Assessing the Neurodevelopmental Impact of Fluoxetine, Citalopram, and Paroxetine on Neural Stem Cell-Derived Neurons
Abstract
:1. Introduction
2. Results
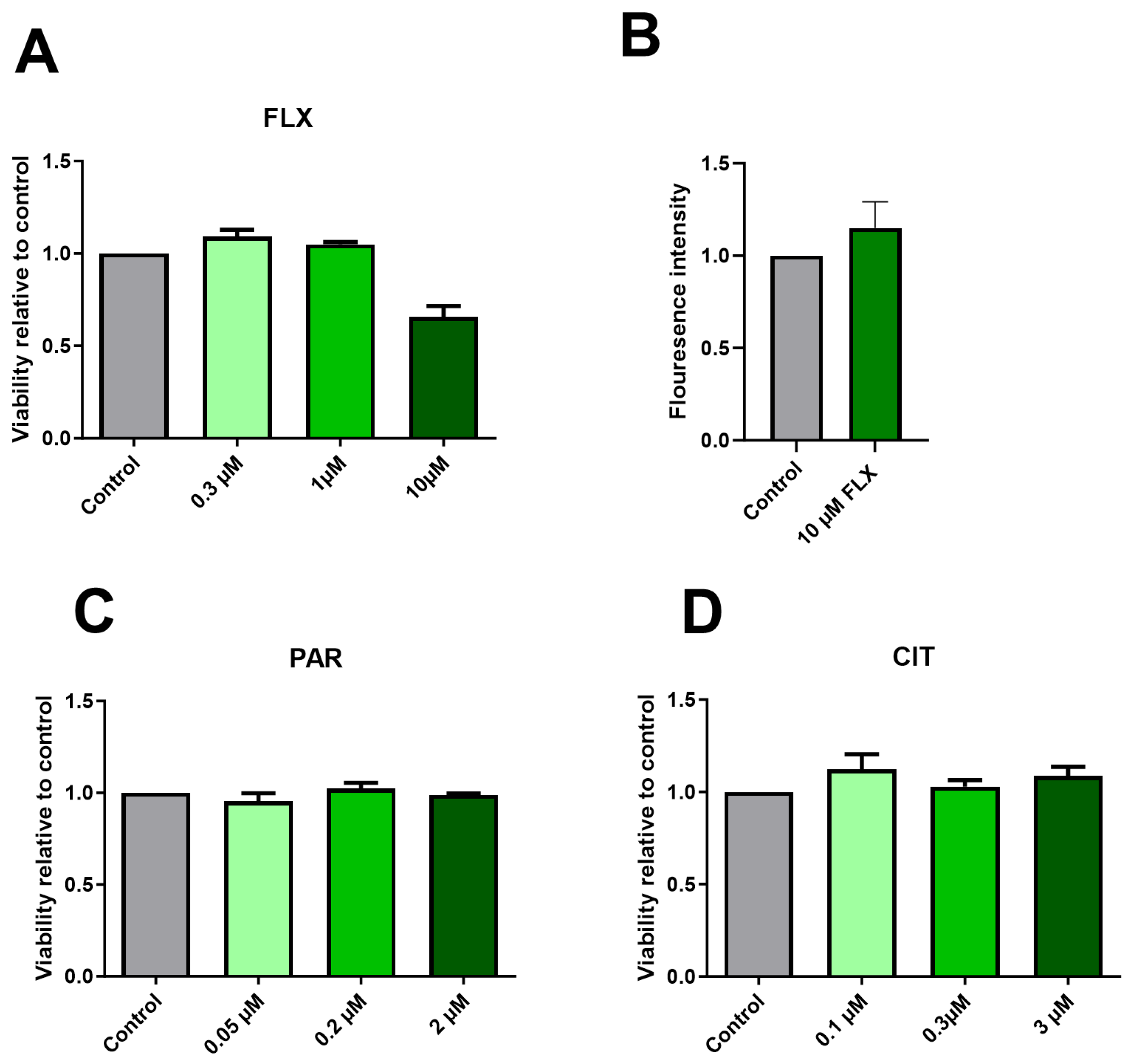
2.1. Effect of SSRIs on Cell Viability
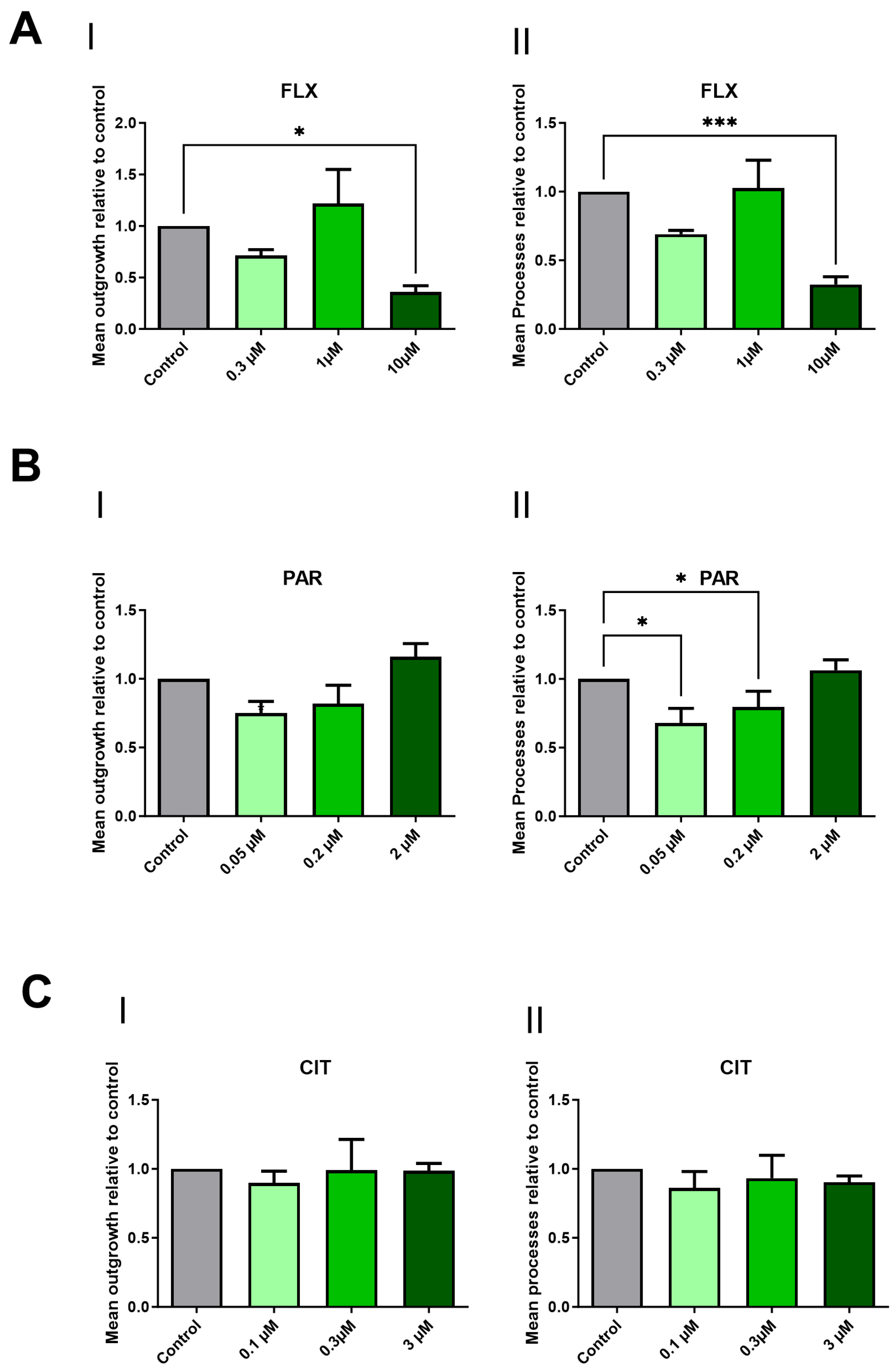
2.2. Paroxetine and Fluoxetine Demonstrate a Reduction in Neurite Outgrowth
2.3. Effect of FLX and PAR on 21 Neurodevelopmental-Associated Transcriptional Markers
2.4. 3H-5HT Uptake Assay Indicated the Absence of SERT
3. Discussion
4. Materials and Methods
- Reagents
4.1. Cell Culture
4.2. Drug Concentrations and Exposure
4.3. Cell Viability Assay Using Resazurin Assay (Alamar Blue)
4.4. Apoptosis Detection Assay
4.5. Analysis of Neurite Outgrowth
4.6. RNA Extraction and cDNA Synthesis
4.7. Reverse Transcription-Quantitative Polymerase Chain Reaction Analysis of 21 Genes Associated with Neurodevelopmental Toxicity or Neuronal Activity
4.8. AmpliSeq Analysis of Maturing Neurons without Exposure
4.9. Uptake Experiments to Measure SERT Activity
4.10. Statistical Analysis and Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-HT | 5-hydroxytryptamine |
| ASD | Autism spectrum disorder |
| cDNA | Complementary DNA |
| CPM | Count per minute |
| CIT | Citalopram |
| DMSO | Dimethyl sulfoxide |
| DNT | Developmental neurotoxicity |
| FC | Fold changes |
| FLX | Fluoxetine |
| hESCs | Human embryonic stem cells |
| hNSCs | Human neural stem cells |
| PAR | Paroxetine |
| RT-qPCR | Reverse transcriptase quantitative polymerase chain reaction |
| SERT | Serotonin transporter |
| SSRI | Selective serotonin reuptake inhibitors |
References
- Alyson Gorun, M.D. Choosing and Discussing SSRIs for Depression in Pregnancy: A Basic Guide for Residents. Am. J. Psychiatry Resid. J. 2018, 13, 3–5. [Google Scholar] [CrossRef]
- Lupu, D.; Sjödin, M.O.D.; Varshney, M.; Lindberg, J.; Loghin, F.; Rüegg, J. Fluoxetine modulates sex steroid levels in vitro. Clujul Med. 2017, 90, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Marinho, L.S.R.; Chiarantin, G.M.D.; Ikebara, J.M.; Cardoso, D.S.; de Lima-Vasconcellos, T.H.; Higa, G.S.V.; Ferraz, M.S.A.; De Pasquale, R.; Takada, S.H.; Papes, F.; et al. The impact of antidepressants on human neurodevelopment: Brain organoids as experimental tools. Semin. Cell Dev. Biol. 2023, 144, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Anderson, I.M.; Edwards, J.G. Guidelines for choice of selective serotonin reuptake inhibitor in depressive illness. Adv. Psychiatr. Treat. 2018, 7, 170–180. [Google Scholar] [CrossRef]
- Hyttel, J. Pharmacological characterization of selective serotonin reuptake inhibitors (SSRIs). Int. Clin. Psychopharmacol. 1994, 9, 19–26. [Google Scholar] [CrossRef]
- Hyttel, J. Comparative pharmacology of selective serotonin re-uptake inhibitors (SSRIs). Nord. J. Psychiatry 1993, 47 (Suppl. S30), 5–12. [Google Scholar] [CrossRef]
- Brumbaugh, J.E.; Ball, C.T.; Crook, J.E.; Stoppel, C.J.; Carey, W.A.; Bobo, W.V. Poor Neonatal Adaptation After Antidepressant Exposure During the Third Trimester in a Geographically Defined Cohort. Mayo Clin. Proc. Innov. Qual. Outcomes 2023, 7, 127–139. [Google Scholar] [CrossRef]
- Levy, M.; Kovo, M.; Miremberg, H.; Anchel, N.; Herman, H.G.; Bar, J.; Schreiber, L.; Weiner, E. Maternal use of selective serotonin reuptake inhibitors (SSRI) during pregnancy—Neonatal outcomes in correlation with placental histopathology. J. Perinatol. 2020, 40, 1017–1024. [Google Scholar] [CrossRef]
- Cole, J.A.; Ephross, S.A.; Cosmatos, I.S.; Walker, A.M. Paroxetine in the first trimester and the prevalence of congenital malformations. Pharmacoepidemiol. Drug Saf. 2007, 16, 1075–1085. [Google Scholar] [CrossRef]
