Insights into Clematis cirrhosa L. Ethanol Extract: Cytotoxic Effects, LC-ESI-QTOF-MS/MS Chemical Profiling, Molecular Docking, and Acute Toxicity Study
Abstract
:1. Introduction
2. Results
2.1. Cytotoxicity Evaluation
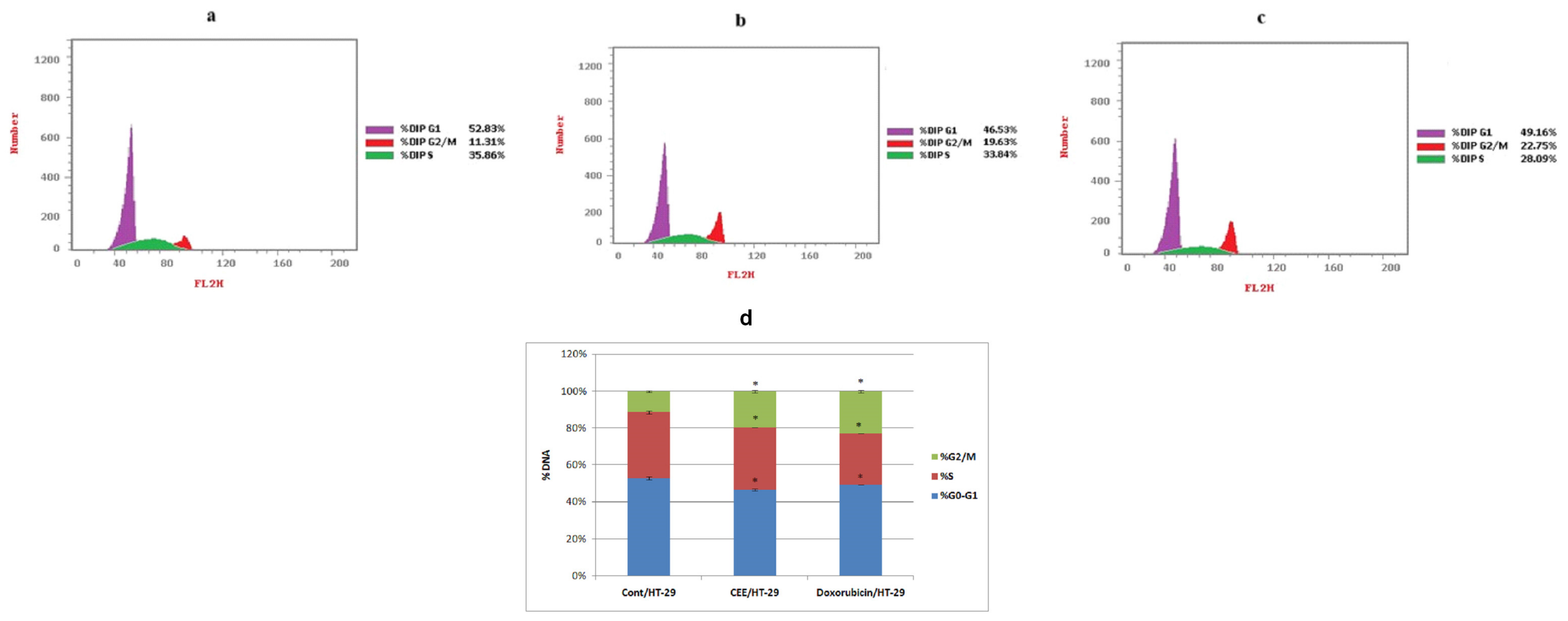
2.1.1. Cell Cycle Analysis
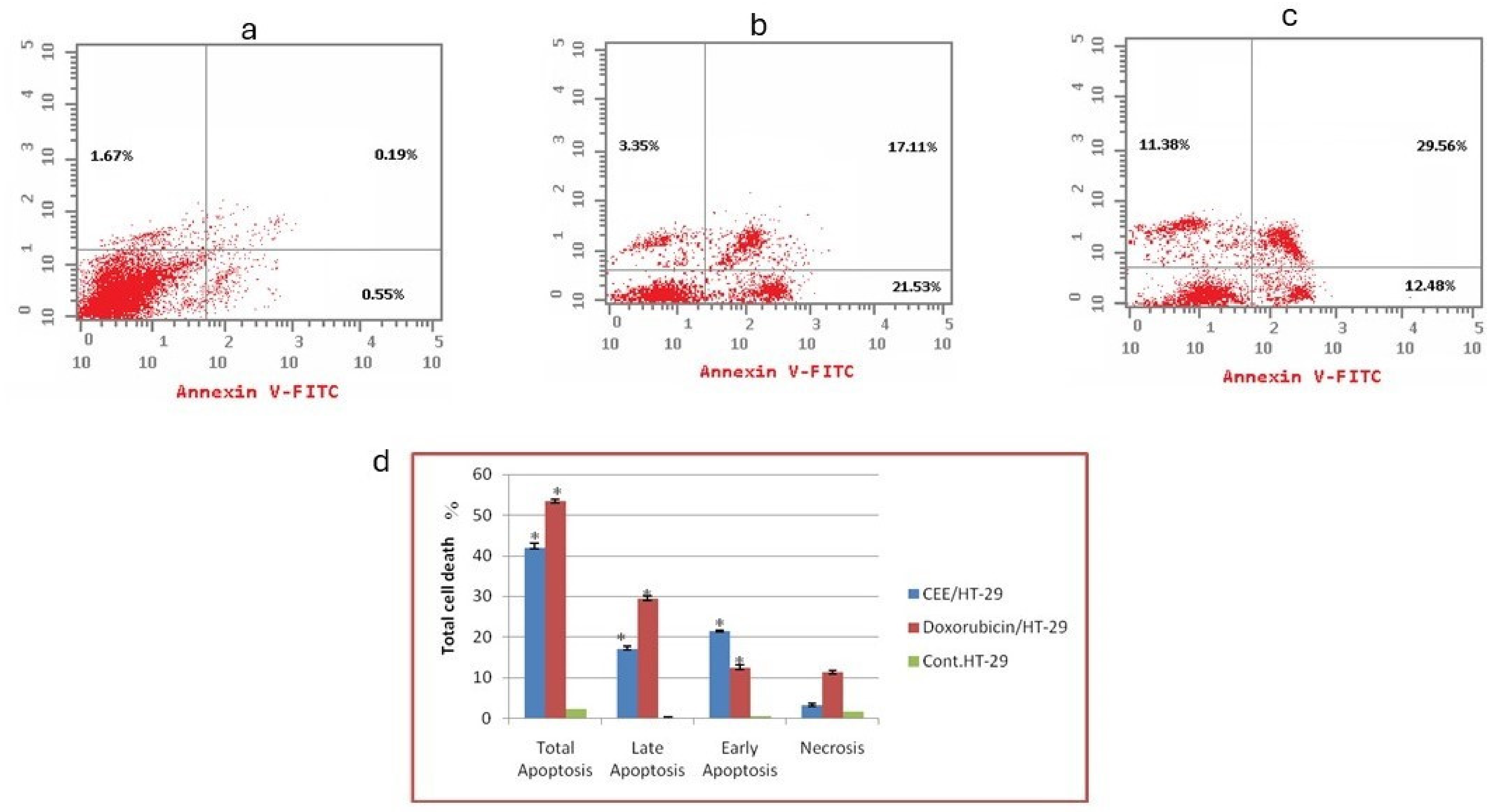
2.1.2. Flow Cytometry-Based Annexin V-FITC/PI Assay for the Detection of Apoptosis
2.1.3. Scratch Wound Healing
2.1.4. mRNA Transcription Levels of VEGF, BAX, BCL-2, and Caspase-3
2.2. LC-ESI-TOF-MS/MS
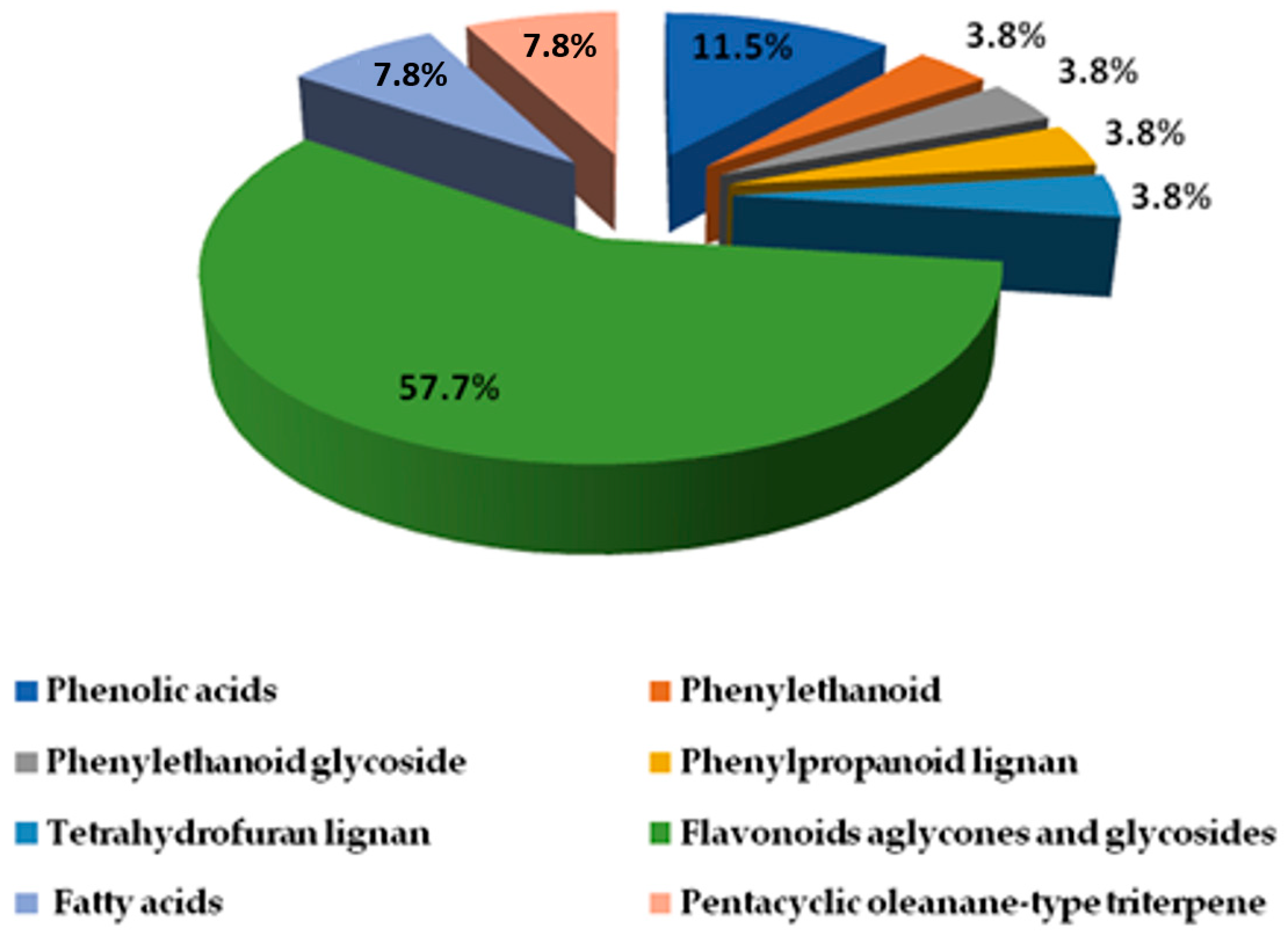
2.2.1. Phenolic Compound Identification
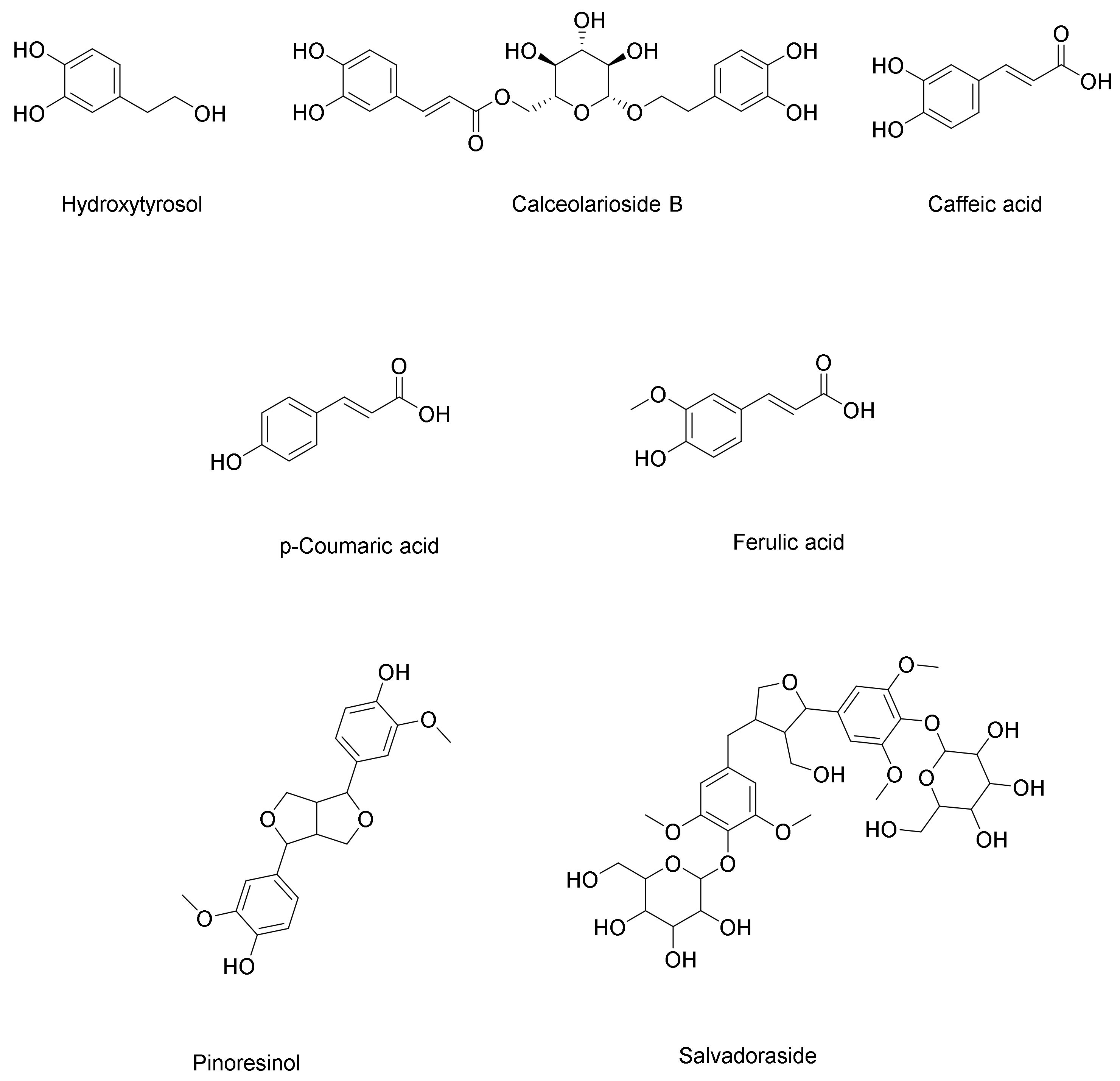
Phenolic Acids, Phenylethanoid, and Phenylpropanoid
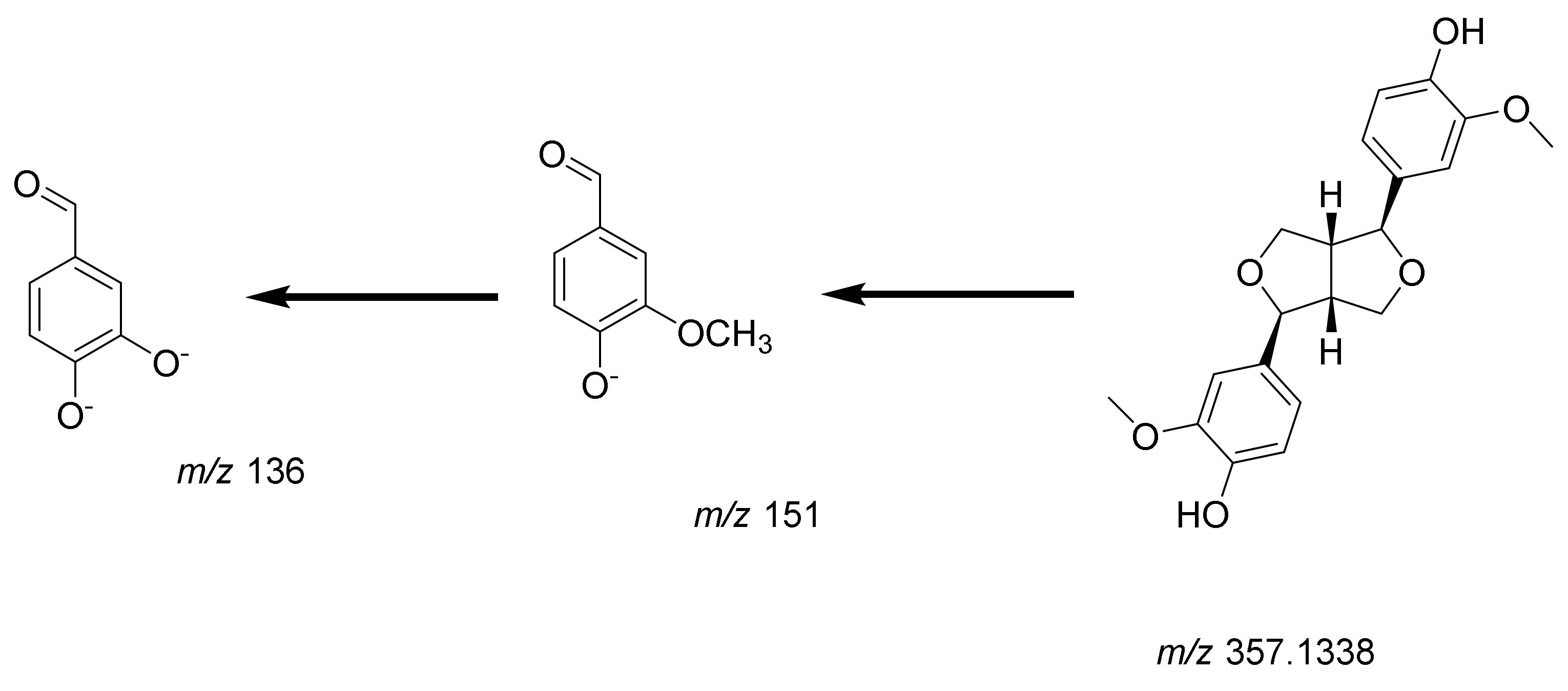
Lignans
2.2.2. Identification of Flavonoids (Aglycones and Glycosides)
2.2.3. Identification of Miscellaneous Compounds
2.3. Docking of Identified Metabolites in Extract of Clematis cirrhosa
2.3.1. VEGFR-2 Inhibition
2.3.2. Caspase-3 Inhibition
2.4. Acute Toxicity
2.4.1. Acute Toxicological Evaluation
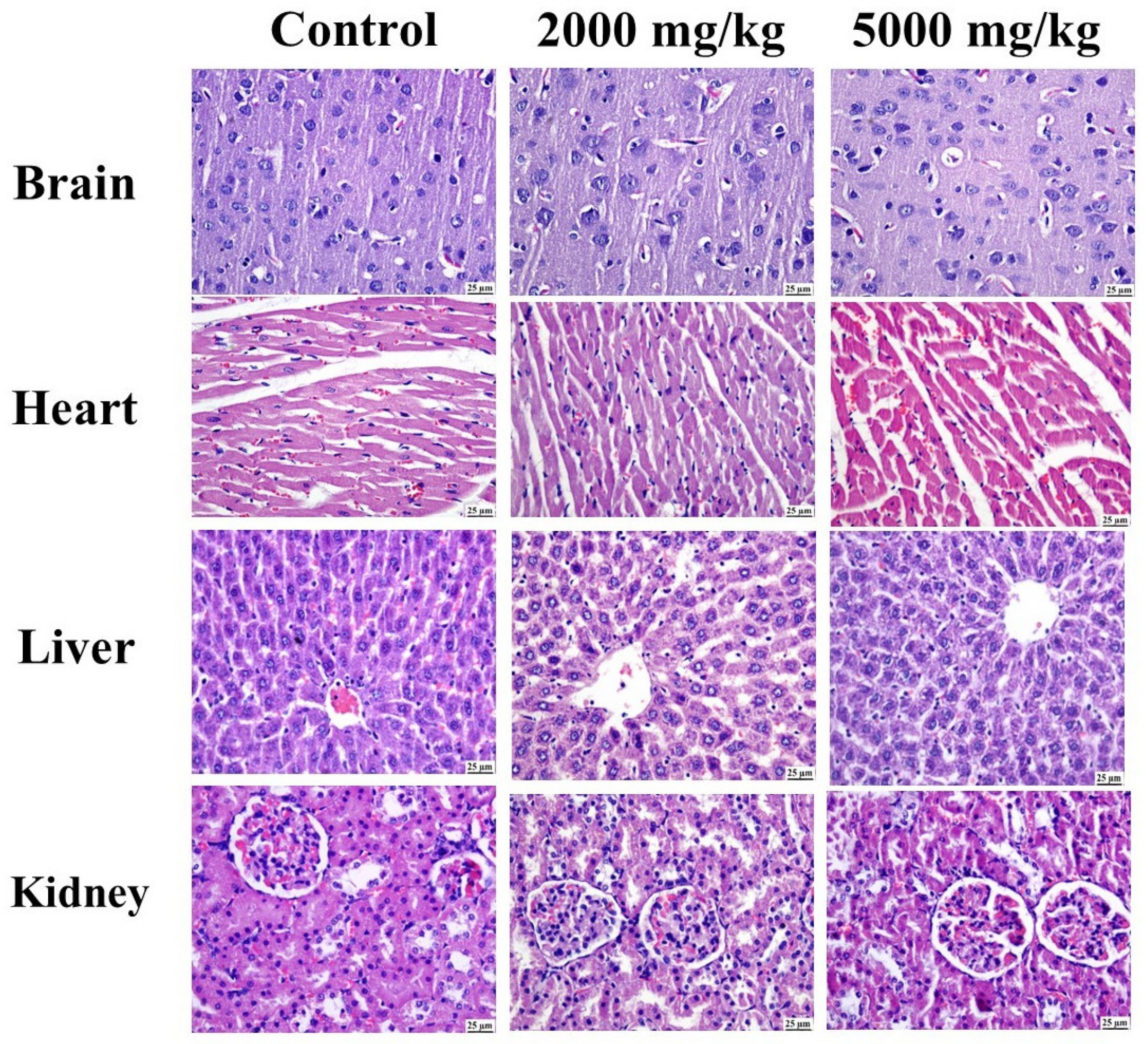
2.4.2. Histopathology
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Materials
4.3. Cytotoxicity Evaluation
4.4. Cell Cycle Analysis
4.5. Flow Cytometry-Based Annexin V–FITC/PI Assay for Detection of Apoptosis
4.6. Scratch Wound Healing
4.7. mRNA Transcription Levels of VEGF, BAX, BCL-2, and Caspase-3
4.8. LC-ESI-QTOF-MS/MS Analysis
4.8.1. Sample Preparation and Injection
4.8.2. LC-ESI-QTOF-MS/MS Conditions
4.8.3. Data Processing
4.9. Molecular Docking Study
4.10. Acute Toxicity Study
4.10.1. Animals
4.10.2. Histopathological Examination
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Observatory: “Cancer Today”; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Campagna, M.-C. Chemotherapy Complications. Hosp. Med. Clin. 2016, 5, 400–412. [Google Scholar] [CrossRef]
- Zi, X.; Zhang, R. Anti-cancer molecular targets of natural products. Curr. Cancer Drug Targets 2013, 13, 485. [Google Scholar] [CrossRef] [PubMed]
- Greenwell, M.; Rahman, P. Medicinal plants: Their use in anticancer treatment. Int. J. Pharm. Sci. Res. 2015, 6, 4103. [Google Scholar] [PubMed]
- Ahmadi, S.; Emamirad, S. Recent progresses and challenges in formulations of vincristine and its derivatives for hinderingcancer cells. Nano Micro Biosyst. 2023, 2, 36–41. [Google Scholar]
- Dhyani, P.; Quispe, C.; Sharma, E.; Bahukhandi, A.; Sati, P.; Attri, D.C.; Szopa, A.; Sharifi-Rad, J.; Docea, A.O.; Mardare, I.; et al. Anticancer potential of alkaloids: A key emphasis to colchicine, vinblastine, vincristine, vindesine, vinorelbine and vincamine. Cancer Cell Int. 2022, 22, 206. [Google Scholar] [CrossRef]
- Teibo, J.O.; Irozuru, C.E.; Teibo, T.K.A.; Omotoso, O.E.; Babalghith, A.O.; Batiha, G.E.-S. Perspective Chapter: Appraisal of Paclitaxel (Taxol) Pros and Cons in the Management of Cancer–Prospects in Drug Repurposing. In Drug Repurposing—Advances, Scopes and Opportunities in Drug Discovery; IntechOpen: London, UK, 2023. [Google Scholar]
- Motyka, S.; Jafernik, K.; Ekiert, H.; Sharifi-Rad, J.; Calina, D.; Al-Omari, B.; Szopa, A.; Cho, W.C. Podophyllotoxin and its derivatives: Potential anticancer agents of natural origin in cancer chemotherapy. Biomed. Pharmacother. 2023, 158, 114145. [Google Scholar] [CrossRef]
- Adak, D.; Ray, P.; Setua, S. Unlocking Therapeutic Precision: “Camptotheca acuminata, a Traditional Chinese Herb Tailored for Phytonano-Cancer Theranostics”. Pharmacol. Res. Chin. Med. 2024, 11, 100447. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Zhao, M.; Qian, K. Optimization of polysaccharides extraction from Clematis huchouensis Tamura and its antioxidant activity. Carbohydr. Polym. 2014, 111, 762–767. [Google Scholar] [CrossRef]
