 4.397 ± 0.569 |
| Neu, 109/L | 2.047 ± 0.684 | 0.665 ± 0.148 | 1.370 ± 0.303 | 1.687 ± 0.505 | 1.857 ± 0.559 |
| Lym, 109/L | 3.697 ± 0.529 | 1.137 ± 1.078 | 2.880 ± 0.690 | 2.477 ± 0.110 | 2.25 ± 0.335 |
| Mon, 109/L | 0.243 ± 0.049 | 0.170 ± 0.069 | 0.155 ± 0.029 | 0.163 ± 0.035 | 0.137 ± 0.035 |
| Eos, 109/L | 0.190 ± 0.010 | 0.113 ± 0.064 | 0.130 ± 0.107 | 0.170 ± 0.050 | 0.153 ± 0.101 |
| Bas, 109/L | 0.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.005 | 0.000 ± 0.000 | 0 ± 0 |
| RBC, 1012/L | 9.060 ± 0.565 | 5.263 ± 3.846 | 9.210 ± 1.740 | 9.227 ± 0.220 | 8.737 ± 1.154 |
| HGB, g/L | 142.667 ± 9.018 | 86.333 ± 6.517 | 155.000 ± 24.069 | 147.333 ± 3.055 | 137.333 ± 16.773 |
| PLT, 109/L | 536.667 ± 104.026 | 338.667 ± 322.274 | 563.000 ± 9.899 | 207.500 ± 19.092 | 940.333 ± 264.462 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chingizova, E.A.; Yurchenko, E.A.; Chingizov, A.R.; Klimovich, A.A.; Pislyagin, E.A.; Menchinskaya, E.S.; Kuzmich, A.S.; Trinh, P.T.H.; Ngoc, N.T.D.; Van, T.T.T.; et al. The Effects of Marine Fungal Asterripeptides A–C on In Vitro and In Vivo Staphylococcus aureus Skin Infection. Pharmaceuticals 2024, 17, 1345. https://doi.org/10.3390/ph17101345
Chingizova EA, Yurchenko EA, Chingizov AR, Klimovich AA, Pislyagin EA, Menchinskaya ES, Kuzmich AS, Trinh PTH, Ngoc NTD, Van TTT, et al. The Effects of Marine Fungal Asterripeptides A–C on In Vitro and In Vivo Staphylococcus aureus Skin Infection. Pharmaceuticals. 2024; 17(10):1345. https://doi.org/10.3390/ph17101345
Chicago/Turabian StyleChingizova, Ekaterina A., Ekaterina A. Yurchenko, Artur R. Chingizov, Anna A. Klimovich, Evgeny A. Pislyagin, Ekaterina S. Menchinskaya, Aleksandra S. Kuzmich, Phan Thi Hoai Trinh, Ngo Thi Duy Ngoc, Tran Thi Thanh Van, and et al. 2024. "The Effects of Marine Fungal Asterripeptides A–C on In Vitro and In Vivo Staphylococcus aureus Skin Infection" Pharmaceuticals 17, no. 10: 1345. https://doi.org/10.3390/ph17101345
APA StyleChingizova, E. A., Yurchenko, E. A., Chingizov, A. R., Klimovich, A. A., Pislyagin, E. A., Menchinskaya, E. S., Kuzmich, A. S., Trinh, P. T. H., Ngoc, N. T. D., Van, T. T. T., Guzhova, I. V., Aminin, D. L., & Yurchenko, A. N. (2024). The Effects of Marine Fungal Asterripeptides A–C on In Vitro and In Vivo Staphylococcus aureus Skin Infection. Pharmaceuticals, 17(10), 1345. https://doi.org/10.3390/ph17101345