- Singal, D.; Chateau, D.; Struck, S.; Lee, J.B.; Dahl, M.; Derksen, S.; Katz, L.Y.; Ruth, C.; Hanlon-Dearman, A.; Brownell, M. In Utero Antidepressants and Neurodevelopmental Outcomes in Kindergarteners. Pediatrics 2020, 145, e20191157. [Google Scholar] [CrossRef]
- Morales, D.R.; Slattery, J.; Evans, S.; Kurz, X. Antidepressant use during pregnancy and risk of autism spectrum disorder and attention deficit hyperactivity disorder: Systematic review of observational studies and methodological considerations. BMC Med. 2018, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Gemmel, M.; Bögi, E.; Ragan, C.; Hazlett, M.; Dubovicky, M.; van den Hove, D.L.; Oberlander, T.F.; Charlier, T.D.; Pawluski, J.L. Perinatal selective serotonin reuptake inhibitor medication (SSRI) effects on social behaviors, neurodevelopment and the epigenome. Neurosci. Biobehav. Rev. 2018, 85, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Sprowles, J.L.N.; Hufgard, J.R.; Gutierrez, A.; Bailey, R.A.; Jablonski, S.A.; Williams, M.T.; Vorhees, C.V. Perinatal exposure to the selective serotonin reuptake inhibitor citalopram alters spatial learning and memory, anxiety, depression, and startle in Sprague-Dawley rats. Int. J. Dev. Neurosci. 2016, 54, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.S.; Gyllenberg, D.; Malm, H.; McKeague, I.W.; Hinkka-Yli-Salomäki, S.; Artama, M.; Gissler, M.; Cheslack-Postava, K.; Weissman, M.M.; Gingrich, J.A.; et al. Association of Selective Serotonin Reuptake Inhibitor Exposure During Pregnancy With Speech, Scholastic, and Motor Disorders in Offspring. JAMA Psychiatry 2016, 73, 1163–1170. [Google Scholar] [CrossRef]
- Vuong, H.E.; Coley, E.J.L.; Kazantsev, M.; Cooke, M.E.; Rendon, T.K.; Paramo, J.; Hsiao, E.Y. Interactions between maternal fluoxetine exposure, the maternal gut microbiome and fetal neurodevelopment in mice. Behav. Brain Res. 2021, 410, 113353. [Google Scholar] [CrossRef]
- de Leeuw, V.C.; Hessel, E.V.S.; Pennings, J.L.A.; Hodemaekers, H.M.; Wackers, P.F.K.; van Oostrom, C.T.M.; Piersma, A.H. Differential effects of fluoxetine and venlafaxine in the neural embryonic stem cell test (ESTn) revealed by a cell lineage map. Neurotoxicology 2020, 76, 1–9. [Google Scholar] [CrossRef]
- Lupu, D.; Varshney, M.K.; Mucs, D.; Inzunza, J.; Norinder, U.; Loghin, F.; Nalvarte, I.; Rüegg, J. Fluoxetine Affects Differentiation of Midbrain Dopaminergic Neurons In Vitro. Mol. Pharmacol. 2018, 94, 1220–1231. [Google Scholar] [CrossRef]
- Svirsky, N.; Levy, S.; Avitsur, R. Prenatal exposure to selective serotonin reuptake inhibitors (SSRI) increases aggression and modulates maternal behavior in offspring mice. Dev. Psychobiol. 2016, 58, 71–82. [Google Scholar] [CrossRef]
- Ames, J.L.; Ladd-Acosta, C.; Fallin, M.D.; Qian, Y.; Schieve, L.A.; DiGuiseppi, C.; Lee, L.C.; Kasten, E.P.; Zhou, G.; Pinto-Martin, J.; et al. Maternal Psychiatric Conditions, Treatment With Selective Serotonin Reuptake Inhibitors, and Neurodevelopmental Disorders. Biol. Psychiatry 2021, 90, 253–262. [Google Scholar] [CrossRef]
- Stapel, B.; Melzer, C.; von der Ohe, J.; Hillemanns, P.; Bleich, S.; Kahl, K.G.; Hass, R. Effect of SSRI exposure on the proliferation rate and glucose uptake in breast and ovary cancer cell lines. Sci. Rep. 2021, 11, 1250. [Google Scholar] [CrossRef]
- Sanchez, C.; Reines, E.H.; Montgomery, S.A. A comparative review of escitalopram, paroxetine, and sertraline: Are they all alike? Int. Clin. Psychopharmacol. 2014, 29, 185. [Google Scholar] [CrossRef] [PubMed]
