Abstract
Worldwide urbanization and subsequent migration have accelerated the emergence and spread of diverse novel human diseases. Among them, diseases caused by viruses could result in epidemics, typified by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which hit the globe towards the end of December 2019. The global battle against SARS-CoV-2 has reignited interest in finding alternative treatments for viral infections. The marine world offers a large repository of diverse and unique bioactive compounds. Over the years, many antiviral compounds from marine organisms have been isolated and tested in vitro and in vivo. However, given the increasing need for alternative treatment, in silico analysis appears to provide a time- and cost-effective approach to identifying the potential antiviral compounds from the vast pool of natural metabolites isolated from marine organisms. In this perspective review, we discuss marine-derived bioactive metabolites as potential therapeutics for all known disease-causing viruses including the SARS-CoV-2. We demonstrate the efficacy of marine-derived bioactive metabolites in the context of various antiviral activities and their in silico, in vitro, and in vivo capacities.
Keywords:
SARS-CoV-2; therapeutics; pandemic; marine metabolites; marine organisms; viruses; viral infection 1. Introduction
Anthropogenic activities, including intensive agriculture and globalization, among others, have eroded biodiversity worldwide [1,2,3], accelerating the emergence and spread of numerous new human diseases [4,5]. Emerging infectious diseases, especially those caused by viruses, pose a threat to global health capable of causing widespread mortality in pandemics or localized outbreaks with high fatality rates [6,7]. Over the past five decades, there has been a continuous discovery of new emerging viruses of zoonotic origin. The first such case was the Ebola virus, which initially occurred in 1976 in Zaire and Sudan, and since then, there has been an ongoing report of Ebola outbreaks [8]. In 1981, the first case of what would become known as AIDS was recorded as Pneumocystis carinii pneumonia, primarily among homosexual males in the United States, heralding the onset of the AIDS epidemic; the causative agent, a retrovirus, was subsequently identified in 1983 [9]. The turn of the millennium witnessed the emergence of novel coronaviruses, with the Severe Acute Respiratory Syndrome (SARS) outbreak originating in Hong Kong in 2003. This was followed by the Middle East Respiratory Syndrome (MERS) in Saudi Arabia in 2012 [10]. In December 2019, the city of Wuhan in China’s Hubei Province became the epicenter for an outbreak of a pneumonia-like illness of unknown etiology, which was later identified as COVID-19, caused by a novel coronavirus designated SARS-CoV-2 [11].
Beyond their profound mortality and socioeconomic impacts, infectious diseases caused by emerging viruses represent escalating threats to global health. Accordingly, developing robust antiviral treatments and pre-emptive measures against potential pandemics has become a global public health priority [5,12]. Currently, vaccines and antiviral drugs are the primary interventions employed for the prevention and treatment of human viral infections. Vaccines are regarded as the most effective method for preventing viral infections [13]. Despite intensive research on a variety of viral pathogens, including the recent coronavirus strains, the repertoire of available antiviral treatments remains limited, compounded by the concerning decline in efficacy over time against certain viruses [14,15,16].
Since the approval of idoxuridine, the first antiviral drug, numerous others have been developed. However, the high mutation rate and genetic diversity of viruses often leads to treatment failure and rapid development of drug resistance [17]. Another significant concern is the cytotoxicity associated with these antiviral agents, which can limit their therapeutic utility [17]. On the other hand, vaccination is heralded as the most potent preventive strategy against viral infections, yet its effectiveness is not uniform across all populations, particularly among older adults, necessitating supplemental antiviral therapies [18]. This was illustrated by a community-wide serosurvey assessing the effectiveness of the BNT162b2 and CoronaVac vaccines against the SARS-CoV-2 Omicron variant over 100 days. The results of the study showed that at 100 days, vaccine effectiveness decreased to 26 and 35% for 3 and 4 doses of BNT162b2 and 6 and 11% for 3 and 4 doses of CoronaVac vaccines [15]. Given these challenges, it is crucial to explore and develop new therapeutic agents, particularly from natural sources such as marine-derived metabolites.
Over the years, natural products have provided resources/ingredients for developing drugs to treat and manage many human diseases. Covering over 70% of the earth’s surface, oceans are home to a wide array of organisms, thus providing a unique source of various metabolites with significant health benefits [19]. Research on marine microorganisms has steadily expanded since it started in the 1960s as a new area of study for natural products [20]. The unique secondary metabolites found in marine organisms with a variety of biological functions have evolved because of ecological stresses such as competition for space, surface fouling, predation, and successful reproduction [21]. For a long time, it was mainly disregarded how crucial these secondary metabolites are in the regulation of pathogenic and parasitic organisms. However, with improved extraction and characterization technologies, secondary metabolites can be sourced from marine organisms (both micro and macro).
Researchers have successfully isolated over 12,000 novel metabolites and continue to discover hundreds of new compounds annually from marine organisms, yielding new and potent natural bioactive ingredients [21,22]. On one hand, the terrestrial environment contains various plant-derived natural ingredients and molecules used as medicines; however, on the other hand, the marine ecosystem offers more untapped species of organisms from which potential bioactive natural compounds may be isolated [23]. These metabolites exhibit many biological activities of great pharmacological potentials, such as antimicrobial, antifungal, antifertility, antibiotic, and anticarcinogenic, and may serve as prophylaxis and treatment of human diseases.
Marine microorganisms, a subclass of marine organisms are recognized for their ability to produce antiviral agents, and they may offer limitless biological resources for obtaining therapeutic medications intended to treat and manage viral diseases in humans, as well as an endless supply of innovative compounds with promising medicinal properties and significant market potential [20]. Marine fungi alone yield between 150 and 200 novel molecules per year, including sesquiterpenoids, polyketides, and alkaloids [24]. Donia and Hamann reported the inhibitory potential of these marine-derived bioactive compounds against herpes simplex virus 1, poliovirus, yellow fever, dengue virus, rhinovirus, vesicular stomatitis virus, influenza viruses, and HIV-1 [21] with Griffithsin (a lectin extracted from red algae), suggested for anti-HIV activity, in clinical trials [19]. Furthermore, the structural engineering of these compounds by adding different functional groups (e.g., amines and ketones) and introducing double bonds, as well as the use of polymeric nanosystems, can bring about improved antiviral properties [25,26]. These agents, targeting various stages of the viral replication cycle, offer promising avenues for both therapeutic intervention and prophylactic measures against viral diseases, including COVID-19.
Thus, this perspective review discusses prospective marine-derived bioactive metabolites that may serve as therapeutic interventions in managing/treating various viral diseases. In addition, promising marine metabolites with potential inhibitory effects specifically against the replication mechanism of SARS-CoV-2 main proteases according to literature studies of molecular docking and simulation (in silico studies), coupled with in vitro and in vivo studies, are further discussed.
2. Metabolites from Marine Organisms
The marine ecosystem remains a repository of taxonomically diverse groups of unexplored micro- and macro-organisms, compared to its terrestrial counterpart. The complex marine habitats are exposed to extreme conditions and ecological pressures, including competition for space, pollution, and predation, which have powered the evolution of an assortment of potential secondary metabolites with different biological activities [21]. Naturally occurring secondary metabolites remain the main source of active ingredients for new therapeutic agents. In this regard, secondary metabolites from marine organisms have attracted immense attention over the years as potential raw materials for new broad-spectrum therapeutics due to the large ecological diversity of biological species contained in the marine environment [27].
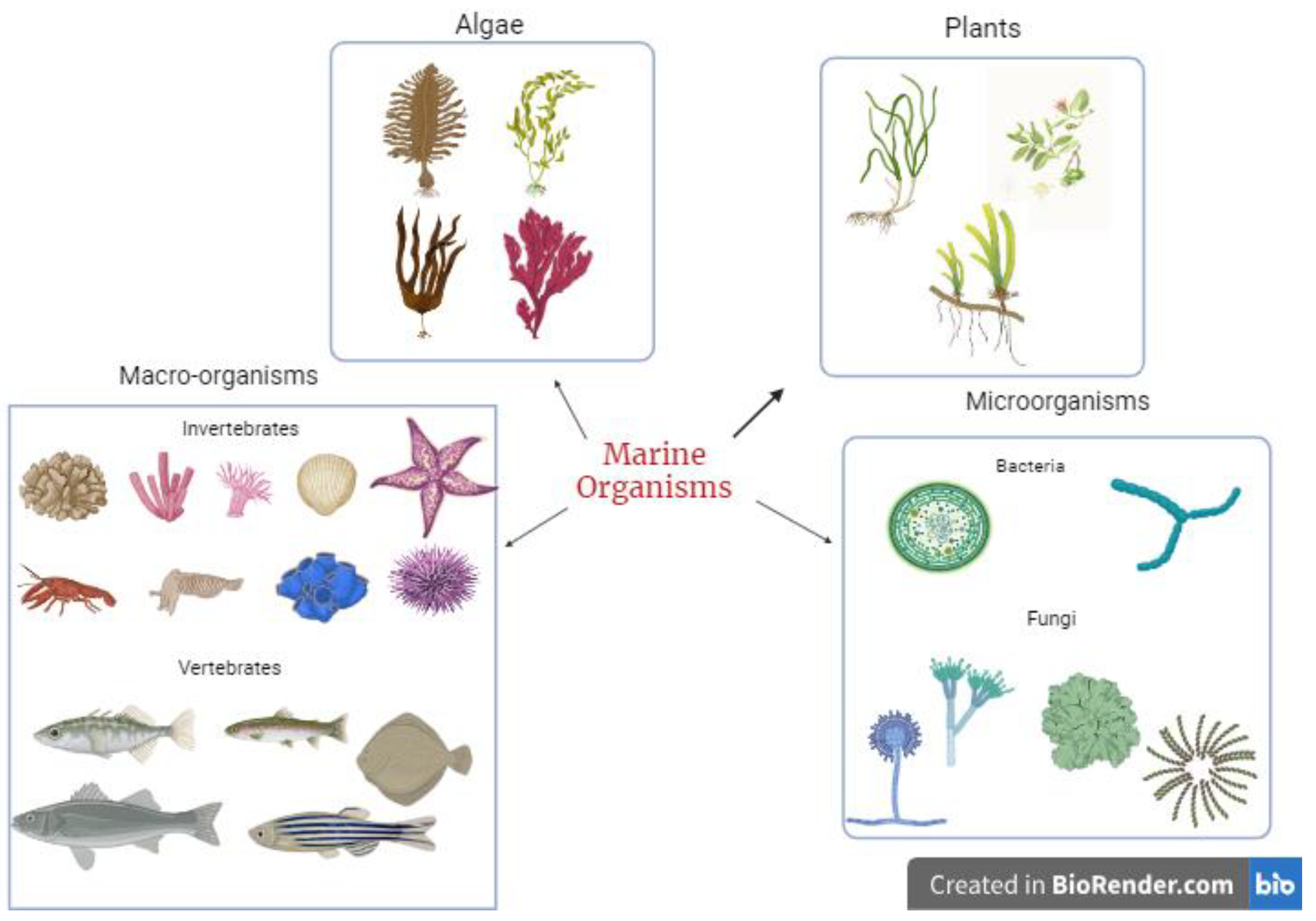
These secondary metabolites are synthesized by marine organisms as a survival and defence mechanism against other organisms, thus making them potential sources of bioactive compounds. With the emergence of various infectious diseases coupled with the menace of antibiotic resistance, marine organisms serve as a rich source of novel bioactive compounds for managing current and future viral diseases. The diversity of marine species (Figure 1) allows all kinds of potent metabolites to be isolated and tested for their potential pharmacological benefits to humans. Some bioactive compounds isolated and identified from marine organisms include terpenes, peptides and proteins, polysaccharides, lipids, alkaloids, and macrolides. The range of these compounds is related to the diverse mechanisms used by marine organisms to increase survival.