- Ferchichi, L.; Chohra, D.; Mellouk, K.; Alsheikh, S.M.; Cakmak, Y.S.; Zengin, G. Chemical composition and antioxidant activity of essential oil from the aerial parts of Clematis cirrhosa L. (Ranunculaceae) growing in Algeria. Ann. Rom. Soc. Cell Biol. 2021, 25, 1314–1324. [Google Scholar]
- Yesilada, E.; Küpeli, E. Clematis vitalba L. aerial part exhibits potent anti-inflammatory, antinociceptive and antipyretic effects. J. Ethnopharmacol. 2007, 110, 504–515. [Google Scholar] [CrossRef]
- Hao, D.C.; Gu, X.J.; Xiao, P.G.; Peng, Y. Chemical and biological research of Clematis medicinal resources. Chin. Sci. Bull. 2013, 58, 1120–1129. [Google Scholar] [CrossRef]
- Zhao, M.; Da-Wa, Z.-M.; Guo, D.-L.; Fang, D.-M.; Chen, X.-Z.; Xu, H.-X.; Gu, Y.-C.; Xia, B.; Chen, L.; Ding, L.-S.; et al. Cytotoxic triterpenoid saponins from Clematis tangutica. Phytochemistry 2016, 130, 228–237. [Google Scholar] [CrossRef]
- Chawla, R.; Kumar, S.; Sharma, A. The genus Clematis (Ranunculaceae): Chemical and pharmacological perspectives. J. Ethnopharmacol. 2012, 143, 116–150. [Google Scholar] [CrossRef]
- El-Oqlah, A.A.; Lahham, J.N. A check list of vascular plants of Ajlun mountain (Jordan). Candollea 1985, 40, 377–387. [Google Scholar]
- Ali-Shtayeh, M.S.; Abu Ghdeib, S.I. Antifungal activity of plant extracts against dermatophytes. Mycoses 1999, 42, 665–672. [Google Scholar] [CrossRef]
- Alruwad, M.I.; Salah, R.; Dine, E.; Gendy, A.M.; Sabry, M.M.; El Hefnawy, H.M. Exploring the Biological and Phytochemical Potential of Jordan’s Flora: A Review and Update of Eight Selected Genera from Mediterranean Region. Molecules 2024, 29, 1160. [Google Scholar] [CrossRef]
- Atmani, D.; Ruiz-Larrea, M.B.; Ruiz-Sanz, J.I.; Lizcano, L.J.; Bakkali, F.; Atmani, D. Antioxidant potential, cytotoxic activity and phenolic content of Clematis flammula leaf extracts. J. Med. Plants Res. 2011, 5, 589–598. [Google Scholar] [CrossRef]
- Hai, W.; Cheng, H.; Zhao, M.; Wang, Y.; Hong, L.; Tang, H.; Tian, X. Two new cytotoxic triterpenoid saponins from theroots of Clematis argentilucida. Fitoterapia 2012, 83, 759–764. [Google Scholar] [CrossRef]
- Alruwad, M.I.; Sabry, M.M.; Gendy, A.M.; El-Dine, R.S.; El Hefnawy, H.M. In Vitro Cytotoxic Potential of Selected Jordanian Flora and Their Associated Phytochemical Analysis. Plants 2023, 12, 1626. [Google Scholar] [CrossRef]
- El-Sayed, M.A.; Al-Gendy, A.A.; Hamdan, D.I.; El-Shazly, A.M. Phytoconstituents, LC-ESI-MS profile, antioxidant and antimicrobial activities of Citrus × limon L. Burm. f. cultivar variegated pinklemon. J. Pharm. Sci. Res. 2017, 9, 375. [Google Scholar]
- Kholkhal, W.; Ilias, F.; Bekhechi, C.; Bekkara, F.A. Eryngium maritimum: A rich medicinal plant of polyphenols and flavonoids compounds with antioxidant, antibacterial and antifungal activities. Curr.Res. J. Biol. Sci. 2012, 4, 437–443. [Google Scholar]
- Sánchez-Rabaneda, F.; Jáuregui, O.; Casals, I.; Andrés-Lacueva, C.; Izquierdo-Pulido, M.; Lamuela-Raventós, R.M. Liquid chromatographic/electrospray ionization tandem mass spectrometric study of the phenolic composition of cocoa (Theobroma cacao). J. Mass. Spectrom. 2003, 38, 35–42. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, H.; Xu, X.; Xue, X.; Liu, M.; Xu, S.; Liu, H.; Gao, Y.; Zhang, H.; Li, X. Compounds identification in Semen cuscutae by ultra-high-performance liquid chromatography (UPLCs) coupled to electrospray ionization mass spectrometry. Molecules 2018, 23, 1199. [Google Scholar] [CrossRef]