- Stahl, S.M. Mechanism of action of serotonin selective reuptake inhibitors: Serotonin receptors and pathways mediate therapeutic effects and side effects. J. Affect. Disord. 1998, 51, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Westenberg, H.; Sandner, C. Tolerability and safety of fluvoxamine and other antidepressants. Int. J. Clin. Pract. 2006, 60, 482–491. [Google Scholar]
- Artigas, F. Serotonin receptors involved in antidepressant effects. Pharmacol. Ther. 2013, 137, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.S.; Kuller, J.A.; Rhee, E.H.J. Antidepressants in Pregnancy: A Review of Commonly Prescribed Medications. Obstet. Gynecol. Surv. 2011, 66, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Karson, C.N.; Newton, J.E.; Livingston, R.; Jolly, J.B.; Cooper, T.B.; Sprigg, J.; Komoroski, R.A. Human brain fluoxetine concentrations. J. Neuropsychiatry Clin. Neurosci. 1993, 5, 322–329. [Google Scholar]
- Kinoshita, M.; Hirayama, Y.; Fujishita, K.; Shibata, K.; Shinozaki, Y.; Shigetomi, E.; Takeda, A.; Le, H.P.N.; Hayashi, H.; Hiasa, M.; et al. Anti-Depressant Fluoxetine Reveals its Therapeutic Effect Via Astrocytes. EBioMedicine 2018, 32, 72–83. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kang, S.R.; Yune, T.Y. Fluoxetine Prevents Oligodendrocyte Cell Death by Inhibiting Microglia Activation after Spinal Cord Injury. J. Neurotrauma 2014, 32, 633–644. [Google Scholar] [CrossRef]
- Dhami, K.S.; Churchward, M.A.; Baker, G.B.; Todd, K.G. Fluoxetine and its metabolite norfluoxetine induce microglial apoptosis. J. Neurochem. 2019, 148, 761–778. [Google Scholar] [CrossRef]
- Po, W.W.; Thein, W.; Khin, P.P.; Khing, T.M.; Han, K.W.W.; Park, C.H.; Sohn, U.D. Fluoxetine Simultaneously Induces Both Apoptosis and Autophagy in Human Gastric Adenocarcinoma Cells. Biomol. Ther. 2020, 28, 202–210. [Google Scholar] [CrossRef]
- Stresser, D.M.; Kopec, A.K.; Hewitt, P.; Hardwick, R.N.; Van Vleet, T.R.; Mahalingaiah, P.K.S.; O’Connell, D.; Jenkins, G.J.; David, R.; Graham, J.; et al. Towards in vitro models for reducing or replacing the use of animals in drug testing. Nat. Biomed. Eng. 2023, 8, 930–935. [Google Scholar] [CrossRef]
- Bal-Price, A.K.; Coecke, S.; Costa, L.; Crofton, K.M.; Fritsche, E.; Goldberg, A.; Grandjean, P.; Lein, P.J.; Li, A.; Lucchini, R.; et al. Advancing the science of developmental neurotoxicity (DNT): Testing for better safety evaluation. Altex 2012, 29, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.; Masjosthusmann, S.; Bartmann, K.; Bendt, F.; Dolde, X.; Dönmez, A.; Förster, N.; Holzer, A.-K.; Hübenthal, U.; Keßel, H.E.; et al. Establishment of a human cell-based in vitro battery to assess developmental neurotoxicity hazard of chemicals. Chemosphere 2023, 311, 137035. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, K.; Lekholm, E.; Ahemaiti, A.; Fredriksson, R. Differentiation of Human Embryonic Stem Cells into Neuron, Cholinergic, and Glial Cells. Stem Cells Int. 2020, 2020, 8827874. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, K.; Philippot, G.; Salomonsson, S.B.; cediel-Ulloa, A.; Gholizadeh, E.; Fredriksson, R. Transcriptomic Characterization of Maturing Neurons from Human Neural Stem Cells Across Developmental Time Points. 2024. to be submitted. [Google Scholar]
- Warkus, E.L.L.; Marikawa, Y. Fluoxetine Inhibits Canonical Wnt Signaling to Impair Embryoid Body Morphogenesis: Potential Teratogenic Mechanisms of a Commonly Used Antidepressant. Toxicol. Sci. 2018, 165, 372–388. [Google Scholar] [CrossRef]
- Hinojosa, M.G.; Johansson, Y.; Cediel-Ulloa, A.; Ivanova, E.; Gabring, N.; Gliga, A.; Forsby, A. Evaluation of mRNA markers in differentiating human SH-SY5Y cells for estimation of developmental neurotoxicity. Neurotoxicology 2023, 97, 65–77. [Google Scholar] [CrossRef]