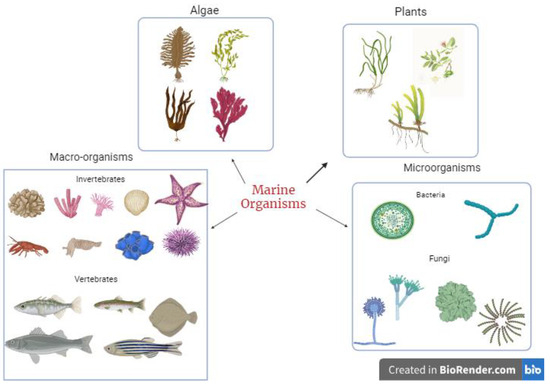
Figure 1.
Sources of bioactive metabolites from marine organisms.
4. SARS-CoV-2 Virology and Mode of Entry
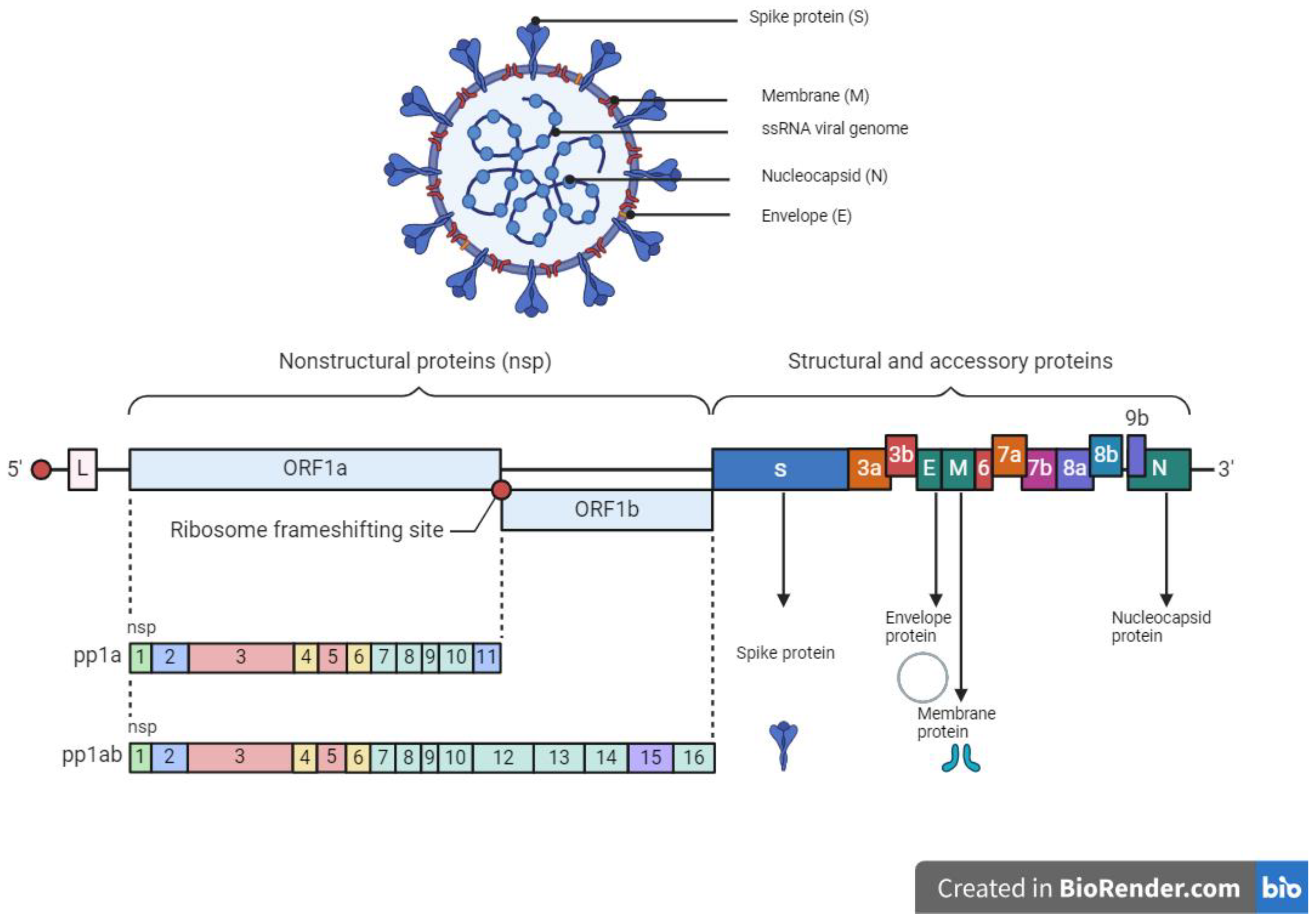
The SARS-CoV-2 which caused the severe acute respiratory coronavirus disease 2019 (COVID-19) belongs to the Coronaviridae family with a zoonotic potential, thus transmitted from humans and other mammals [134]. It shares a similar genomic sequence with the original SARS-CoV (~79.5% similarity) and BatCoV RaTG13 (~96% similarity). SARS-CoV-2 is a 50–200 nM positive-sense single-stranded enveloped RNA virus (+ssRNA) with a genome size of 28–30 kb [135,136,137]. The genome of SARS-CoV-2 consists of over 29,000 bases and codes for 29 proteins. Of the 29 proteins, the viral genome encodes 4 structural proteins and 16 non-structural replicate polyproteins which play a crucial role in the viral replication complex (Figure 2). The structural proteins include the spike (S) glycoprotein which binds to the host ACE2 receptor to initiate infection, small envelope (E) glycoprotein, and membrane (M) glycoprotein distributed along the viral envelope, and nucleocapsid (N) phosphoprotein which is an RNA-binding protein that facilitates the packaging of the genome and protects the viral genome [137,138]. Structural proteins are important for infection and replication in the host cell, thus making them ideal candidates or targets for antiviral therapies. The non-structural proteins (NsPs) are synthesized as long polypeptides which release the RNA-dependent RNA polymerase (RdRp), Nsp12, when activated by the main protease (MP), Nsp5. MP can be targeted by antiviral drugs against SARS-CoV-2, given its key role in virus replication and transcription [139,140].
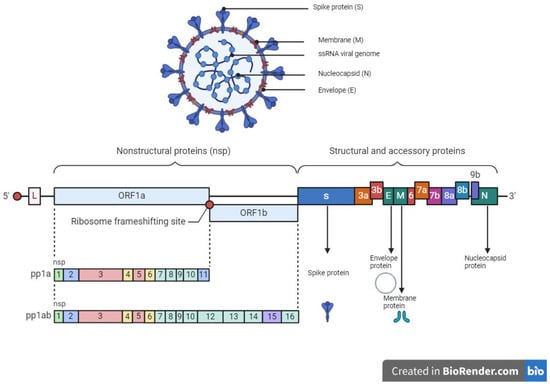
Figure 2.
Structure and genome organization of SARS-CoV-2.
Infection of host cells by SARS-CoV-2 transmission occurs via endocytosis, which involves the interaction of host cell surface receptors (fusion) with endosomal components. The spike proteins are essential for the entry of the virus into a host cell. Particularly, the S1/S2 subunit facilitates attachment to the cell and subsequent fusion. It requires initial priming by the transmembrane protease, serine 2 (TMPRSS2), cysteine protease, and cathepsin L (CatL). In the host cell, the angiotensin-converting enzyme 2 (ACE2), a type I membrane receptor protein found in the lungs and arteries [137], serves as the binding site for SARS-CoV. The SARS-CoV-2 virion attaches to the enzymatic domain of ACE2 on the surface of cells via the receptor-binding domain (RBD) of the S1 unit, and subsequently, the cell TMPRSS2 opens the S protein, allowing for the fusion of the S2 subunit and ACE2 [141]. This results in endocytosis and the translocation of both the virus and enzyme into endosomes [142]. The virus subsequently escapes when the pH of the endosome drops or is cleaved by cathepsin, thus releasing its RNA into the cell cytoplasm. The virus then replicates and spreads new copies of the virus to infect more cells [136,141].
Therapeutic Target Site to Inhibit SARS-CoV-2 Entry and Replication
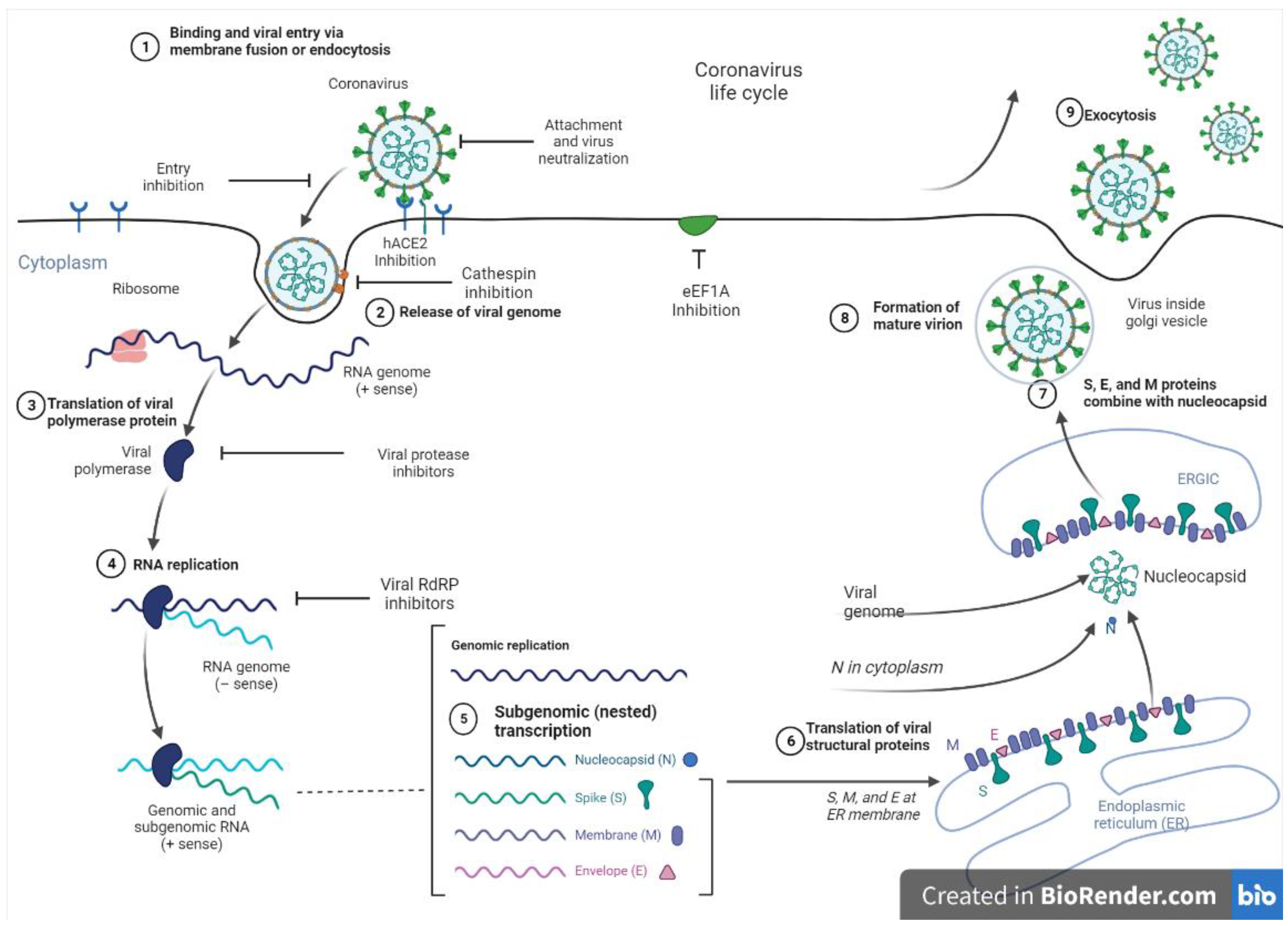
At present, vaccines, monoclonal antibodies, peptides, small molecule drugs, and interferon therapies serve as viable options to manage SARS-CoV-2. Nevertheless, targeting the replication machinery of the virus remains a promising therapeutic approach. In this regard, viral proteases are suitable targets, as these enzymes play critical roles in the replication of the virus by cleaving proproteins after translation into the host cell cytosol during viral protein maturation [143]. Figure 3 shows the entry and replication cycle of SARS-CoV-2 entry with inhibition sites of some marine-derived metabolites. SARS-CoV-2 spike (S) glycoprotein binds to the ACE2 receptor on the host cell surface, and the virus subsequently enters the cells via endocytosis to release its positive-sense ribonucleic acid (RNA) into the host cell. The viral genomic RNA is then transcribed and translated to produce non-structural proteins (nsps), including replicase polyproteins (RNA-dependent RNA polymerase and helicase), which then creates an RdRp complex. Within the RdRp complex, subgenomic transcription and RNA replication occur to synthesize negative-strand guide RNA (gRNA) and a set of subgenomic RNAs for viral replication and transcription. Subgenomic RNAs are synthesized and translated into viral structural proteins such as the spike (S), nucleocapsid (N), membrane (M), and envelope (E). After viral structural proteins are translated, S, E, and M proteins are processed in the Endoplasmic Reticulum-Golgi (ERG) intermediate compartment of the host cell. In the cytoplasm, nucleocapsids assemble and bud into the lumen of the ERG intermediate compartment. Finally, the mature virus inside the Golgi vesicle is exocytosed from the infected cell. Through ACE-2 receptors, a mature virus can infect the lung, endothelium, intestine, heart, testis, and kidney [140,144].