- Yuan, J.; Hao, L.J.; Wu, G.; Wang, S.; Duan, J.A.; Xie, G.Y.; Qin, M.J. Effects of drying methods on the phytochemicals contents and antioxidant properties of chrysanthemum flower heads harvested at two developmental stages. J. Funct. Foods 2015, 19, 786–795. [Google Scholar] [CrossRef]
- Xiao, X.; Xu, L.; Hu, H.; Yang, Y.; Zhang, X.; Peng, Y.; Xiao, P. DPPH radical scavenging and postprandial hyperglycemia inhibition activities and flavonoid composition analysis of hawk tea by UPLC-DAD and UPLC-Q/TOFMSE. Molecules 2017, 22, 1622. [Google Scholar] [CrossRef]
- Masike, K.; Mhlongo, M.I.; Mudau, S.P.; Nobela, O.; Ncube, E.N.; Tugizimana, F.; George, M.J.; Madala, N.E. Highlighting mass spectrometric fragmentation differences and similarities between hydroxycinnamoyl-quinic acids and hydroxycinnamoyl-isocitric acids. Chem. Cent. J. 2017, 11, 29. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, H.; Wu, H.; Pan, Y.; Wang, K.; Jin, Y.; Zhang, C. Characterization and quantification by LC-MS/MS of the chemical components of the heating products of the flavonoids extract in pollen typhae for transformation rule exploration. Molecules 2015, 20, 18352–18366. [Google Scholar] [CrossRef]
- Sobral, F.; Calhelha, R.C.; Barros, L.; Dueñas, M.; Tomás, A.; Santos-Buelga, C.; Vilas-Boas, M.; Ferreira, I.C.F.R. Flavonoid composition and antitumor activity of bee bread collected in northeast Portugal. Molecules 2017, 22, 248. [Google Scholar] [CrossRef]
- Michel, T.; Khlif, I.; Kanakis, P.; Termentzi, A.; Allouche, N.; Halabalaki, M.; Skaltsounis, A.-L. UHPLC-DAD-FLD and UHPLC-HRMS/MS based metabolic profiling and characterization of different Olea europaea organs of Koroneiki and Chetoui varieties. Phytochem. Lett. 2015, 11, 424–439. [Google Scholar] [CrossRef]
- Wang, J.; Jia, Z.; Zhang, Z.; Wang, Y.; Liu, X.; Wang, L.; Lin, R. Analysis of chemical constituents of Melastoma dodecandrum Lour. by UPLC-ESI-Q-Exactive Focus-MS/MS. Molecules 2017, 22, 476. [Google Scholar] [CrossRef]
- Carvalho, A.A.; dos Santos, L.R.; de Freitas, J.S.; de Sousa, R.P.; de Farias, R.R.S.; Júnior, G.M.V.; Rai, M.; Chaves, M.H. First report of flavonoids from leaves of Machaerium acutifolium by DI-ESI-MS/MS. Arab. J. Chem. 2022, 15, 103765. [Google Scholar] [CrossRef]
- Li, D.; Park, S.-H.; Shim, J.-H.; Lee, H.-S.; Tang, S.-Y.; Park, C.-S.; Park, K.-H. In vitro enzymatic modification of puerarin to puerarin glycosides by maltogenic amylase. Carbohydr. Res. 2004, 339, 2789–2797. [Google Scholar] [CrossRef] [PubMed]
- Mahrous, F.S.M.; Mohammed, H.; Sabour, R. LC-ESI-QTOF-MS/MS of Holoptelea integrifolia (Roxb.) Planch. Leaves and In silico study of phenolic compounds’ antiviral activity against the HSV1 virus. Azhar Int. J. Pharm. Med. Sci. 2021, 1, 91–101. [Google Scholar] [CrossRef]
- Jaiswal, R.; Karar, M.G.E.; Gadir, H.A.; Kuhnert, N. Identification and characterisation of phenolics from Ixora coccinea L. (Rubiaceae) by liquid chromatography multi-stage mass spectrometry. Phytochem. Anal. 2014, 25, 567–576. [Google Scholar] [CrossRef]
- Zhou, C.; Luo, Y.; Lei, Z.; Wei, G. UHPLC-ESI-MS Analysis of purified flavonoids fraction from stem of Dendrobium denneanum Paxt. and its preliminary study in inducing apoptosis of HepG2 Cells. Evid.-Based Complement. Altern. Med. 2018, 2018, 8936307. [Google Scholar] [CrossRef]
- Iwashina, T.; Smirnov, S.V.; Damdinsuren, O.; Kondo, K. Flavonoids from Reaumuria soongarica (Tamaricaceae) in Mongolia. Bull. Natl. Mus. Nat. Sci. Ser. B 2012, 38, 189–195. [Google Scholar]