- Huang, C.; van Wijnen, A.J.; Im, H.-J. Serotonin Transporter (5-Hydroxytryptamine Transporter, SERT, SLC6A4) and Sodium-dependent Reuptake Inhibitors as Modulators of Pain Behaviors and Analgesic Responses. J. Pain 2024, 25, 618–631. [Google Scholar] [CrossRef]
- Li, S.; Zhang, L.; Huang, R.; Xu, T.; Parham, F.; Behl, M.; Xia, M. Evaluation of chemical compounds that inhibit neurite outgrowth using GFP-labeled iPSC-derived human neurons. Neurotoxicology 2021, 83, 137–145. [Google Scholar] [CrossRef]
- Ryan, K.R.; Sirenko, O.; Parham, F.; Hsieh, J.-H.; Cromwell, E.F.; Tice, R.R.; Behl, M. Neurite outgrowth in human induced pluripotent stem cell-derived neurons as a high-throughput screen for developmental neurotoxicity or neurotoxicity. Neurotoxicology 2016, 53, 271–281. [Google Scholar] [CrossRef]
- Stiegler, N.V.; Krug, A.K.; Matt, F.; Leist, M. Assessment of chemical-induced impairment of human neurite outgrowth by multiparametric live cell imaging in high-density cultures. Toxicol. Sci. 2011, 121, 73–87. [Google Scholar] [CrossRef]
- Johansson, Y.; Andreassen, M.; Hartsch, M.; Wagner, S.; Forsby, A. Attenuated neuronal differentiation caused by acrylamide is not related to oxidative stress in differentiated human neuroblastoma SH-SY5Y cells. Food Chem. Toxicol. 2024, 187, 114623. [Google Scholar] [CrossRef]
- Tate, K.; Kirk, B.; Tseng, A.; Ulffers, A.; Litwa, K. Effects of the Selective Serotonin Reuptake Inhibitor Fluoxetine on Developing Neural Circuits in a Model of the Human Fetal Cortex. Int. J. Mol. Sci. 2021, 22, 10457. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yin, H.; Feng, N.; Wang, L.; Wang, X. Inhibitory effects of antidepressant fluoxetine on cloned Kv2.1 potassium channel expressed in HEK293 cells. Eur. J. Pharmacol. 2020, 878, 173097. [Google Scholar] [CrossRef] [PubMed]
- Schaz, U.; Föhr, K.J.; Liebau, S.; Fulda, S.; Koelch, M.; Fegert, J.M.; Boeckers, T.M.; Ludolph, A.G. Dose-dependent modulation of apoptotic processes by fluoxetine in maturing neuronal cells: An in vitro study. World J. Biol. Psychiatry 2011, 12, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Hui, J.; Zhang, J.; Kim, H.; Tong, C.; Ying, Q.; Li, Z.; Mao, X.; Shi, G.; Yan, J.; Zhang, Z.; et al. Fluoxetine Regulates Neurogenesis In Vitro Through Modulation of GSK-3β/β-Catenin Signaling. Int. J. Neuropsychopharmacol. 2015, 18, pyu099. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.T.; Hsu, F.T.; Liu, Y.C.; Chen, C.H.; Hsu, L.C.; Lin, S.S. Fluoxetine Induces Apoptosis through Extrinsic/Intrinsic Pathways and Inhibits ERK/NF-κB-Modulated Anti-Apoptotic and Invasive Potential in Hepatocellular Carcinoma Cells In Vitro. Int. J. Mol. Sci. 2019, 20, 757. [Google Scholar] [CrossRef]
- Ishima, T.; Futamura, T.; Ohgi, Y.; Yoshimi, N.; Kikuchi, T.; Hashimoto, K. Potentiation of neurite outgrowth by brexpiprazole, a novel serotonin–dopamine activity modulator: A role for serotonin 5-HT1A and 5-HT2A receptors. Eur. Neuropsychopharmacol. 2015, 25, 505–511. [Google Scholar] [CrossRef]
- Chen, M.-K.; Peng, C.-C.; Maner, R.S.; Zulkefli, N.D.; Huang, S.-M.; Hsieh, C.-L. Geniposide ameliorated fluoxetine-suppressed neurite outgrowth in Neuro2a neuroblastoma cells. Life Sci. 2019, 226, 1–11. [Google Scholar] [CrossRef]
- DeGiosio, R.A.; Grubisha, M.J.; MacDonald, M.L.; McKinney, B.C.; Camacho, C.J.; Sweet, R.A. More than a marker: Potential pathogenic functions of MAP2. Front. Mol. Neurosci. 2022, 15, 974890. [Google Scholar] [CrossRef]