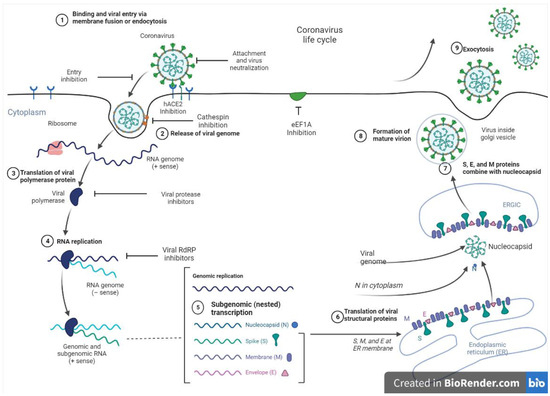
Figure 3.
SARS-CoV-2 entry and replication cycle with potential inhibition sites for marine metabolites.
Overall, the ACE2 protein, transmembrane protease serine 2 (TMPRSS2), papain-like protease (PL2pro), and main protease (Mpro)/chymotrypsin-like protease (3CLpro), which are crucial for viral replication and proliferation in the human host, are all potential targets under investigation for therapeutic interventions against SARS-CoV-2 [134,135,137,143,145]. PL2pro and 3CLpro/Mpro cleave large polyproteins of SARS-CoV-2 before being proteolytically processed to generate the individual proteins required for viral replication [141,142]. 3CLpro/Mpro plays a leading role in transcription, releasing replicative proteins, including the viral RNA polymerase and helicase proteins [146,147]. 3CLpro/Mpro is the main protease found only in the coronavirus family and is considered the most suitable target for virus inhibition as it cleaves the coronavirus polyprotein at eleven conserved sites. It is worth noting that glycan-protein interactions are important during viral binding to the host cell, considering that glycosylation of the S-protein shields the proteins from immune recognition. Hence, disrupting S-protein glycosylation significantly impairs viral entry, thus serving as another potential target for vaccine development and therapeutic interventions [137]. Furthermore, neuropilin 1 (NRP1), a host protein, aids virus entry, making it an attractive target [148]. In addition, RNA-dependent RNA polymerase (RdRp), which catalyzes the replication of the viral RNA genome, is a probable target as well [149]. Also, more focus has been placed on decreasing the levels of ACE2, a part of the renin–angiotensin system that regulates blood pressure, given that it is the main entry point of the virus in humans. However, this may not be a good approach since it can alter the central pressure control system and cause stroke or other medical conditions [148].
5. Potential In Silico and Pre-Clinical Studies of Marine-Derived Metabolites against Target Sites of SARS-CoV-2 as Therapeutics
Various in vitro and in vivo anti-SARS-CoV and anti-MERS-CoV studies have been carried out using a wide myriad of bioactive compounds during the previous outbreaks. Considering that they share some similarities with SARS-CoV-2, some of these bioactive compounds may be repurposed to screen for their potential against SARS-CoV-2. Designing an efficient broad-spectrum antiviral therapy against coronaviruses is an efficient way to counter various mutant strains of SARS-CoV-2, which hinder the effectiveness of the current vaccines [150]. Natural bioactive compounds can be used to design new antiviral drugs against viral infections, coupled with boosting the innate immune system. However, there are challenges involved considering the diversity of natural metabolites, chemical intricacies, and different extraction methodologies [135].
To save time in screening bioactive compounds for potential activity, a virtual or computational screening approach is recommended. In this regard, in silico techniques such as molecular docking, molecular dynamics simulations, and network pharmacology are useful for the preliminary identification of natural compounds that can directly inhibit target proteins [5,135,151]. Molecular docking evaluates the binding and interaction between the specified molecules (i.e., marine-derived metabolites) and the target protein(s), whereas network pharmacology employs computationally simulated drug-targeted interactions to identify potential inhibitors for a particular target and mode of action [152,153]. Additionally, the process evaluates the stability of the predicted protein–ligand complex considering factors such as the nature of the solvent [154]. By including biological circumstances, such as structural motions and the 3D structure of the targets, more reliable affinity values of the metabolites are estimated [155]. Nevertheless, most studies about network pharmacology for SARS-CoV-2 are related to existing traditional drugs with limited studies available for marine-derived drugs [156,157].
5.1. In Silico, In Vitro, and In Vivo Studies of Major Classes of Metabolites against Entry and Replication of SARS-CoV-2
Diverse and unique polysaccharides, proteins, lipids, terpenoids, flavonoids, steroids, and alkaloids with virucidal activities have been extracted from marine organisms [158]. Some of these metabolites and their derivatives are reported to be protease inhibitors that can inhibit DNA and RNA viruses, and thus may serve as potential protease inhibitors against SARS-CoV-2 [5].
5.1.1. Polysaccharides
Marine-derived polysaccharides are considered important biological macromolecules with unique and diverse structures and are considered valuable resources for drug discovery and design [159]. Moreover, marine-derived polysaccharides are cheaply available in nature, non-toxic, safe, biocompatible, and biodegradable [160].
- a.
- Sulfated Polysaccharide (SP)
Found in the cell walls of marine microbes, sulfated polysaccharides (SP) are naturally occurring water-soluble complex polymers extracted using water as a solvent [159]. Others have speculated that SP-derived therapy may be used to manage COVID-19 disease because it prevents/inhibits adherence of the S-protein to the heparin sulfate co-receptor and thus decreases viral infection by acting as a decoy in host tissues [159,160]. Various concentrations of fucoidan (RPI-27 (151) and RPI-28 (152)) extracted from Saccharina japonica showed antiviral activity against SARS-CoV-2 in Vero cells. RPI-27 (151) significantly inhibited SARS-CoV-2 infection in Vero cells (EC50 = 0.08 μM) compared to RPI-28 (152) (EC50 = 1.2 μM) [161].
Also, carrageenans (CGNs), a group of sulfated D-series polysaccharides with α-galactose residues and possessing negatively charged sulfate ester groups are extracted from marine seaweeds. Thus, carrageenans can interact with the positively charged membrane of SARS-CoV-2 and inhibit entry through the nasal cavity [162]. Morokutti-Kurz et al. [162] tested different SPs for their ability to inhibit viral entry and attachment in SARS-CoV-2 Spike pseudotyped lentivirus (SSPL). They found that ι-carrageenan (153) inhibited SSPL cell entry in a dose-dependent manner (IC50 = 5.3 μM), whereas at 10 μg/mL, ι-carrageenan (153) exerted 80% inhibitory activity against SSPL, in which 100 μg/mL of κ- and λ-carrageenan (154) (155) were needed each to achieve similar inhibitory activity. Further analysis corroborated the inhibitory effect of ι-carrageenan 153 (3.75 μM) in Vero B4 cells against wild-type SARS-CoV-2 PR-1 [162]. A carrageenan-based anti-SARS-CoV-2 nasal spray is currently the subject of clinical trials in the USA, and related studies are also being conducted in the UK [163].
Glycosaminoglycans are another class of sulfated polysaccharides with potential SARS-CoV-2 inhibitory properties. Song et al. [164] checked the inhibitory properties of sulfated glycosaminoglycans (SCSP) (156) isolated from sea cucumber Stichopus japonicus and observed that SCSP exhibited the highest inhibitory activity (IC50 of 9.10 μg/mL) compared to fucoidan from brown algae, and chondroitin sulfate C from sharks (CS). They further demonstrated that SCSP can bind specifically to the S glycoprotein to inhibit entry of SARS-CoV-2 into host cells using pseudotype virus with S glycoprotein of SARS-CoV-2. The authors postulated that the binding of SCSP was facilitated by the high structural flexibility. Flexibility is necessary for the binding of polysaccharides to the S glycoprotein [164,165]. Another sulfated glycosaminoglycan (156) from the bacteria, Pseudomonas sp. was reported to have a high binding energy with Mpro at −7.98 kcal/mol in silico [5].
In an in vitro study, Jang et al. [166] reported that λ-CGN (154) purified from marine red algae suppressed cell entry of SARS-CoV-2 glycoproteins-derived pseudoviruses in a dose-dependent manner. Furthermore, Yim et al. [167] reported virus entry of SARS-CoV-2 was inhibited by crude sulfated fucoidan extracts (85) obtained from different seaweeds including Undaria pinnatifida sporophyll, Laminaria japonica, Hizikia fusiforme, Sargassum horneri, Codium fragile, and Porphyra tenera, and Haliotis discus hannai [167].
- b.
- Non-sulfated polysaccharides
Aside from sulfated polysaccharides, marine algae also contains other glucans, such as laminarin and alginate β-glucan, which are non-sulfated. A molecular docking and dynamics analysis showed that Laminarin (157) has a high binding affinity to the SARS-CoV-2s-glycoprotein and Mpro (−7.83 and −7.81 kcal/mol, respectively). On the other hand, β-glucan (158) also exhibited a strong binding affinity towards Mpro (−7.83 kcal/mol) [160]. The brown seaweed species Laminaria hyperborea, Laminaria digitata, Macrocystis pyrifera, and Ascophyllum nodosum from the class Phaeophyceae, can be used to extract the linear polysaccharide alginates [168]. With regards to alginate, Cano-Vincent et al. [169] previously demonstrated that calcium alginate (159) biomaterial can inactivate enveloped viruses such as bacteriophage phi 6 (94.92% viral inactivation) and SARS-CoV-2 Delta variant (96.94% viral inactivation). The authors opine that the negatively charged groups of calcium alginate (159) facilitate binding to viral envelopes [5]. In addition, You et al. [170] identified a novel polysaccharide, CLSP-2 (160) from edible seaweed C. lentillifera, which significantly inhibited SARS-CoV-2 infection in HeLa cells at ≥12.5 μg/mL with an IC50 of 48.48 μg/mL.
5.1.2. Proteins
- a.
- Peptides
Bioactive peptides, arising from the hydrolysis of proteins, possess unique amino acid sequences that confer on them various biological activities. Marine organisms are a cheap source of proteins for acquiring bioactive peptides. Yao et al. [171] reported that oligopeptides (2–8 amino acids long) (161) arising from in silico hydrolysis of proteins from salmon, squid, tuna, mackerel, and pomfret exhibited high binding affinity to SARS-CoV-2 Mpro and monoamine oxidase A. Peptides that interrupt the binding of SARS-CoV-2 spike proteins to ACE are particularly enticing candidates against SARS-CoV-2 cell entry. For instance, peptides (sequences GDLGKTTTVSNWSPPKYKDTP (162) and VW (163)) obtained from Thunnus obesus and Undaria pinnatifida have been shown to stably bind to both hACE2 (−246.50 and −117.65 kcal/mol) and spike RBD-ACE2 complex (−223.60 and −123.42 kcal/mole) [23]. The binding of these peptides may disrupt the interaction of SARS-CoV-s2 spike proteins with ACE2 and thus prevent cell entry of the virus. Also, peptides (Asp-Trp (164) and Val-Tyr (165)) isolated from tilapia viscera hydrolysate exhibited great binding affinity to four SARS-CoV-2 components including Mpro, S-glycoprotein, RBD-ACE2, and deubiquitinase inhibitors [172].