- Ceccacci, S.; DeLucia, A.; Tito, A.; Tortora, A.; Falanga, D.; Arciello, S.; Ausanio, G.; Di Cicco, C.; Monti, M.C.; Apone, F. An Oenothera biennis Cell Cultures Extract Endowed with Skin Anti-Ageing Activity Improves Cell Mechanical Properties. Metabolites 2021, 11, 527. [Google Scholar] [CrossRef]
- Eissa, M.A.; Hashim, Y.Z.H.Y.; El-Kersh, D.M.; Abd-Azziz, S.S.S.; Salleh, H.M.; Isa, M.L.M.; Warif, N.M.A. Metabolite Profiling of Aquilaria malaccensis leaf extract using Liquid Chromatography-Q-TOF-Mass spectrometry and investigation of its potential antilipoxygenase activity in-vitro. Processes 2020, 8, 202. [Google Scholar] [CrossRef]
- Grayer, R.J.; Kite, G.C.; Abou-Zaid, M.; Archer, L.J. The application of atmospheric pressure chemical ionization liquid chromatography–mass spectrometry in the chemotaxonomic study of flavonoids: Characterisation of flavonoids from Ocimum gratissimum var. gratissimum. Phytochem. Anal. Int. J. Plant Chem. Biochem. Tech. 2000, 11, 257–267. [Google Scholar]
- Chohra, D.; Ferchichi, L.; Cakmak, Y.S.; Zengin, G.; Alsheikh, S.M. Phenolic Profiles, Antioxidant Activities and Enzyme Inhibitory Effects of an Algerian Medicinal Plant (Clematis Cirrhosa L.). South African J. Bot. 2020, 132, 164–170. [Google Scholar] [CrossRef]
- Neji, S.B.; Bouaziz, M. Production of biologically active hydroxy tyrosol rich extract via catalytic conversion of tyrosol. RSC Adv. 2022, 12, 2595–2602. [Google Scholar] [CrossRef] [PubMed]
- El-sayed, M.; Abbas, F.A.; Refaat, S.; El-Shafae, A.M.; Fikry, E. UPLC-ESI-MS/MS Profile of The Ethyl Acetate Fraction of Aerial Parts of Bougainvillea ‘Scarlett O’Hara’ Cultivated in Egypt. Egypt. J. Chem. 2021, 64, 793–806. [Google Scholar] [CrossRef]
- Chen, G.; Li, X.; Saleri, F.; Guo, M. Analysis of flavonoids in Rhamnus davurica and its antiproliferative activities. Molecules 2016, 21, 1275. [Google Scholar] [CrossRef] [PubMed]
- Kırmızıbekmez, H.; İnan, Y.; Reis, R.; Sipahi, H.; Gören, A.C.; Yeşilada, E. Phenolic compounds from the aerial parts of Clematis viticella L. and their in vitro anti-inflammatory activities. Nat. Prod. Res. 2019, 33, 2541–2544. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, Y.; Liang, H.; Song, H.; Zhao, J.; Liu, L.; Zeng, J.; Sun, L.; Ma, S.; Meng, D. A Novel Multidimensional Strategy to Evaluate Belamcanda Chinensis (L) DC and Iris Tectorum Maxim Based on Plant Metabolomics, Digital Reference Standard Analyzer and Biological Activities Evaluation. Chin. Med. 2021, 16, 85. [Google Scholar] [CrossRef]
- Ding, Q.; Yang, L.; Yang, H.W.; Jiang, C.; Wang, Y.F. Cytotoxic and antibacterial triterpenoids derivatives from Clematis ganpiniana. J. Ethnopharmacol. 2009, 126, 382–385. [Google Scholar] [CrossRef]
- Taylor, W.R.; Stark, G.R. Regulation of the G2/M transition by P53. Oncogene 2001, 20, 1803–1815. [Google Scholar] [CrossRef]
- Weinberg, R.A.; Hanahan, D. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar]
- Shibuya, M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: A crucial target for anti-and pro-angiogenic therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Dillekås, H.; Rogers, M.S.; Straume, O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019, 8, 5574–5576. [Google Scholar] [CrossRef]