- Gatford, N.J.F.; Deans, P.J.M.; Duarte, R.R.R.; Chennell, G.; Sellers, K.J.; Raval, P.; Srivastava, D.P. Neuroligin-3 and neuroligin-4X form nanoscopic clusters and regulate growth cone organization and size. Hum. Mol. Genet. 2021, 31, 674–691. [Google Scholar] [CrossRef]
- Muellerleile, J.; Vnencak, M.; Ippolito, A.; Krueger-Burg, D.; Jungenitz, T.; Schwarzacher, S.W.; Jedlicka, P. Neuroligin-3 Regulates Excitatory Synaptic Transmission and EPSP-Spike Coupling in the Dentate Gyrus In Vivo. Mol. Neurobiol. 2022, 59, 1098–1111. [Google Scholar] [CrossRef]
- Gudi, V.; Gai, L.; Herder, V.; Tejedor, L.S.; Kipp, M.; Amor, S.; Sühs, K.-W.; Hansmann, F.; Beineke, A.; Baumgärtner, W.; et al. Synaptophysin Is a Reliable Marker for Axonal Damage. J. Neuropathol. Exp. Neurol. 2017, 76, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Harris, G.; Smirnova, L.; Zufferey, V.; Sá, R.; Baldino Russo, F.; Baleeiro Beltrao Braga, P.C.; Chesnut, M.; Zurich, M.G.; Hogberg, H.T.; et al. Antidepressant Paroxetine Exerts Developmental Neurotoxicity in an iPSC-Derived 3D Human Brain Model. Front. Cell. Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, V.; Stowe, Z.N.; Altshuler, L.L.; Hwang, S.; Lee, E.; Haynes, D. Placental passage of antidepressant medications. Am. J. Psychiatry 2003, 160, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.P.; Bury, T.B.S.; Donovan, E.E.; Lambert, O.J.; Palmucci, J.R.; Adamczak, S.K. Exposure to SSRI-type antidepressants increases righting time in the marine snail Ilyanassa obsoleta. Environ. Sci. Pollut. Res. 2017, 24, 725–731. [Google Scholar] [CrossRef]
- van der Most, M.A.; Estruch, I.M.; van den Brink, N.W. Contrasting dose response relationships of neuroactive antidepressants on the behavior of C. elegans. Ecotoxicol. Environ. Saf. 2023, 250, 114493. [Google Scholar] [CrossRef]
- Bossus, M.C.; Guler, Y.Z.; Short, S.J.; Morrison, E.R.; Ford, A.T. Behavioural and transcriptional changes in the amphipod Echinogammarus marinus exposed to two antidepressants, fluoxetine and sertraline. Aquat. Toxicol. 2014, 151, 46–56. [Google Scholar] [CrossRef]
- Xu, F.; Luk, C.; Richard, M.P.; Zaidi, W.; Farkas, S.; Getz, A.; Lee, A.; Van Minnen, J.; Syed, N.I. Antidepressant fluoxetine suppresses neuronal growth from both vertebrate and invertebrate neurons and perturbs synapse formation between Lymnaea neurons. Eur. J. Neurosci. 2010, 31, 994–1005. [Google Scholar] [CrossRef]
- Barakat, A.K.; Scholl, C.; Steffens, M.; Brandenburg, K.; Ising, M.; Lucae, S.; Holsboer, F.; Laje, G.; Kalayda, G.V.; Jaehde, U.; et al. Citalopram-induced pathways regulation and tentative treatment-outcome-predicting biomarkers in lymphoblastoid cell lines from depression patients. Transl. Psychiatry 2020, 10, 210. [Google Scholar] [CrossRef]
- Mennickent, S.; Fierro, R.; Vega, M.; De Diego, M.; Godoy, C.G. Quantitative determination of fluoxetine in human serum by high performance thin layer chromatography. J. Sep. Sci. 2010, 33, 2206–2210. [Google Scholar] [CrossRef]
- Deák, F.; Lasztóczi, B.; Pacher, P.; Petheö, G.L.; Valéria, K.; Spät, A. Inhibition of voltage-gated calcium channels by fluoxetine in rat hippocampal pyramidal cells. Neuropharmacology 2000, 39, 1029–1036. [Google Scholar] [CrossRef]
- Levine, B.; Zhang, X.; Smialek, J.; Kunsman, G.; Frontz, M. Citalopram Distribution in Postmortem Cases. J. Anal. Toxicol. 2001, 25, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.-P.; Zou, M.; Wang, J.-Y.; Zhu, J.-J.; Lai, J.-M.; Zhou, L.-L.; Chen, S.-F.; Zhang, X.; Zhu, J.-H. Paroxetine ameliorates lipopolysaccharide-induced microglia activation via differential regulation of MAPK signaling. J. Neuroinflamm. 2014, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Zelleroth, S.; Nylander, E.; Örtenblad, A.; Stam, F.; Nyberg, F.; Grönbladh, A.; Hallberg, M. Structurally different anabolic androgenic steroids reduce neurite outgrowth and neuronal viability in primary rat cortical cell cultures. J. Steroid Biochem. Mol. Biol. 2021, 210, 105863. [Google Scholar] [CrossRef] [PubMed]