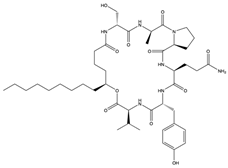
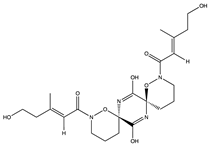
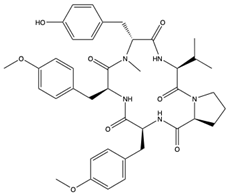
Furthermore, plitidepsin (dehydrodidemnin B) (166), a cyclic depsipeptide originally isolated from the tunicate Aplidium albicans, is reported to exhibit up to 90% inhibitory activity (at 0.88 nM) against SARS-CoV-2, which is 27.5 times higher compared to remdesivir [173]. Further in vivo studies with mouse models infected with SARS-CoV-2 show that plitidepsin (166) reduced the viral load significantly. Also, Ding et al. [174] reported that cyclic dipeptides (167) isolated from Aspergillus versicolor DY180635, an endophyte of the sea crab (Chiromantes haematocheir), exhibited good binding affinity to SARS-CoV-2 Mpro.
Similarly, didemnins, another category of cyclic depsipeptides, can bind to Mpro [175]. For instance, didemnins A, B, and C (168–170), isolated from Caribbean tunicate (Trididemnum solidum) showed high binding affinity (-11.82 kcal/mol, -10.27 kcal/mol, and -9.26 kcal/mol, respectively) to SARS-CoV-2 Mpro [175]. The authors reported that didemnin B (169) interacted with the active site of Mpro through hydrogen bonding with the Glu166, an important residue necessary for the hydrolytic activity of the enzyme. The cyclotheonamide peptides, pseudotheonamide C (171), and D (172) isolated from the marine sponge, Theonella swinhoei, likewise were able to bind to the serine protease (TMPRSS2) with binding energies of −11.6 kcal/mol and −10.7 kcal/mol, respectively [134]. Additionally, a modified depsipeptide gallinamide A (also known as symplostatin 4) (173) has also been shown to have SARS-CoV-2 inhibitory properties [176]. Isolated from the marine cyanobacteria of the Schizothrix genus, gallinamide A (173) inhibited cathepsin L, a lysosomal cysteine protease with an EC50 of 28 nM, and decreased viral load in VeroE6 cells, with an IC90 of 88 nM [176].
- b.
- Lectins
Lectins from marine organisms have been garnering interest lately, especially those from algae [158]. Lectins are carbohydrate-binding non-immunoglobulin-type proteins that recognize specific sugar groups on other molecules [158,177]. Compared to lectins from other sources, marine lectins recognize and bind to a wide variety of sugar moieties including sugar monomers and oligosaccharides [149,158]. Considering that SARS-CoV-2 uses spike glycoproteins to bind and facilitate on the cell surface glycans of potential hosts to initiate entry into cells, this makes them a perfect target for lectins.
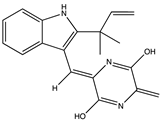
Griffithsin (81) is a lectin found in the red-algae Griffithsia sp., which has a strong specificity for mannose residues of viral glycol proteins [149,158,178]. They have the potential to interrupt the self-assembly of viruses during replication. It has been reported that treatment of SARS-CoV-2-infected rats with 10 mg/kg (b.w.)/day of Griffithsin (81) resulted in a 100% survival rate compared to the nontreated group [149,158,178]. Griffithsin (81) has been shown to inhibit the s-protein-mediated adhesion of the RBD to hACE2 with an IC50 of 0.3 μM [179]. Consequently, Griffithsin (81) significantly inhibited SARS-CoV-2 pseudovirus infection in a dose-dependent manner in vitro, with an IC50 of 293 nmol/L. Treatment of cells with Griffithsin (81) before or at the early stages of infection (0–0.5 h) resulted in up to 80% inhibition of SARS-CoV-2 compared to 32% inhibition when administered 8 h after infection [179]. This suggests that Griffithsin (81) is effective against the virus at the initial stages of infection.
- c.
- Protein-bound pigments
Some studies have also highlighted protein-bound pigments as potential inhibitors of SARS-CoV-2 infection. Phycobilins are light-capturing tetrapyrrole chromophores found in certain cyanobacteria, rhodophytes, chloroplasts of red algae, glaucophytes, and some cryptomonads. In recent times, these molecules have been widely studied for their antioxidant and antiviral activities [149,150]. In silico studies have shown that phycocyanobilins (PCB) (174), a group of blue phycobilins, have a high binding energy of −8.6 and −9.3 kcal/mol for PCB-Mpro and PCB-RdRp, respectively [180]. Pendyala et al. [150] further showed that PCB (174) can bind to Mpro and PLpro via polar interactions with specific binding pockets of amino acids such as G143 (38.5), N119, S46, and Y54 for Mpro, and D164(C), R166(C), D164(A), and G271(A) for PLpro. Petit et al. [181] also reported that PCB (174) obtained from Arthrospira sp. also exhibited strong binding affinity to SARS-CoV-2 S-glycoprotein using molecular docking studies. They also reported that both van der Waals attractions and hydrogen bonding contributed to the binding of PCB to spike RBD in silico. The molecular docking studies further revealed that PCB interacted with several amino acid residues of the spike RBD including TYR453, GLN493, TYR495, PHE497, ASN501, TYR505, SER494, GLN498, and GLY496 via different bonds [181].
As with PCB (174), in silico analysis predicted that other phycobilins such as phycourobilin (PUB) (175), phycoerythrobilin (PEB) (176), and phycoviolobilin (PVB) (177) can bind to SARS-CoV-2 Mpro and PLpro with similar affinity (between −8.2 and −10.0 kcal/mol) [181,182]. Another pigment that can bind to components of SARS-CoV-2 is C-phycocyanin (178), a phycobiliprotein from the blue-green algae Arthrospira platensis. Raj et al. [183] reported that C-phycocyanin (178) competes with ATP for binding to the active site of nsp-12.
5.1.3. Lipids
Lipids are involved extensively in the life cycle of SARS-CoV-2. They form the basis of host cell and viral membranes and act as the initial point of interaction between the virus and its potential host [158].
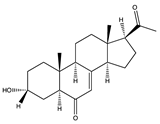
The receptor binding domain (RBD) of SARS-CoV-2 has been revealed to have three fatty acid binding pockets (FABP), which are lined by hydrophobic amino acids forming a bent tube that serves as an anchor for free fatty acids (FFA) [184]. Linoleic acid (LA) (179), an omega 6 (ω-6) PUFA, has been reported to fit into the FABP and occupy all pockets [184]. The binding of LA to the S protein induces the protein to adopt a stable closed S conformation, resulting in a reduced interaction with ACE2 [184]. Similarly, long-chain omega 3 (ω-3) PUFAs, including docosahexaenoic acid (DHA) (180) and eicosapentaenoic acid (EPA) (181), bind to the FABP and induce the closed conformation of the spike protein, to an even greater extent than LA (179) [155]. ω-3 PUFAs therefore have the potential to interrupt the interaction of ACE2 and RBD, thus reducing viral entry. Additionally, increased intake of ω-3 PUFAs decreases inflammation and coagulation caused by COVID-19 [155].
Another group of natural metabolites with a prominent role in cell–cell interactions are cerebrosides. These metabolites also carry out cell regulation and signal transduction. The molecular docking and dynamics analysis by Zahran et al. [185] revealed that cerebrosides such as A1 (182) and C1 (183) (from the Korean sponge Haliclona renier), LAMA-1 and penicilloside B (184) (from the Egyptian Penicillium chrysogenum), and asperiamide B (185) (from the Chinese-Sea-water-derived fungus Aspergillus niger) exhibit binding affinity to hACE2 (−7.1 to −7.6, kcal/mol). Moreover, Tassakka et al. [186] found that FAs/lipids were the prominent metabolites both in ethanolic and ethyl acetate extracts of Halymenia durvillei, which inhibited the activity of Mpro.
5.2. In Silico Studies of Other Secondary Metabolites (Phytochemicals) with Potential Antiviral and Therapeutic Properties against SARS-CoV-2
5.2.1. Polyphenols
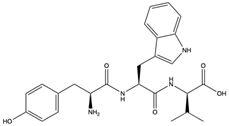
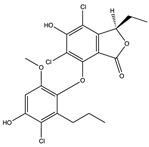
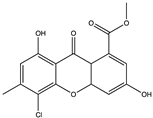
SARS-CoV-1 and 2 infections generate reactive oxygen species (ROS) which are known to cause oxidative damage, inflammation, lung infection, and epithelial tissue degeneration. 3CLpro/Mpro activates the NF-kB-dependent reporter gene which causes ROS generation in the HL-CZ cells, leading to a disruption of the oxidation–reduction processes of the cell [178]. Marine organisms are a rich source of antioxidants and several other secondary metabolites classified as broad-spectrum compounds, which can be used as therapies to manage SARS-CoV-2 infection together with antivirals. Polyphenols such as phloroglucinol oligomers and phlorotannins are a type of tannin found in brown algae and have shown promising antiviral action [181]. In silico analysis showed that the phlorotannin dieckol (86) can bind to the Spike RBD of SARS-CoV-2 high affinity (−8.1 kcal/mol) [181]. Aatif et al. [187] also reported that dieckol (86) from Ecklonia cava exhibited similar binding affinity (−8.326 kcal/mol) towards the RBD of the spike protein. Eckol (186) and trifucol (188) are other phlorotannins obtained from the brown alga Ecklonia cava and Himanthalia elongate. Eckol (186) has a high binding affinity, in silico, to the Mpro (−8.19 kcal/mol), while trifucol (187) binds to both the S-glycoprotein (−7.5 kcal/mol) and the Mpro (−6.3 kcal/mol) [160].
Out of 770 compounds, Gentile et al. [134] reported that dieckol (86), 8,8-bieckol (188), 6,6-bieckol (189), and other phlorotannins were among 17 compounds that were predicted to interact with SARS-CoV-2 Mpro. Heptafuhalol A (190), a phlorotannin fuhalol from Sargassum spinuligerum also exhibited strong binding affinity towards Mpro (−14.60 kcal/mol) by interacting with some amino acid residues within the protease receptor including Thr24, Ser46, Asn142, Glu166, and Pro168. Furthermore, other fuhalol and phlorthol phlorotannin including phlorethopentafuhalol A (191) and B (192), pseudopentafuhalol C (193), hydroxypentafuhalol A (194), pentaphlorethol B (195), aeruginosin 98B (196), resinoside B (197), pentaphlorethol A (198), and tunichrome An2 (199) also have promising SARS-CoV-2 protease inhibitory potentials. Also, the flavonoids -apigenin-7-O-neohesperidoside (rhoifolin) (200), luteolin-7-rutinoside (201), and resinoside B (197) (from the brown alga Sargassum spinuligerum) bind to SARS-CoV-2 Mpro with great affinity (−12.4 kcal/mol) [134]. Others have reported that apigenin (202) can interact with Mpro via H-bonds between the aromatic region and residues Leu141, Glu166, and Thr190, as well as π-stacking interaction with Gln189 [134]. Using molecular docking, Vijayaraj et al. [5] also showed that esculetin ethyl ester (203), a derivative of phenylpropanoid coumarin, from the marine sponge Axinella cf. corrugata can bind to ARS-CoV-2 Mpro (−8.42 kcal/mol). Esculetin ethyl ester (203) has specifically been shown to effectively inhibit SARS-CoV recombinant 3CLpro/Mpro with an ID50 value of 46 µmol L−1 [146].