- Peng, H.; Lv, H.; Wang, Y.; Liu, Y.-H.; Li, C.-Y.; Meng, L.; Chen, F.; Bao, J.-K. Clematis montana lectin, a novel mannose-binding lectin from traditional Chinese medicine with antiviral and apoptosis-inducing activities. Peptides 2009, 30, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Xia, T.-S.; Shi, L.; Xu, L.; Chen, W.; Zhu, Y.; Ding, Q. Rhamnose β-hederin inhibits migration and invasion of human breast cancer cell line MDA-MB-231. Biochem. Biophys. Res. Commun. 2018, 495, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Shi, L.; Wu, J.; Zhou, X.; Li, X.; Sun, X.; Zhu, L.; Xia, T.; Ding, Q. A hederagenin saponin isolated from Clematis ganpiniana induces apoptosis in breast cancer cells via the mitochondrial pathway. Oncol. Lett. 2018, 15, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.-L.; Wang, X.-L.; Luo, X.-L.; Liu, S.-Y.; Xu, P.; Hu, J.-F.; Liu, X.H. Boehmenan, a lignan from the Chinese medicinal plant Clematis armandii, inhibits A431 cell growth via blocking p70S6/S6 kinase pathway. Integr. Cancer Ther. 2017, 16, 351–359. [Google Scholar] [CrossRef]
- Lee, T.H.; Huang, N.K.; Lai, T.C.; Yang, A.T.Y.; Wang, G.J. Anemonin, from Clematis crassifolia, potent and selective inducible nitric oxide synthase inhibitor. J. Ethnopharmacol. 2008, 116, 518–527. [Google Scholar] [CrossRef]
- Yan, L.H.; Xu, L.Z.; Lin, J.; Zou, Z.M.; Zhao, B.H.; Yang, S.L. Studies on lignan constituents of Clematis parviloba. Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi = China J. Chin. Mater. Medica 2008, 33, 1839–1843. [Google Scholar]
- Guo, X.; Wang, G.; Li, J.; Li, J.; Sun, X. Analysis of Floral Color Differences between Different Ecological Conditions of Clematis tangutica (Maxim.) Korsh. Molecules 2023, 28, 462. [Google Scholar] [CrossRef]
- Hung, T.M.; Thuong, P.T.; Bae, K.-H. Antioxidant effect of flavonoids isolated from the root of Clematis trichotoma Nakai. Korean J. Med. Crop Sci. 2005, 13, 227–232. [Google Scholar]
- Zhang, L.; Luo, X.; Tian, J. Chemical constituents from Clematis terniflora. Chem. Nat. Compd. 2007, 43, 128–131. [Google Scholar] [CrossRef]
- Hashimoto, M.; Iwashina, T.; Kitajima, J.; Matsumoto, S. Flavonol glycosides from Clematis cultivars and taxa, and their contribution to yellow and white flower colors. Bull. Natl. Mus. Nat. Sci. Ser. B 2008, 34, 127–134. [Google Scholar]
- Watanabe, K.; Kanno, S.-I.; Tomizawa, A.; Yomogida, S.; Ishikawa, M. Acacetin induces apoptosis in human T cell leukemia Jurkat cells via activation of a caspase cascade. Oncol. Rep. 2012, 27, 204–209. [Google Scholar] [PubMed]
- Rezaei-Seresht, H.; Cheshomi, H.; Falanji, F.; Movahedi-Motlagh, F.; Hashemian, M.; Mireskandari, E. Cytotoxic activity of caffeic acid and gallic acid against MCF-7 human breast cancer cells: An in silico and in vitro study. Avicenna J. Phytomed. 2019, 9, 574. [Google Scholar] [PubMed]
- Hernandes, L.C.; Machado, A.R.T.; Tuttis, K.; Ribeiro, D.L.; Aissa, A.F.; Dévoz, P.P.; Antunes, L.M.G. Caffeic acid and chlorogenic acid cytotoxicity, genotoxicity and impact on global DNA methylation in human leukemic cell lines. Genet. Mol. Biol. 2020, 43, e20190347. [Google Scholar] [CrossRef]
- Karimvand, M.N.; Kalantar, H.; Khodayar, M.J. Cytotoxic and apoptotic effects of ferulic acid on renal carcinoma cell line (ACHN). Jundishapur J. Nat. Pharm. Prod. 2020, 15, e81969. [Google Scholar] [CrossRef]
- Szwajgier, D.; Baranowska-Wojcik, E.; Borowiec, K. Phenolic acids exert anticholinesterase and cognition-improving effects. Curr. Alzheimer Res. 2018, 15, 531–543. [Google Scholar] [CrossRef]
- Hemmati, S.; Schmidt, T.J.; Fuss, E. (+)-Pinoresinol/(−)-lariciresinol reductase from Linum perenne Himmelszelt involved in the biosynthesis of justicidin B. FEBS Lett. 2007, 581, 603–610. [Google Scholar] [CrossRef]