- Delp, J.; Cediel-Ulloa, A.; Suciu, I.; Kranaster, P.; van Vugt-Lussenburg, B.M.A.; Munic Kos, V.; van der Stel, W.; Carta, G.; Bennekou, S.H.; Jennings, P.; et al. Neurotoxicity and underlying cellular changes of 21 mitochondrial respiratory chain inhibitors. Arch. Toxicol. 2021, 95, 591–615. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Hägglund, M.G.A.; Sreedharan, S.; Nilsson, V.C.O.; Shaik, J.H.A.; Almkvist, I.M.; Bäcklin, S.; Wrange, Ö.; Fredriksson, R. Identification of SLC38A7 (SNAT7) Protein as a Glutamine Transporter Expressed in Neurons. J. Biol. Chem. 2011, 286, 20500–20511. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Borghi, R.; Trivisano, M.; Specchio, N.; Tartaglia, M.; Compagnucci, C. Understanding the pathogenetic mechanisms underlying altered neuronal function associated with CAMK2B mutations. Neurosci. Biobehav. Rev. 2023, 152, 105299. [Google Scholar] [CrossRef]
- Nicole, O.; Pacary, E. CaMKIIβ in Neuronal Development and Plasticity: An Emerging Candidate in Brain Diseases. Int. J. Mol. Sci. 2020, 21, 7272. [Google Scholar] [CrossRef]
- Rodríguez-Palmero, A.; Boerrigter, M.M.; Gómez-Andrés, D.; Aldinger, K.A.; Marcos-Alcalde, Í.; Popp, B.; Everman, D.B.; Lovgren, A.K.; Arpin, S.; Bahrambeigi, V.; et al. DLG4-related synaptopathy: A new rare brain disorder. Genet. Med. 2021, 23, 888–899. [Google Scholar] [CrossRef]
- Jang, D.H.; Chae, H.; Kim, M. Autistic and Rett-like features associated with 2q33.3-q34 interstitial deletion. Am. J. Med. Genet. Part A 2015, 167, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Westphal, D.S.; Andres, S.; Makowski, C.; Meitinger, T.; Hoefele, J. MAP2—A Candidate Gene for Epilepsy, Developmental Delay and Behavioral Abnormalities in a Patient With Microdeletion 2q34. Front. Genet. 2018, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Kooblall, K.G.; Stevenson, M.; Heilig, R.; Stewart, M.; Wright, B.; Lockstone, H.; Buck, D.; Fischer, R.; Wells, S.; Lines, K.E.; et al. Identification of cellular retinoic acid binding protein 2 (CRABP2) as downstream target of nuclear factor I/X (NFIX): Implications for skeletal dysplasia syndromes. JBMR Plus 2024, 8, ziae060. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.S.; Riess, A.; Moog, U.; Briggs, T.A.; Chandler, K.E.; Rauch, A.; Stampfer, M.; Steindl, K.; Gläser, D.; Joset, P.; et al. FOXP2 variants in 14 individuals with developmental speech and language disorders broaden the mutational and clinical spectrum. J. Med. Genet. 2017, 54, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Namiki, J.; Shibata, S.; Mastuzaki, Y.; Okano, H. The neural stem/progenitor cell marker nestin is expressed in proliferative endothelial cells, but not in mature vasculature. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2010, 58, 721–730. [Google Scholar] [CrossRef]
- Zhu, F.; Feng, M.; Sinha, R.; Murphy, M.P.; Luo, F.; Kao, K.S.; Szade, K.; Seita, J.; Weissman, I.L. The GABA receptor GABRR1 is expressed on and functional in hematopoietic stem cells and megakaryocyte progenitors. Proc. Natl. Acad. Sci. USA 2019, 116, 18416–18422. [Google Scholar] [CrossRef]
- Blatt, G.J.; Fatemi, S.H. Alterations in GABAergic Biomarkers in the Autism Brain: Research Findings and Clinical Implications. Anat. Rec. 2011, 294, 1646–1652. [Google Scholar] [CrossRef]