5.2.2. Alkaloids
Since the pentacyclic congener ptilomycalin A was discovered to have antiviral properties, polycyclic guanidine alkaloids (PGAs) have generated much interest. PGAs are found in Poecilosclerida sponges such as Batzella, Crambe, and Ptilocaulis, and some starfishes such as Fromia monilis and Celerina heffernani. In silico analysis of fifteen structurally divergent PGAs against five different proteins of SARS-CoV-2 by El-Demerdash et al. [27] revealed a superior binding affinity of the pentacyclic guanidinic scaffolds crambescidin- 786 (204) and 826 (205) to different proteases. Crambescidin 786 (204) has exceptionally good binding affinities towards Mpro (–8.05 kcal/mol), nucleocapsid phosphoprotein (−6.49 kcal/mol), and nsp10 (−9.06 kcal/mol). Crambescidin 826 (205) showed similar binding affinity against Mpro (−7.99 kcal/mol), in addition to S proteins (−6.95 kcal/mol), and nucleocapsid phosphoprotein (−8.01 kcal/mol).
Zahran et al. [185] carried out molecular docking studies of 15 metabolites selected based on their physicochemical properties to investigate their potential effect against the SARS-CoV-2 targets Mpro, methyltransferase (nsp16), RNA-dependent RNA polymerase, RdRp (nsp12) spike protein, and human ACE2 (hACE2). Among them, the glycoside, tirandamycins analogue isotirandamycin B (206) and derivatives tirandamycin A (207) and B (208) extracted from Streptomyces sp. exhibited significant binding to SARS-CoV-2 methyltransferase nsp16/10 (−8.4, −8.5 and -8.3 kcal/mol, respectively), Mpro (−7.8, −7.9 and −7.8 kcal/mol, respectively) and RdRp (−7.6, −8.1 and −7.8 kcal/mol, respectively), demonstrating that they are potential SARS-CoV-2 inhibitors. In addition, alteramide A (209), a tetracyclic alkaloid extracted from Pseudoalteromonas sp., showed a strong binding to RdRp (−9 kcal/mol), nsp16/10 (−8.2kcal/mol), Mpro (−7.1 kcal/mol), and s-protein (−7.4 kcal/mol).
The in silico study by Khan et al. [188] of five marine compounds showed the alkaloids isofistularin-3 (C31H30Br6N4O1) (210) and chimyl alcohol (1-O-hexadecylglycerol) (C19H40O) (211), isolated from Desmapsamma anchorata, exhibit high binding affinity towards Mpro. Specifically, isofistularin-3 (210) formed hydrogen and hydrophobic interactions with Thr24, Leu27, His41, Phe140, Cys145, His163, Met165, Pro168, and His172, present in the active site and its surroundings. Aspergicin (212), an antibacterial alkaloid obtained by mixed fermentation of two marine-derived mangrove epiphytic Aspergillus fungi, likewise showed a strong binding affinity towards hACE2 (−17.66 kcal/mol) [189]. Also, callophysin A (213), an indole alkaloid from red alga Callophycus oppositifolius, showed significant binding properties towards Mpro (−8.776 kcal/mol) [190].
Caulerpin (214) is another low toxic bis-indole alkaloid found in distinct species of marine algae, especially the Caulpera genus. It has been isolated from the green macroalgae (Caulerpa racemose), the red algae (Chondria armata), and the brown algae (Sargassum platycarpum). Caulerpin (214) and some of its derivatives are known to possess a lot of biological properties. Ahmed et al. [147] carried out a molecular docking analysis of caulerpin (214) and its analogs against the SARS-CoV-2 Mpro and spike protein. They showed that the derivatives had a higher binding affinity towards Mpro and s-protein than chemical drugs like lopinavir, simeprevir, hydroxychloroquine, chloroquine, and amprenavir.
Abdelrheem et al. [191] also showed that caulerpin (214) can bind the SARS-CoV-2 Mpro high affinity (–9.28 kcal/mol) by forming three H-bond interactions with LYS 137, GLU 288, and LYS 5 of 6LU7 amino acid residues. Gaudêncio and Pereira, [192] also reported that benzo[f]pyrano [4,3-b]chromene, notoamide I (a prenylated indole alkaloid from Aspergillus sp.) (215), emindole SB beta-mannoside (an indole diterpene from Dichotomomyces cejpii) (216), and two bromoindole derivatives, bromodeoxytopsentin (217) and dibromodeoxytopsentin (218) (which are bisindole alkaloids), can bind SARS-CoV-2 Mpro (binding energies; −8.4, −8.4, −8.2, −7.6 and 7.6 kcal/mol, respectively). Also, 14-debromoaraplysillin I (219) (from the marine sponge Psammaplysilla purpurea), a phenethylamine monoamine alkaloid, had a binding affinity of −111.52 against RdRp [193].
5.2.3. Terpenes
Ilimaquinone (220), a prenylquinone terpene and a member of the monohydroxy-1,4-benzoquinones isolated from the marine sponge Hippospongia metachromia, exhibited promising affinity towards active binding pockets of major SARS-CoV-2 proteins including 3CLpro/Mpro (−7.1 kcal/mol), 6M0J (−6.9 kcal/mol), PLpro (- 8.1 kcal/mol), Nsp10 (−7.6 kcal/mol), Nsp14 (−8.1 kcal/mol), and Nsp13 (−8.2 kcal/mol) [154]. Sepay et al. [194] screened more than fifty natural products from various sources for their binding potentials towards SARS-CoV-2 and reported that the terpenoid (T3) (221) from marine sponge Cacospongia mycofijiensis showed the best binding score at −9.1 kcal/mol. Also, the terpenoids T1 (222), T2 (223), and T4 (224), which are all geometric T3 (222), showed good binding scores as well (−8.6, −8.20, and −8.08 kcal/mol, respectively). The author postulated that the higher hydrophobicity and lower flexibility of these terpenoids augmented their affinities towards SARS-CoV-2 Mpro.
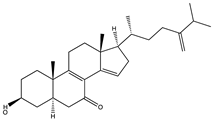
Dictyosphaeric acid A (225) (a polyketide decalactone from the green alga Dictyosphaeria versluyii) and excavatolide M (226) (a briarane-type diterpene from the coral Briareum excavatum) showed inhibitory activities against SARS-CoV-2 by disrupting the interaction of TMPRSS2-SPPIs [195]. However, the drug ability test showed that excavatolide M (226) is toxic and not suitable for use as a drug [195]. The diterpenoid, hamigeran b (227) from the marine sponge Hamigera tarangaensis, also showed high binding affinity towards Mpro (−7.98 kcal/mol) [5]. Also, the terpenoid fasciospongide A (228) (from the sponge Fasciospongia sp.) and epolactaene (229) (from the fungus Penicillium sp.) have been shown to bind to 3CLpro/Mpro with high affinity, −104.37 and −102.9 kJ/mol, respectively [193]. The terpenoid steroid moniloside A (230) (from the starfish Formia monilis) also showed a strong binding affinity (−63.12 kJ/mol) toward RdRp [193].
5.2.4. Other Metabolites
The phytocompounds of Corallina officinalis, Caulerpa racemos, Colpomenia sinuosa, Gracilaria edulis Gracilaria corticata, Sargassum wightii, and Ulva fasciata were studied for their numerous antiviral metabolites as potential inhibitors of SARS-CoV-2 ACE2-bound omicron B.1.1.529 spike protein trimer [196]. The molecular docking analysis showed that the glycoside, caffeic acid hexoside (231), and phloretin (232) from S. wightii can bind to crucial residues ASN417, SER496, TYR501, and HIS505 of spike protein, which support angiotensin-converting enzyme II receptor interaction [196]. Also, the lipid sterol cholestan-3-ol, 2-methylene-, (3beta, 5 alpha) (CMBA) (233) from C. officinalis showed a strong binding potential (−6.0 kcal) towards the omicron RBD mutated residues LEU452 and ALA484.
ACE2 produced by marine organisms can also serve as a receptor binding domain to the SARS-CoV-2 spike glycoprotein to suppress its transmission. For instance, ACE2 (234) of Delphinapterus leucas (Beluga whale) had a binding affinity of −988.5 kcal/mol towards the SARS-CoV-2 spike glycoprotein which is comparable to the hACE2 binding affinity (−946.4 kcal/mol) to the spike glycoprotein. Thus, ACE2 and ACE2-like structures from the marine biota could be used as decoys for viral binding [197]. Fayed et al. [198] screened several marine compounds for their pharmacophore potentials against SARS-CoV-2 Mpro (6lu7 and 6y2f), spike glycoprotein, and RNA Polymerase, and reported that compounds with a flavonoid core, acyl indole, and pyrrole carboxamide alkaloids performed better. The co-crystallized ligands of Mpro showed perfect overlay with the pyrroles sceptrin (235) and debromo sceptrin (237). Among all the target proteins, thalassiolin (A-C) (237–239) had the best binding and similarity values. Also, ACE2 and Mpro were shown to interact well with compounds isolated from marine sponges including microspinosamide (240) (−16.8 and 13.7 kcal/mol), neamphamide A (241) (−13.7 and 13.1 kcal/mol), mirabamide A (242) (−11.3 and 10.3 kcal/mol), and sterol clathsterol (243) (−10.5 and 10.1 kcal/mol) [199]. However, the drug-likeness test of all compounds was below Lipinski’s rule of 5, even though all the compounds had shown potential inhibition against the HIV-1 virus.
Structurally, the natural inorganic polyphosphate (polyP), considered a physiological, metabolic energy (ATP)-providing and morphogenetically active linear polymer of orthophosphate released from human blood platelets, is expressed in every cell including marine bacteria and sponges [200,201]. Polyphosphate (polyP) helps in the mediation of blood clots due to interaction with the protease coagulation factor VII; however, its production is reduced in COVID-19 patients due to a deficiency in platelet count [201]. Müller et al. [202] and Neufurth et al. [200] found that polyp (244) blocks the binding of the receptor binding domain (RBD), thus preventing the binding of the spike protein to host ACE-2 receptor at concentrations ranging from 1 to 100 µg/mL with 70% effectiveness at 10 µg/mL. Neufurth et al. [200] proposed that the 15 phosphate units of polyP (244) interacted with the basic residues, Arg, Lys, and His on the spike protein. Müller et al. [202] also reported polyp (244) increased ATP production, cell attachment, and expression of the membrane-tethered mucin MUC1 and the secreted mucin MUC5AC genes in the mucus layer, thus enhancing the barrier against inhaled pathogens such as the coronavirus SARS-CoV-2 [202,203].