- Gao, C.; Li, X.; Yu, S.; Liang, L. Inhibition of Cancer Cell Growth by Oleanolic Acid in Multidrug Resistant Liver Carcinoma Is Mediated via Suppression of Cancer Cell Migration and Invasion, Mitochondrial Apoptosis, G2/M Cell Cycle Arrest and Deactivation of JNK/p38 Signalling Pathway. J. BUON 2019, 24, 1964–1969. [Google Scholar]
- Wang, K.; Liu, X.; Liu, Q.; Ho, I.H.; Wei, X.; Yin, T.; Zhan, Y.; Zhang, W.; Zhang, W.; Chen, B.; et al. Hederagenin potentiated cisplatin-and paclitaxel-mediated cytotoxicity by impairing autophagy in lung cancer cells. Cell Death Dis. 2020, 11, 611. [Google Scholar] [CrossRef]
- Ekins, S.; Mestres, J.; Testa, B. In silico pharmacology for drug discovery: Applications to targets and beyond. Br. J. Pharmacol. 2007, 152, 21–37. [Google Scholar] [CrossRef]
- Jordan, S.A.; Cunningham, D.G.; Marles, R.J. Assessment of herbal medicinal products: Challenges, and opportunities to increase the knowledge base for safety assessment. Toxicol. Appl. Pharmacol. 2010, 243, 198–216. [Google Scholar] [CrossRef]
- Ernest, S.N.; Kpemissi, M. Protective Effect of Bioactive Fractions of C. dalzielii Against Weight Gain in Mice Fed with a High-Fat Diet. Int. J. Sci. Res. 2019, 10, 34144–34153. [Google Scholar]
- Kumar, N.; Rai, A.; Reddy, N.D.; Raj, P.V.; Jain, P.; Deshpande, P.; Mathew, G.; Kutty, N.G.; Udupa, N.; Rao, C.M. Silymarin liposomes improves oral bioavailability of silybin besides targeting, and immune cells. Pharmacol. Rep. 2014, 66, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.Z.; Cui, L.; Guo, Y.J.; Rong, Y.M.; Lu, X.H.; Sun, M.Y.; Zhang, L.; Tian, J. In vivo study of four preparative extracts of Clematis terniflora DC. For antinociceptive activity and anti-inflammatory activity in rat model of carrageenan-induced chronicnon-bacterial prostatitis. J. Ethnopharmacol. 2011, 134, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- OECD. Guideline 407: Repeated dose 28-day oral toxicity in rodents. In Guidelines for Testing of Chemicals; Organization for Economic Co-operation and Development (OECD): Paris, France, 2008. [Google Scholar]
- Khurana, L.; ElGindi, M.; Tilstam, P.V.; Pantouris, G. Elucidating the role of an immunomodulatory protein in cancer: From protein expression to functional characterization. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 629, pp. 307–360. [Google Scholar]
- El Gizawy, H.A.; El-Haddad, A.E.; Attia, Y.M.; Fahim, S.A.; Zafer, M.M.; Saadeldeen, A.M. In Vitro Cytotoxic Activity and Phytochemical Characterization (UPLC/T-TOF-MS/MS) of the Watermelon (Citrullus lanatus) Rind Extract. Molecules 2022, 27, 2480. [Google Scholar] [CrossRef]
- Chenoweth, D.M. Chemical Tools for Imaging, Manipulating, and Tracking Biological Systems: Diverse Methods for Prokaryotic and Eukaryotic Systems; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Mishra, D.; Mishra, A.; Singh, M.P. In Silico insight to identify Potential Inhibitors of BUB1B from Mushroom bioactive compounds to Prevent Breast cancer metastasis. bioRxiv 2022. [Google Scholar] [CrossRef]
- Mishra, D.; Mishra, A.; Rai, S.N.; Vamanu, E.; Singh, M.P. Identification of prognostic biomarkers for suppressing tumorigenesis and metastasis of Hepatocellular carcinoma through transcriptome analysis. Diagnostics 2023, 13, 965. [Google Scholar] [CrossRef]
- Mishra, D.; Mishra, A.; Nand Rai, S.; Vamanu, E.; Singh, M.P. Demystifying the role of prognostic biomarkers in breastcancer through integrated transcriptome and pathway enrichment analyses. Diagnostics 2023, 13, 1142. [Google Scholar] [CrossRef]