- Agarwala, S.; Ramachandra, N.B. Role of CNTNAP2 in autism manifestation outlines the regulation of signaling between neurons at the synapse. Egypt. J. Med. Hum. Genet. 2021, 22, 22. [Google Scholar] [CrossRef]
- Siddiqui, T.; Cosacak, M.I.; Popova, S.; Bhattarai, P.; Yilmaz, E.; Lee, A.J.; Min, Y.; Wang, X.; Allen, M.; İş, Ö.; et al. Nerve growth factor receptor (Ngfr) induces neurogenic plasticity by suppressing reactive astroglial Lcn2/Slc22a17 signaling in Alzheimer’s disease. npj Regen. Med. 2023, 8, 33. [Google Scholar] [CrossRef]
- de Leeuw, V.C.; van Oostrom, C.T.M.; Wackers, P.F.K.; Pennings, J.L.A.; Hodemaekers, H.M.; Piersma, A.H.; Hessel, E.V.S. Neuronal differentiation pathways and compound-induced developmental neurotoxicity in the human neural progenitor cell test (hNPT) revealed by RNA-seq. Chemosphere 2022, 304, 135298. [Google Scholar] [CrossRef]
- Boyé, K.; Geraldo, L.H.; Furtado, J.; Pibouin-Fragner, L.; Poulet, M.; Kim, D.; Nelson, B.; Xu, Y.; Jacob, L.; Maissa, N.; et al. Endothelial Unc5B controls blood-brain barrier integrity. Nat. Commun. 2022, 13, 1169. [Google Scholar] [CrossRef] [PubMed]
- Salpietro, V.; Dixon, C.L.; Guo, H.; Bello, O.D.; Vandrovcova, J.; Efthymiou, S.; Maroofian, R.; Heimer, G.; Burglen, L.; Valence, S.; et al. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat Commun 2019, 10, 3094. [Google Scholar] [CrossRef] [PubMed]
- Coombs, I.D.; Ziobro, J.; Krotov, V.; Surtees, T.-L.; Cull-Candy, S.G.; Farrant, M. A gain-of-function GRIA2 variant associated with neurodevelopmental delay and seizures: Functional characterization and targeted treatment. Epilepsia 2022, 63, e156–e163. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.H.; D’Arcangelo, G. The Reelin Gene and Its Functions in Brain Development. In Reelin Glycoprotein: Structure, Biology and Roles in Health and Disease; Fatemi, S.H.; D’Arcangelo, G. Springer: New York, NY, USA, 2008; pp. 1–13. [Google Scholar] [CrossRef]
- Joly-Amado, A.; Kulkarni, N.; Nash, K.R. Reelin Signaling in Neurodevelopmental Disorders and Neurodegenerative Diseases. Brain Sci. 2023, 13, 1479. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Liu, Z.; Guo, S.; Han, Y.; Wang, X.; Ren, W.; Chen, J.; Zhen, H.; Nie, C.; Xing, K.-K.; et al. Astrocytic Neuroligin-3 influences gene expression and social behavior, but is dispensable for synapse number. Mol. Psychiatry 2024, 1–13. [Google Scholar] [CrossRef]
- Bay, H.; Haghighatfard, A.; Karimipour, M.; Seyedena, S.Y.; Hashemi, M. Expression alteration of Neuroligin family gene in attention deficit and hyperactivity disorder and autism spectrum disorder. Res. Dev. Disabil. 2023, 139, 104558. [Google Scholar] [CrossRef]
- Xu, J.; Du, Y.-l.; Xu, J.-w.; Hu, X.-g.; Gu, L.-f.; Li, X.-m.; Hu, P.-h.; Liao, T.-l.; Xia, Q.-q.; Sun, Q.; et al. Neuroligin 3 Regulates Dendritic Outgrowth by Modulating Akt/mTOR Signaling. Front. Cell. Neurosci. 2019, 13, 518. [Google Scholar] [CrossRef]
- Onay, H.; Kacamak, D.; Kavasoglu, A.N.; Akgun, B.; Yalcinli, M.; Kose, S.; Ozbaran, B. Mutation analysis of the NRXN1 gene in autism spectrum disorders. Balk. J. Med. Genet. BJMG 2016, 19, 17–22. [Google Scholar] [CrossRef]
- Südhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef]
- Qin, Y.; Du, Y.; Chen, L.; Liu, Y.; Xu, W.; Liu, Y.; Li, Y.; Leng, J.; Wang, Y.; Zhang, X.-Y.; et al. A recurrent SHANK1 mutation implicated in autism spectrum disorder causes autistic-like core behaviors in mice via downregulation of mGluR1-IP3R1-calcium signaling. Mol. Psychiatry 2022, 27, 2985–2998. [Google Scholar] [CrossRef]