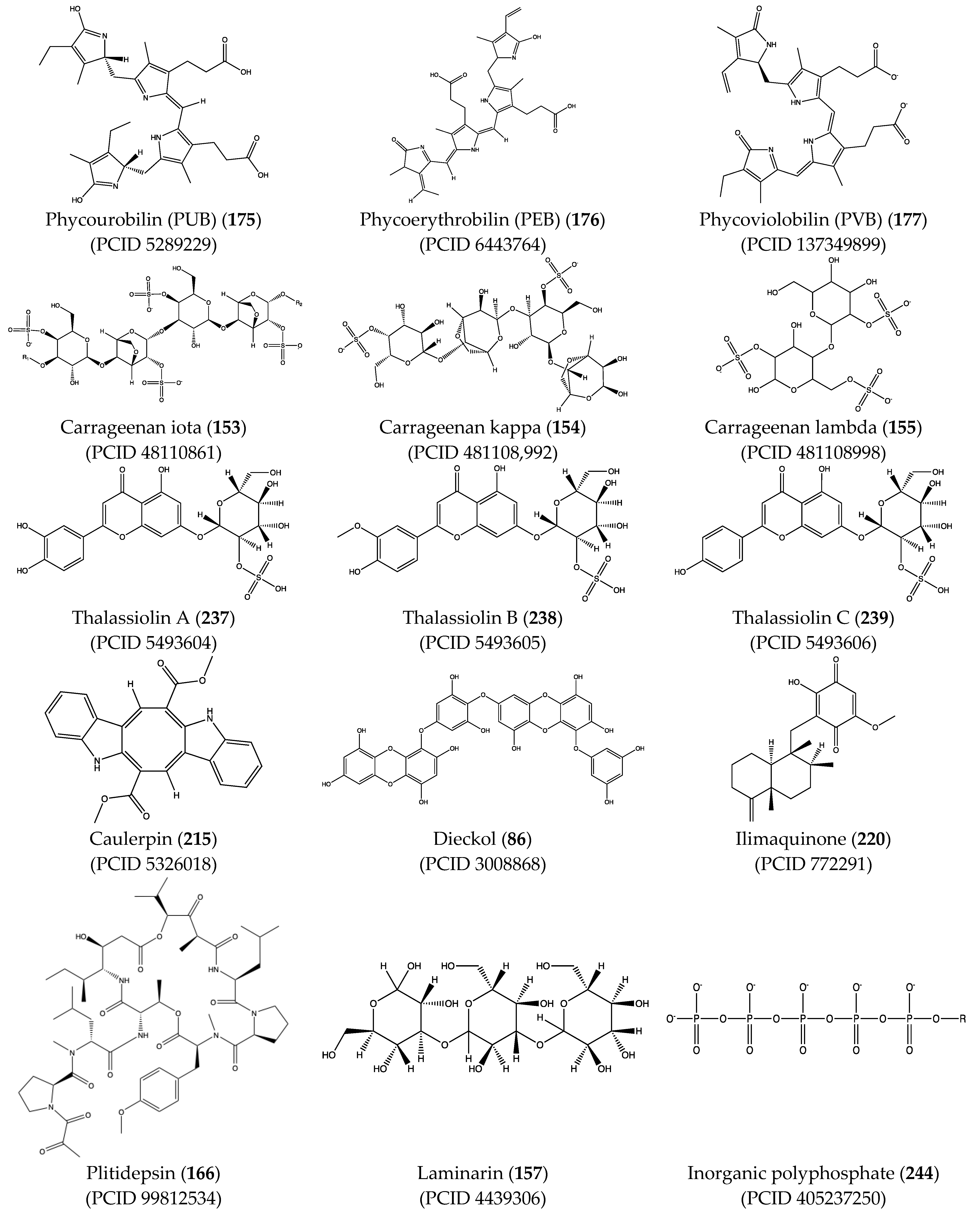
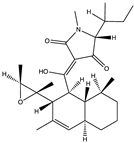
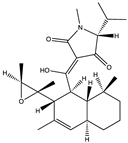
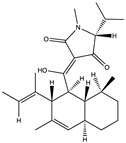
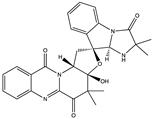
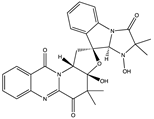
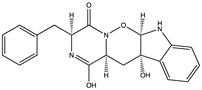
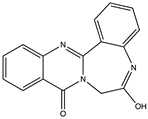
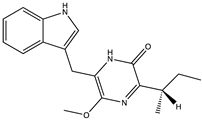
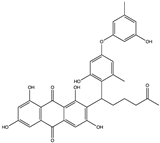
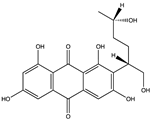
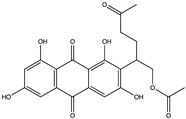
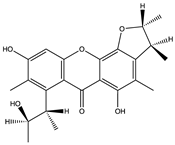
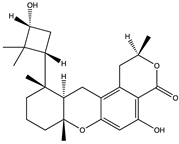
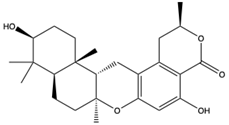
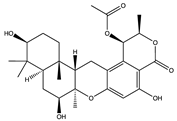
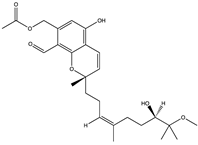
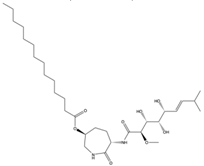
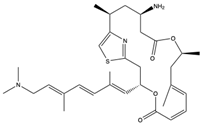
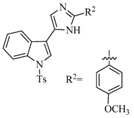
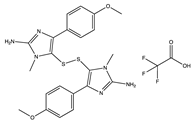
Structural characteristics of some selected compounds from marine organisms with SARS-CoV-2 inhibitory properties are shown in Figure 4 and Figure S8. Additionally, Table S1 shows some selected marine compounds with potential inhibitory properties against SARS-CoV-2 in silico.
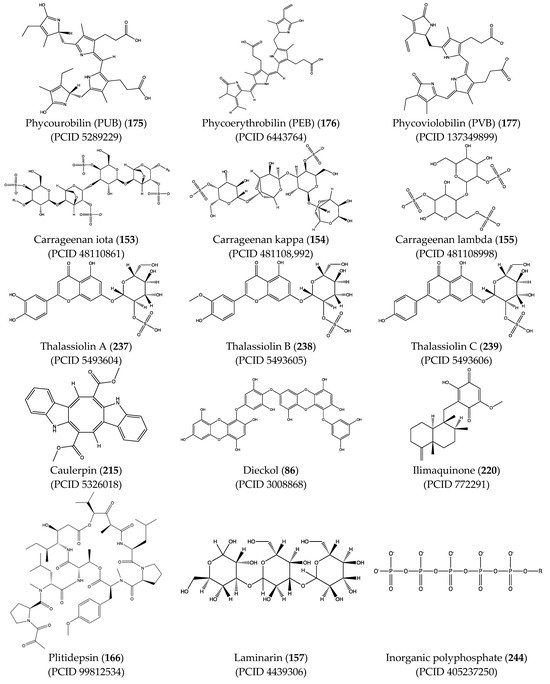
Figure 4.
Chemical structure of some selected compounds from marine organisms with SARS-CoV-2 inhibitory properties.
6. A Promising Future for Marine Bioactive Metabolites to Tackle SARS-CoV-2
Due to the vast and diverse organisms with naturally occurring metabolites found in bodies of water, researchers are increasingly turning to the oceans, rivers, and seas for new natural compounds with antiviral potentials to help create the basis for novel therapeutics. Compounds of various structural classes, including polysaccharides, terpenes, steroids, alkaloids, and peptides that inhibit both RNA and DNA viruses have been isolated from marine micro- and macro-organisms. There is much hope to discover novel resources from marine organisms, which would serve as potential drug leads that would, in addition to controlling viral replication, help manage the symptoms presented by viral diseases. Such compounds could either block the penetration of viruses into the host cells, inhibit viral fusion to host proteins, or inhibit the activity of major viral proteins such as those involved in replication. However, drug development is cost-intensive. Even with significant efforts being made in designing novel SARS-CoV-2 inhibitors from marine organisms, with some (such as plitidepsin (166)) already under clinical trials, most available studies appear exceptionally preliminary and based on computer-aided findings that employed molecular docking, molecular dynamics simulation techniques, and network pharmacology. The scarcity of comprehensive pre-clinical research involving various cell lines and animal models underscores the need for in-depth future investigations. Such studies should examine the compounds identified through in silico analysis for their potential drug-like properties, laying the groundwork for subsequent clinical trials.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph17030328/s1, Figure S1: Chemical structures of identified compounds from marine microorganisms (Bacteria and fungi) that inhibit viruses; Figure S2: Eutypellazines A-L isolated from Eutypella sp. found in deep sea sediment collected from the South Atlantic Ocean; Figure S3: Chemical structures of identified compounds from marine microorganisms (marine algae) that inhibit viruses; Figure S4: Chemical structures of identified compounds from marine plants that inhibit viruses; Figure S5: Chemical structures of identified compounds from marine macro-organisms (invertebrates) that inhibit viruses; Figure S6: Phospholipase A2 (AP-PLA-2) from Echinoderm (starfish) found in Moluccas Islands, eastern Indonesia; Figure S7: Stellattapeptins A and B isolated from Stellatta sp. found in North-western Australia; Figure S8: Chemical structures of identified compounds from marine organisms with inhibitory properties against SARS-CoV-2; Table S1. Selected marine compounds with potential inhibitory properties against SARS-CoV-2 in silico. References [5,60,93,105,134,144,146,150,175,180,181,188,195,198] are cited in the Supplementary Materials.
Author Contributions
Q.N.O., F.O.A. and O.N.K., conceptualization; Q.N.O. and F.O.A., writing—original draft; O.N.K., P.A., Á.S.-A., V.N.U. and C.O.R.O., writing—reviewing and editing of the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Authors Q.N.O. and F.O.A., gratefully acknowledge the research funding from the Ministry of Science and Higher Education of the Russian Federation (Ural Federal University Program of Development within the Priority-2030 Program).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mahajan, M.; Bhardwaj, K. Anthropogenic Ecological Changes and Spill Over of Viruses—A Review. Curr. World Environ. 2021, 16, 594–599. [Google Scholar] [CrossRef]
- Lawler, O.K.; Allan, H.L.; Baxter, P.W.J.; Castagnino, R.; Tor, M.C.; Dann, L.E.; Hungerford, J.; Karmacharya, D.; Lloyd, T.J.; López-Jara, M.J.; et al. The COVID-19 Pandemic Is Intricately Linked to Biodiversity Loss and Ecosystem Health. Lancet Planet. Health 2021, 5, e840–e850. [Google Scholar] [CrossRef] [PubMed]
- Domingo, J.L.; Rovira, J. Effects of Air Pollutants on the Transmission and Severity of Respiratory Viral Infections. Environ. Res. 2020, 187, 109650. [Google Scholar] [CrossRef] [PubMed]
- Neiderud, C.-J. How Urbanization Affects the Epidemiology of Emerging Infectious Diseases. Infect. Ecol. Epidemiol. 2015, 5, 27060. [Google Scholar] [CrossRef]
- Vijayaraj, R.; Altaff, K.; Rosita, A.S.; Ramadevi, S.; Revathy, J. Bioactive Compounds from Marine Resources against Novel Corona Virus (2019-NCoV): In Silico Study for Corona Viral Drug. Nat. Prod. Res. 2020, 35, 5525–5529. [Google Scholar] [CrossRef] [PubMed]
- Irving, W.L. Ebola Virus Transmission. Int. J. Exp. Pathol. 1995, 76, 225–226. [Google Scholar] [PubMed]
- Salmón-Mulanovich, G.; Vásquez, A.; Albújar, C.; Guevara, C.; Laguna-Torres, A.; Salazar, M.; Zamalloa, H.; Cáceres, M.; Gómez-Benavides, J.; Pacheco, V.; et al. Human Rabies and Rabies in Vampire and Nonvampire Bat Species, Southeastern Peru, 2007. Emerg. Infect. Dis. 2009, 15, 1308–1310. [Google Scholar] [CrossRef] [PubMed]
- Breman, J.G.; Heymann, D.L.; Lloyd, G.; McCormick, J.B.; Miatudila, M.; Murphy, F.A.; Muyembé-Tamfun, J.-J.; Piot, P.; Ruppol, J.-F.; Sureau, P.; et al. Discovery and Description of Ebola Zaire Virus in 1976 and Relevance to the West African Epidemic During 2013–2016. J. Infect. Dis. 2016, 214, S93–S101. [Google Scholar] [CrossRef]
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, F.; Wang, R.; Guan, K.; Jiang, T.; Xu, G.; Sun, J.; Chang, C. The Deadly Coronaviruses: The 2003 SARS Pandemic and the 2020 Novel Coronavirus Epidemic in China. J. Autoimmun. 2020, 109, 102434. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-T.; Hsu, W.-C.; Lin, C.-C. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 2014, 4, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ma, A.; Wang, X.; Yang, C.; Chen, X.; Li, G.; Qiu, F. Research Progress on the Antiviral Activities of Natural Products and Their Derivatives: Structure–Activity Relationships. Front. Chem. 2022, 10, 1005360. [Google Scholar] [CrossRef]
- Kamzeeva, P.N.; Aralov, A.V.; Alferova, V.A.; Korshun, V.A. Recent Advances in Molecular Mechanisms of Nucleoside Antivirals. Curr. Issues Mol. Biol. 2023, 45, 6851–6879. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.J.; Cheng, S.M.S.; Leung, K.; Lee, C.K.; Hachim, A.; Tsang, L.C.H.; Yam, K.W.H.; Chaothai, S.; Kwan, K.K.H.; Chai, Z.Y.H.; et al. Real-World COVID-19 Vaccine Effectiveness against the Omicron BA.2 Variant in a SARS-CoV-2 Infection-Naive Population. Nat. Med. 2023, 29, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, I.J.d.S.; Silva, L.R.; da Silva-Júnior, E.F. Challenges in Designing Antiviral Agents. In Viral Infections and Antiviral Therapies; Dhara, A.K., Nayak, A.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 169–209. [Google Scholar]
- Zhao, J.-H.; Wang, Y.-W.; Yang, J.; Tong, Z.-J.; Wu, J.-Z.; Wang, Y.-B.; Wang, Q.-X.; Li, Q.-Q.; Yu, Y.-C.; Leng, X.-J.; et al. Natural Products as Potential Lead Compounds to Develop New Antiviral Drugs over the Past Decade. Eur. J. Med. Chem. 2023, 260, 115726. [Google Scholar] [CrossRef] [PubMed]