- Lee, C.; Lee, S.; Shin, S.G.; Hwang, S. Real-time PCR determination of rRNA gene copy number: Absolute and relative quantification assays with Escherichia coli. Appl. Microbiol. Biotechnol. 2008, 78, 371–376. [Google Scholar] [CrossRef]
- Mohammed, H.A.; Khan, R.A.; Abdel-Hafez, A.A.; Abdel-Aziz, M.; Ahmed, E.; Enany, S.; Mahgoub, S.; Al-Rugaie, O.; Alsharidah, M.; Aly, M.S.; et al. Phytochemical profiling, in vitro and in silicoanti-microbial and anti-cancer activity evaluations and Staph GyraseB and h-TOP-IIβ receptor-docking studies of major constituents of Zygophyllum coccineum L. Aqueous-ethanolic extract and its subsequent fra. Molecules 2021, 26, 577. [Google Scholar] [CrossRef]
- Saleh, A.M.; Mahdy, H.A.; El-Zahabi, M.A.; Mehany, A.B.M.; Khalifa, M.M.; Eissa, I.H. Design, synthesis, in silico studies, and biological evaluation of novel pyrimidine-5-carbonitrile derivatives as potential anti-proliferative agents, VEGFR-2 inhibitors and apoptotic inducers. RSC Adv. 2023, 13, 22122–22247. [Google Scholar] [CrossRef]
- Upadhyay, P.; Shukla, R.; Mishra, S.K. Acute and sub-acute toxicity study of hydro-alcoholic leaves extract of Reinwardtia indica in rats. Biomed. Pharmacother. 2019, 111, 36–41. [Google Scholar] [CrossRef]







| Peak No. | Rt (m) | Mol. Ion (m/z) [M-H]− | Molecular Formula | Error | Identified Compound | Class | MS2 Fragments | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.80 | 461.1058/- | C22H22O11 | −5.61 | Chrysoeriol-O-hexoside | Methoxyflavon-O-glycoside | 446.1667, 299.0528, 284.1156 | [22] |
| 2 | 8.31 | 153/- | C8H10O3 | −1.76 | Hydroxytyrosol | Phenylethanoid | 123.0452 | [23] |
| 3 | 8.49 | 477.1614/- | C23H26O11 | 2.75 | Calceolarioside B | Phenylethanoid glycoside | 315.1071, 179.035, 161.9485 | [24] |
| 5 | 9.23 | 289.0713/- | C15H14O6 | 0.3 | * Catechin | Flavanol | 245.0823, 205.0511 | [25] |
| 6 | 9.25 | 163.0402/- | C9H8O3 | 4.18 | p-Coumaric acid | Hydroxycinnamic acid | 119.9177 | [26] |
| 7 | 9.87 | 625.1382/- | C27H30O17 | −3.64 | Quercetin-3,7-O-dihexoside | Flavonol-O-glycoside | 463.1413, 301.1013 | [27] |
| 8 | 10.2 | 755.2126/- | C33H40O20 | −1.15 | Manghaslin | Flavonol-O-glycoside | 591.1035, 301.0366 | [28] |
| 9 | 10.47 | 743.2789/- | C34H48O18 | 3.58 | Salvadoraside | Phenylpropanoid lignan | 581.1535, 419.1818, 389.1821 | [29] |
| 10 | 10.59 | 357.1338/- | C20H22O6 | −0.04 | Pinoresinol | Tetrahydrofuran lignan | 151.1139, 136.1222 | [30] |
| 11 | 10.74 | 447.0912/- | C21H20O11 | −3.44 | Quercetin-O-rhamnopyranoside (Vincetoxicoside B) | Flavonol-O-glycoside | 301.1383 | [31] |
| 12 | 10.74 | 447.098/- | C21H20O11 | −3.21 | Orientin | Flavone-C-glycoside | 357.0708, 327.0097, 285.2595 | [32] |
| 13 | 11.14 | 577.1563/- | C27H30O14 | 2.72 | Kaempferol-3,7-O-dirhamnoside | Flavonol-O-glycoside | 431.0993, 285.1348 | [33] |
| 14 | 11.44 | 463.0869/- | C21H20O12 | −1.62 | Isoquercitrin | Flavonol-O-glycoside | 301.1385, 151.0776 | [27] |
| 15 | 11.61 | 593.150/- | C27H30O15 | −1.09 | Kaempferol-O-rutinoside | Flavonol-O-glycoside | 285.1159 | [34] |
| 16 | 12.16 | 193.0496/- | C10H10O4 | −2.51 | * Ferulic acid | Hydroxycinnamic acid | 134.0364 | [35] |
| 18 | 13.34 | 269.0454/- | C15H10O5 | 1.49 | Genistein | Isoflavone | 241.081, 225.9265, 197.9058, 143 | [36] |
| 19 | 13.45 | 301.0355/- | C15H10O7 | 0.9 | Quercetin | Flavonol | 273.1587, 179.0358, 151.9203, 107.0858 | [37] |
| 20 | 13.5 | 285.0417/- | C15H10O6 | 6.27 | Luteolin | Flavone | 241.0697, 175.9944, 151.0242, 133.1026 | [38] |
| 22 | 19.08 | 295.2266/- | C18H32O3 | −2.44 | Hydroxy octadecadienoic acid | Fatty acid | 277.2170, 171.8337 | [39] |