- Sala, C.; Vicidomini, C.; Bigi, I.; Mossa, A.; Verpelli, C. Shank synaptic scaffold proteins: Keys to understanding the pathogenesis of autism and other synaptic disorders. J. Neurochem. 2015, 135, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Keményová, P.; Filčíková, D.; Szapuová, Ž.; Kováč, A.; Babinská, K.; Ostatníková, D. Plasma levels of glial cell marker S100B in children with autism. Physiol. Res. 2019, 68, S315–S323. [Google Scholar] [CrossRef] [PubMed]
- Abboud, T.; Rohde, V.; Mielke, D. Mini review: Current status and perspective of S100B protein as a biomarker in daily clinical practice for diagnosis and prognosticating of clinical outcome in patients with neurological diseases with focus on acute brain injury. BMC Neurosci. 2023, 24, 38. [Google Scholar] [CrossRef] [PubMed]
- Melin, M.; Carlsson, B.; Anckarsater, H.; Rastam, M.; Betancur, C.; Isaksson, A.; Gillberg, C.; Dahl, N. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology 2006, 54, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Yoo, H.J.; Cho, I.H.; Park, M.; Kim, S.A. Association with tryptophan hydroxylase 2 gene polymorphisms and autism spectrum disorders in Korean families. Neurosci. Res. 2012, 73, 333–336. [Google Scholar] [CrossRef]
- Veenstra-VanderWeele, J.; Muller, C.L.; Iwamoto, H.; Sauer, J.E.; Owens, W.A.; Shah, C.R.; Cohen, J.; Mannangatti, P.; Jessen, T.; Thompson, B.J.; et al. Autism gene variant causes hyperserotonemia, serotonin receptor hypersensitivity, social impairment and repetitive behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 5469–5474. [Google Scholar] [CrossRef]
- Dark, C.; Homman-Ludiye, J.; Bryson-Richardson, R.J. The role of ADHD associated genes in neurodevelopment. Dev. Biol. 2018, 438, 69–83. [Google Scholar] [CrossRef]
- Gizer, I.R.; Ficks, C.; Waldman, I.D. Candidate gene studies of ADHD: A meta-analytic review. Hum. Genet. 2009, 126, 51–90. [Google Scholar] [CrossRef]
- Harper, C.B.; Mancini, G.M.S.; van Slegtenhorst, M.; Cousin, M.A. Altered synaptobrevin-II trafficking in neurons expressing a synaptophysin mutation associated with a severe neurodevelopmental disorder. Neurobiol. Dis. 2017, 108, 298–306. [Google Scholar] [CrossRef]
- Michetti, C.; Falace, A.; Benfenati, F.; Fassio, A. Synaptic genes and neurodevelopmental disorders: From molecular mechanisms to developmental strategies of behavioral testing. Neurobiol. Dis. 2022, 173, 105856. [Google Scholar] [CrossRef]
- Shen, Y.-C.; Tsai, H.-M.; Ruan, J.-W.; Liao, Y.-C.; Chen, S.-F.; Chen, C.-H. Genetic and functional analyses of the gene encoding synaptophysin in schizophrenia. Schizophr. Res. 2012, 137, 14–19. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hosseini, K.; Cediel-Ulloa, A.; AL-Sabri, M.H.; Forsby, A.; Fredriksson, R. Assessing the Neurodevelopmental Impact of Fluoxetine, Citalopram, and Paroxetine on Neural Stem Cell-Derived Neurons. Pharmaceuticals 2024, 17, 1392. https://doi.org/10.3390/ph17101392
Hosseini K, Cediel-Ulloa A, AL-Sabri MH, Forsby A, Fredriksson R. Assessing the Neurodevelopmental Impact of Fluoxetine, Citalopram, and Paroxetine on Neural Stem Cell-Derived Neurons. Pharmaceuticals. 2024; 17(10):1392. https://doi.org/10.3390/ph17101392
Chicago/Turabian StyleHosseini, Kimia, Andrea Cediel-Ulloa, Mohamed H. AL-Sabri, Anna Forsby, and Robert Fredriksson. 2024. "Assessing the Neurodevelopmental Impact of Fluoxetine, Citalopram, and Paroxetine on Neural Stem Cell-Derived Neurons" Pharmaceuticals 17, no. 10: 1392. https://doi.org/10.3390/ph17101392
APA StyleHosseini, K., Cediel-Ulloa, A., AL-Sabri, M. H., Forsby, A., & Fredriksson, R. (2024). Assessing the Neurodevelopmental Impact of Fluoxetine, Citalopram, and Paroxetine on Neural Stem Cell-Derived Neurons. Pharmaceuticals, 17(10), 1392. https://doi.org/10.3390/ph17101392