- Demicheli, V.; Jefferson, T.; Di Pietrantonj, C.; Ferroni, E.; Thorning, S.; Thomas, R.E.; Rivetti, A. Vaccines for Preventing Influenza in the Elderly. Cochrane Database Syst. Rev. 2018, 2, CD004876. [Google Scholar] [CrossRef]
- Riccio, G.; Ruocco, N.; Mutalipassi, M.; Costantini, M.; Zupo, V.; Coppola, D.; de Pascale, D.; Lauritano, C. Ten-Year Research Update Review: Antiviral Activities from Marine Organisms. Biomolecules 2020, 10, 1007. [Google Scholar] [CrossRef]
- Teng, Y.-F.; Xu, L.; Wei, M.-Y.; Wang, C.-Y.; Gu, Y.-C.; Shao, C.-L. Recent Progresses in Marine Microbial-Derived Antiviral Natural Products. Arch. Pharm. Res. 2020, 43, 1215–1229. [Google Scholar] [CrossRef]
- Donia, M.; Hamann, M.T. Marine Natural Products and Their Potential Applications as Anti-Infective Agents. Lancet Infect. Dis. 2003, 3, 338–348. [Google Scholar] [CrossRef]
- Almeida, M.C.; Resende, D.I.S.P.; da Costa, P.M.; Pinto, M.M.M.; Sousa, E. Tryptophan Derived Natural Marine Alkaloids and Synthetic Derivatives as Promising Antimicrobial Agents. Eur. J. Med. Chem. 2021, 209, 112945. [Google Scholar] [CrossRef] [PubMed]
- Festa, M.; Sansone, C.; Brunet, C.; Crocetta, F.; Di Paola, L.; Lombardo, M.; Bruno, A.; Noonan, D.M.; Albini, A. Cardiovascular Active Peptides of Marine Origin with ACE Inhibitory Activities: Potential Role as Anti-Hypertensive Drugs and in Prevention of SARSCoV-2 Infection. Int. J. Mol. Sci. 2020, 21, 8364. [Google Scholar] [CrossRef] [PubMed]
- Moghadamtousi, S.; Nikzad, S.; Kadir, H.; Abubakar, S.; Zandi, K. Potential Antiviral Agents from Marine Fungi: An Overview. Mar. Drugs 2015, 13, 4520–4538. [Google Scholar] [CrossRef] [PubMed]
- Žigrayová, D.; Mikušová, V.; Mikuš, P. Advances in Antiviral Delivery Systems and Chitosan-Based Polymeric and Nanoparticulate Antivirals and Antiviral Carriers. Viruses 2023, 15, 647. [Google Scholar] [CrossRef]
- Nepali, K.; Sharma, R.; Sharma, S.; Thakur, A.; Liou, J.-P. Beyond the Vaccines: A Glance at the Small Molecule and Peptide-Based Anti-COVID19 Arsenal. J. Biomed. Sci. 2022, 29, 65. [Google Scholar] [CrossRef] [PubMed]
- El-Demerdash, A.; Metwaly, A.M.; Hassan, A.; El-Aziz, T.M.A.; Elkaeed, E.B.; Eissa, I.H.; Arafa, R.K.; Stockand, J.D. Comprehensive Virtual Screening of the Antiviral Potentialities of Marine Polycyclic Guanidine Alkaloids against SARS-CoV-2 (COVID-19). Biomolecules 2021, 11, 460. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Nollet, L.M.L. (Ed.) Marine Microorganisms Extraction and Analysis of Bioactive Compounds; CRC Press: Boca Raton, FL, USA, 2016; ISBN 9781498702560. [Google Scholar]
- Ameen, F.; AlNadhari, S.; Al-Homaidan, A.A. Marine Microorganisms as an Untapped Source of Bioactive Compounds. Saudi J. Biol. Sci. 2021, 28, 224–231. [Google Scholar] [CrossRef]
- Raveh, A.; Delekta, P.C.; Dobry, C.J.; Peng, W.; Schultz, P.J.; Blakely, P.K.; Tai, A.W.; Matainaho, T.; Irani, D.N.; Sherman, D.H.; et al. Discovery of Potent Broad Spectrum Antivirals Derived from Marine Actinobacteria. PLoS ONE 2013, 8, e82318. [Google Scholar] [CrossRef]
- Suthindhiran, K.; Sarath Babu, V.; Kannabiran, K.; Ishaq Ahmed, V.P.; Sahul Hameed, A.S. Anti-Fish Nodaviral Activity of Furan-2-Yl Acetate Extracted from Marine Streptomyces spp. Nat. Prod. Res. 2011, 25, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Wang, S.; Huang, H.; Li, H.; Wang, W.; Li, W. Generation of Methylated Violapyrones with Improved Anti-Influenza A Virus Activity by Heterologous Expression of a Type III PKS Gene in a Marine Streptomyces Strain. Bioorg. Med. Chem. Lett. 2018, 28, 2865–2868. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.; Carlsson, M.; Uvell, H.; Islam, K.; Edlund, K.; Cullman, I.; Altermark, B.; Mei, Y.F.; Elofsson, M.; Willassen, N.P.; et al. Isolation and Characterization of Anti-Adenoviral Secondary Metabolites from Marine Actinobacteria. Mar. Drugs 2014, 12, 799. [Google Scholar] [CrossRef] [PubMed]
- Al-Nahas, M.O.; Darwish, M.M.; Ali, A.E.; Amin, M.A. Characterization of an Exopolysaccharide-Producing Marine Bacterium, Isolate Pseudoalteromonas sp. AM. Afr. J. Microbiol. Res. 2011, 5, 3823–3831. [Google Scholar] [CrossRef]
- Manimaran, M.; Rajkumar, T.; Vimal, S.; Taju, G.; Abdul Majeed, S.; Sahul Hameed, A.S.; Kannabiran, K. Antiviral Activity of 9(10H)-Acridanone Extracted from Marine Streptomyces fradiae Strain VITMK2 in Litopenaeus vannamei Infected with White Spot Syndrome Virus. Aquaculture 2018, 488, 66–73. [Google Scholar] [CrossRef]
- Huang, H.; Song, Y.; Zang, R.; Wang, X.; Ju, J. Octyl Substituted Butenolides from Marine-Derived Streptomyces koyangensis. Nat. Prod. Res. 2019, 35, 2602–2607. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xi, L.; Liu, P.; Wang, Y.; Wang, W.; Huang, Y.; Zhu, W. Diketopiperazine Derivatives from the Marine-Derived Actinomycete Streptomyces sp. FXJ7.328. Mar. Drugs 2013, 11, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Song, Y.; Li, X.; Wang, X.; Ling, C.; Qin, X.; Zhou, Z.; Li, Q.; Wei, X.; Ju, J. Abyssomicin Monomers and Dimers from the Marine-Derived Streptomyces koyangensis SCSIO 5802. J. Nat. Prod. 2018, 81, 1892–1898. [Google Scholar] [CrossRef]
- Imhoff, J.F. Natural Products from Marine Fungi—Still an Underrepresented Resource. Mar. Drugs 2016, 14, 19. [Google Scholar] [CrossRef]
- Liu, S.; Su, M.; Song, S.-J.; Jung, J. Marine-Derived Penicillium Species as Producers of Cytotoxic Metabolites. Mar. Drugs 2017, 15, 329. [Google Scholar] [CrossRef]
- Nicoletti, R.; Trincone, A. Bioactive Compounds Produced by Strains of Penicillium and Talaromyces of Marine Origin. Mar. Drugs 2016, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Chen, Z.; Liu, P.; Wang, Y.; Xin, Z.; Zhu, W. New Rubrolides from the Marine-Derived Fungus Aspergillus Terreus OUCMDZ-1925. J. Antibiot. 2013, 67, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, X.; Du, L.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Sorbicatechols A and B, Antiviral Sorbicillinoids from the Marine-Derived Fungus Penicillium Chrysogenum PJX-17. J. Nat. Prod. 2014, 77, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Guo, W.; Wang, Q.; Zhang, L.; Zhu, M.; Zhu, T.; Gu, Q.; Wang, W.; Li, D. Aspulvinones from a Mangrove Rhizosphere Soil-Derived Fungus Aspergillus Terreus Gwq-48 with Anti-Influenza A Viral (H1N1) Activity. Bioorg. Med. Chem. Lett. 2013, 23, 1776–1778. [Google Scholar] [CrossRef]
- Kong, F.D.; Ma, Q.Y.; Huang, S.Z.; Wang, P.; Wang, J.F.; Zhou, L.M.; Yuan, J.Z.; Dai, H.F.; Zhao, Y.X. Chrodrimanins K-N and Related Meroterpenoids from the Fungus Penicillium sp. SCS-KFD09 Isolated from a Marine Worm, Sipunculus Nudus. J. Nat. Prod. 2017, 80, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.Q.; Lin, X.P.; Wang, Z.; Zhou, X.F.; Qin, X.C.; Kaliyaperumal, K.; Zhang, T.Y.; Tu, Z.C.; Liu, Y. Asteltoxins with Antiviral Activities from the Marine Sponge-Derived Fungus Aspergillus sp. SCSIO XWS02F40. Molecules 2015, 21, 34. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wang, Y.; Liu, P.; Fu, P.; Zhu, T.; Wang, W.; Zhu, W. Indole-Diterpenoids with Anti-H1N1 Activity from the Aciduric Fungus Penicillium Camemberti OUCMDZ-1492. J. Nat. Prod. 2013, 76, 1328–1336. [Google Scholar] [CrossRef]
- He, F.; Bao, J.; Zhang, X.Y.; Tu, Z.C.; Shi, Y.M.; Qi, S.H. Asperterrestide A, a Cytotoxic Cyclic Tetrapeptide from the Marine-Derived Fungus Aspergillus Terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef]
- Sun, Y.L.; Wang, J.; Wang, Y.F.; Zhang, X.Y.; Nong, X.H.; Chen, M.Y.; Xu, X.Y.; Qi, S.H. Cytotoxic and Antiviral Tetramic Acid Derivatives from the Deep-Sea-Derived Fungus Trichobotrys Effuse DFFSCS021. Tetrahedron 2015, 71, 9328–9332. [Google Scholar] [CrossRef]
- Yu, G.; Zhou, G.; Zhu, M.; Wang, W.; Zhu, T.; Gu, Q.; Li, D. Neosartoryadins A and B, Fumiquinazoline Alkaloids from a Mangrove-Derived Fungus Neosartorya Udagawae HDN13-313. Org. Lett. 2016, 18, 244–247. [Google Scholar] [CrossRef]
- Li, J.; Hu, Y.; Hao, X.; Tan, J.; Li, F.; Qiao, X.; Chen, S.; Xiao, C.; Chen, M.; Peng, Z.; et al. Raistrickindole A, an Anti-HCV Oxazinoindole Alkaloid from Penicillium Raistrickii IMB17-034. J. Nat. Prod. 2019, 82, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Y.; Hao, X.; Li, S.; Jia, J.; Guan, Y.; Peng, Z.; Bi, H.; Xiao, C.; Cen, S.; et al. Broad-Spectrum Antiviral Natural Products from the Marine-Derived Penicillium sp. IMB17-046. Molecules 2019, 24, 2821. [Google Scholar] [CrossRef]