| 23 | 19.87 | 471.348/- | C30H48O4 | 1.2 | Hederagenin | Pentacyclic oleanane-type triterpene | 405.2010, 393.1453 | [40] |
| 25 | 21.8 | 271.2274/- | C16H32O3 | 0.3 | Hydroxy palmitic acid | Fatty acid | 253.2179, 225.2179 | [39] |
| 26 | 22.1 | 455.3531/- | C30H48O3 | 1.27 | Oleanolic acid | Pentacyclic oleanane-type triterpene | 407.1748 | [41] |
| Peak No. | Rt (m) | Mol. Ion (m/z) [M+H]+ | Molecular Formula | Error | Identified Compound | Class | MS2 Fragments | Ref. |
|---|---|---|---|---|---|---|---|---|
| 4 | 8.83 | -/181.0505 | C9H8O4 | 2.85 | * Caffeic acid | Hydroxycinnamic acid | 163.0386 | [43] |
| 17 | 13.25 | -/417.121 | C21H20O9 | 5.86 | Puerarin | Isoflavone glycoside | 399.8033, 297.1073, 351.2390, 267.1618 | [40] |
| 21 | 18.33 | -/463.133 | C22H22O11 | 8.96 | Tectoridin | Isoflavone glycoside | 301.1407, 286.2915 | [44] |
| 24 | 20.04 | -/285.0736 | C16H12O5 | 7.02 | Acacetin | Flavone | 270.2440, 242.0699, 153.1299, 133.0979 | [45] |
| Ligand | RMSD Value (Å) | Docking Score (kcal/mol) | Interactions | |
|---|---|---|---|---|
| Hydrogen Bonds | PiInteractions | |||
| Calceolarioside B | 1.48 | −9.20 | 9 | 9 |
| Quercetin-3,7-O-diglucoside | 1.57 | −6.44 | 8 | 3 |
| Manghaslin | 1.92 | −9.68 | 8 | 9 |
| Quercetin 7-O-alpha-L-rhamnopyranoside | 1.04 | −8.37 | 6 | 9 |
| Luteolin | 1.33 | −7.18 | 6 | 8 |
| Sorafenib | 1.36 | −8.50 | 5 | 19 |
| Ligand | RMSD Value (Å) | Docking Score (kcal/mol) | Interactions | |
|---|---|---|---|---|
| Hydrogen Bonds | PiInteractions | |||
| Tectoridin | 1.75 | −7.59 | 6 | 3 |
| Quercetin-3,7-O-diglucoside | 1.47 | −8.62 | 6 | 5 |
| Manghaslin | 1.78 | −8.38 | 4 | 5 |
| Salvadoraside | 1.73 | −9.88 | 11 | 0 |
| Kaempferol-3,7-O-alpha-L-dirhamnoside | 1.90 | −8.58 | 7 | 4 |
| Gene | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| VEGF | 5′-TTGCCTTGCTGCTCTACCTCCA-3′ | 5′-GATGGCAGTAGCTGCGCTGATA-3′ |
| BAX | 5′-TCAGGATGCGTCCACCAAGAAG-3′ | 5′-TGTGTCCACGGCGGCAATCATC-3′ |
| BCL-2 | 5′-ATCGCCCTGTGGATGACTGAGT-3′ | 5′-GCCAGGAGAAATCAAACAGAGGC-3′ |
| Caspase-3 | 5′-GGAAGCGAATCAATGGACTCTGG-3′ | 5′-GCATCGACATCTGTACCAGACC-3′ |
| GAPDH | 5′-GTCTCCTCTGACTTCAACAGCG-3′ | 5′-ACCACCCTGTTGCTGTAGCCAA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alruwad, M.I.; Salah El Dine, R.; Gendy, A.M.; Saleh, A.M.; Khalaf, M.A.; El Hefnawy, H.M.; Sabry, M.M. Insights into Clematis cirrhosa L. Ethanol Extract: Cytotoxic Effects, LC-ESI-QTOF-MS/MS Chemical Profiling, Molecular Docking, and Acute Toxicity Study. Pharmaceuticals 2024, 17, 1347. https://doi.org/10.3390/ph17101347
Alruwad MI, Salah El Dine R, Gendy AM, Saleh AM, Khalaf MA, El Hefnawy HM, Sabry MM. Insights into Clematis cirrhosa L. Ethanol Extract: Cytotoxic Effects, LC-ESI-QTOF-MS/MS Chemical Profiling, Molecular Docking, and Acute Toxicity Study. Pharmaceuticals. 2024; 17(10):1347. https://doi.org/10.3390/ph17101347
Chicago/Turabian StyleAlruwad, Manal I., Riham Salah El Dine, Abdallah M. Gendy, Abdulrahman M. Saleh, Mohamed A. Khalaf, Hala M. El Hefnawy, and Manal M. Sabry. 2024. "Insights into Clematis cirrhosa L. Ethanol Extract: Cytotoxic Effects, LC-ESI-QTOF-MS/MS Chemical Profiling, Molecular Docking, and Acute Toxicity Study" Pharmaceuticals 17, no. 10: 1347. https://doi.org/10.3390/ph17101347
APA StyleAlruwad, M. I., Salah El Dine, R., Gendy, A. M., Saleh, A. M., Khalaf, M. A., El Hefnawy, H. M., & Sabry, M. M. (2024). Insights into Clematis cirrhosa L. Ethanol Extract: Cytotoxic Effects, LC-ESI-QTOF-MS/MS Chemical Profiling, Molecular Docking, and Acute Toxicity Study. Pharmaceuticals, 17(10), 1347. https://doi.org/10.3390/ph17101347