- Huang, Z.; Nong, X.; Ren, Z.; Wang, J.; Zhang, X.; Qi, S. Anti-HSV-1, Antioxidant and Antifouling Phenolic Compounds from the Deep-Sea-Derived Fungus Aspergillus Versicolor SCSIO 41502. Bioorg. Med. Chem. Lett. 2017, 27, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Q.; Li, X.M.; Li, X.; Li, H.L.; Meng, L.H.; Wang, B.G. New Citrinin Analogues Produced by Coculture of the Marine Algal-Derived Endophytic Fungal Strains Aspergillus Sydowii EN-534 and Penicillium Citrinum EN-535. Phytochem. Lett. 2018, 25, 191–195. [Google Scholar] [CrossRef]
- Jin, Y.; Qin, S.; Gao, H.; Zhu, G.; Wang, W.; Zhu, W.; Wang, Y. An Anti-HBV Anthraquinone from Aciduric Fungus Penicillium sp. OUCMDZ-4736 under Low PH Stress. Extremophiles 2018, 22, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Janardhan, A.; Kumar, A.P.; Viswanath, B.; Saigopal, D.V.R.; Narasimha, G. Production of Bioactive Compounds by Actinomycetes and Their Antioxidant Properties. Biotechnol. Res. Int. 2014, 2014, 217030. [Google Scholar] [CrossRef]
- Janardhan, A.; Kumar, A.P.; Viswanath, B.; Gopal, D.S.; Narasimha, G. Antiviral and Larvicidal Properties of Novel Bioactive Compounds Produced from Marine Actinomycetes. Russ. J. Mar. Biol. 2018, 44, 424–428. [Google Scholar] [CrossRef]
- Niu, S.; Liu, D.; Shao, Z.; Proksch, P.; Lin, W. Eutypellazines A–M, Thiodiketopiperazine-Type Alkaloids from Deep Sea Derived Fungus Eutypella sp. MCCC 3A00281. RSC Adv. 2017, 7, 33580–33590. [Google Scholar] [CrossRef]
- Luo, M.; Zang, R.; Wang, X.; Chen, Z.; Song, X.; Ju, J.; Huang, H. Natural Hydroxamate-Containing Siderophore Acremonpeptides A-D and an Aluminum Complex of Acremonpeptide D from the Marine-Derived Acremonium Persicinum SCSIO 115. J. Nat. Prod. 2019, 82, 2594–2600. [Google Scholar] [CrossRef]
- Liang, X.; Nong, X.H.; Huang, Z.H.; Qi, S.H. Antifungal and Antiviral Cyclic Peptides from the Deep-Sea-Derived Fungus Simplicillium Obclavatum EIODSF 020. J. Agric. Food Chem. 2017, 65, 5114–5121. [Google Scholar] [CrossRef]
- Chen, X.; Si, L.; Liu, D.; Proksch, P.; Zhang, L.; Zhou, D.; Lin, W. Neoechinulin B and Its Analogues as Potential Entry Inhibitors of Influenza Viruses, Targeting Viral Hemagglutinin. Eur. J. Med. Chem. 2015, 93, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.L.; Wei, M.Y.; Chen, H.Y.; Guan, F.F.; Wang, C.Y.; Shao, C.L. (+)- and (−)-Pestaloxazine A, a Pair of Antiviral Enantiomeric Alkaloid Dimers with a Symmetric Spiro[Oxazinane-Piperazinedione] Skeleton from Pestalotiopsis sp. Org. Lett. 2015, 17, 4216–4219. [Google Scholar] [CrossRef]
- Ma, X.; Nong, X.H.; Ren, Z.; Wang, J.; Liang, X.; Wang, L.; Qi, S.H. Antiviral Peptides from Marine Gorgonian-Derived Fungus Aspergillus sp. SCSIO 41501. Tetrahedron Lett. 2017, 58, 1151–1155. [Google Scholar] [CrossRef]
- Jia, Y.L.; Guan, F.F.; Ma, J.; Wang, C.Y.; Shao, C.L. Pestalotiolide A, a New Antiviral Phthalide Derivative from a Soft Coral-Derived Fungus Pestalotiopsis sp. Nat. Prod. Sci. 2015, 21, 227–230. [Google Scholar] [CrossRef]
- Zhao, Y.; Si, L.; Liu, D.; Proksch, P.; Zhou, D.; Lin, W. Truncateols A–N, New Isoprenylated Cyclohexanols from the Sponge-Associated Fungus Truncatella Angustata with Anti-H1N1 Virus Activities. Tetrahedron 2015, 71, 2708–2718. [Google Scholar] [CrossRef]
- Liu, F.A.; Lin, X.; Zhou, X.; Chen, M.; Huang, X.; Yang, B.; Tao, H. Xanthones and Quinolones Derivatives Produced by the Deep-Sea-Derived Fungus Penicillium sp. SCSIO Ind16F01. Molecules 2017, 22, 1999. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Si, L.; Liu, D.; Zhou, A.; Zhang, Z.; Shao, Z.; Wang, S.; Zhang, L.; Zhou, D.; Lin, W. Spiromastilactones: A New Class of Influenza Virus Inhibitors from Deep-Sea Fungus. Eur. J. Med. Chem. 2016, 108, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.H.; Zhang, H.B.; Zhong, M.J.; Ma, L.Y.; Liu, D.S.; Liu, W.Z.; Ren, H. Potential Antiviral Xanthones from a Coastal Saline Soil Fungus Aspergillus Iizukae. Mar. Drugs 2018, 16, 449. [Google Scholar] [CrossRef]
- Yu, M.L.; Guan, F.F.; Cao, F.; Jia, Y.L.; Wang, C.Y. A New Antiviral Pregnane from a Gorgonian-Derived Cladosporium sp. Fungus. Nat. Prod. Res. 2018, 32, 1260–1266. [Google Scholar] [CrossRef]
- Pang, X.; Lin, X.; Wang, J.; Liang, R.; Tian, Y.; Salendra, L.; Luo, X.; Zhou, X.; Yang, B.; Tu, Z.; et al. Three New Highly Oxygenated Sterols and One New Dihydroisocoumarin from the Marine Sponge-Derived Fungus Cladosporium sp. SCSIO41007. Steroids 2018, 129, 41–46. [Google Scholar] [CrossRef]
- Cao, X.; Shi, Y.; Wu, X.; Wang, K.; Huang, S.; Sun, H.; Dickschat, J.S.; Wu, B. Talaromyolides A-D and Talaromytin: Polycyclic Meroterpenoids from the Fungus Talaromyces sp. CX11. Org. Lett. 2019, 21, 6539–6542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, Y.; Jia, C.; Lang, J.; Niaz, S.I.; Li, J.; Yuan, J.; Yu, J.; Chen, S.; Liu, L. Antiviral and Anti-Inflammatory Meroterpenoids: Stachybonoids A–F from the Crinoid-Derived Fungus Stachybotrys Chartarum 952. RSC Adv. 2017, 7, 49910–49916. [Google Scholar] [CrossRef]
- Qin, C.; Lin, X.; Lu, X.; Wan, J.; Zhou, X.; Liao, S.; Tu, Z.; Xu, S.; Liu, Y. Sesquiterpenoids and Xanthones Derivatives Produced by Sponge-Derived Fungus Stachybotry sp. HH1 ZSDS1F1-2. J. Antibiot. 2014, 68, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, F.; Liu, Y.; Liu, Y.; Li, K.; Yang, X.; Liu, S.; Zhou, X.; Yang, J. Spirostaphylotrichin X from a Marine-Derived Fungus as an Anti-Influenza Agent Targeting RNA Polymerase PB2. J. Nat. Prod. 2018, 81, 2722–2730. [Google Scholar] [CrossRef] [PubMed]
- Mensah, E.O.; Kanwugu, O.N.; Panda, P.K.; Adadi, P. Marine Fucoidans: Structural, Extraction, Biological Activities and Their Applications in the Food Industry. Food Hydrocoll. 2023, 142, 108784. [Google Scholar] [CrossRef]
- Hayashi, K.; Lee, J.-B.; Atsumi, K.; Kanazashi, M.; Shibayama, T.; Okamoto, K.; Kawahara, T.; Hayashi, T. In Vitro and in Vivo Anti-Herpes Simplex Virus Activity of Monogalactosyl Diacylglyceride from Coccomyxa sp. KJ (IPOD FERM BP-22254), a Green Microalga. PLoS ONE 2019, 14, e0219305. [Google Scholar] [CrossRef] [PubMed]
- Raposo, M.F.D.J.; De Morais, A.M.M.B.; De Morais, R.M.S.C. Influence of Sulphate on the Composition and Antibacterial and Antiviral Properties of the Exopolysaccharide from Porphyridium Cruentum. Life Sci. 2014, 101, 56–63. [Google Scholar] [CrossRef]
- Kim, M.; Yim, J.H.; Kim, S.Y.; Kim, H.S.; Lee, W.G.; Kim, S.J.; Kang, P.S.; Lee, C.K. In Vitro Inhibition of Influenza A Virus Infection by Marine Microalga-Derived Sulfated Polysaccharide p-KG03. Antivir. Res. 2012, 93, 253–259. [Google Scholar] [CrossRef]
- Gastineau, R.; Pouvreau, J.B.; Hellio, C.; Morançais, M.; Fleurence, J.; Gaudin, P.; Bourgougnon, N.; Mouget, J.L. Biological Activities of Purified Marennine, the Blue Pigment Responsible for the Greening of Oysters. J. Agric. Food Chem. 2012, 60, 3599–3605. [Google Scholar] [CrossRef]
- Song, L.; Chen, X.; Liu, X.; Zhang, F.; Hu, L.; Yue, Y.; Li, K.; Li, P. Characterization and Comparison of the Structural Features, Immune-Modulatory and Anti-Avian Influenza Virus Activities Conferred by Three Algal Sulfated Polysaccharides. Mar. Drugs 2015, 14, 4. [Google Scholar] [CrossRef]
- Wang, S.; Wang, W.; Hou, L.; Qin, L.; He, M.; Li, W.; Mao, W. A Sulfated Glucuronorhamnan from the Green Seaweed Monostroma Nitidum: Characteristics of Its Structure and Antiviral Activity. Carbohydr. Polym. 2020, 227, 115280. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Briseño, J.A.; Cruz-Suarez, L.E.; Sassi, J.F.; Ricque-Marie, D.; Zapata-Benavides, P.; Mendoza-Gamboa, E.; Rodríguez-Padilla, C.; Trejo-Avila, L.M. Sulphated Polysaccharides from Ulva Clathrata and Cladosiphon Okamuranus Seaweeds Both Inhibit Viral Attachment/Entry and Cell-Cell Fusion, in NDV Infection. Mar. Drugs 2015, 13, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kim, J.H.; Kwon, J.M.; Kwon, H.J.; Jeong, H.J.; Kim, Y.M.; Kim, D.; Lee, W.S.; Ryu, Y.B. Dieckol, a SARS-CoV 3CL(pro) Inhibitor, Isolated from the Edible Brown Algae Ecklonia Cava. Bioorg. Med. Chem. 2013, 21, 3730–3737. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.R.; Robaina, M.C.S.; Mendes, G.S.; Silva, T.S.L.; Gestinari, L.M.S.; Pamplona, O.S.; Yoneshigue-Valentin, Y.; Kaiser, C.R.; Romanos, M.T.V. Antiviral Activity of Extracts from Brazilian Seaweeds against Herpes Simplex Virus. Rev. Bras. Farmacogn. 2012, 22, 714–723. [Google Scholar] [CrossRef]
- Hamdy, A.H.A.; Mettwallya, W.S.A.; El Fotouh, M.A.; Rodriguez, B.; El-Dewany, A.I.; El-Toumy, S.A.A.; Hussein, A.A. Bioactive Phenolic Compounds from the Egyptian Red Sea Seagrass Thalassodendron Ciliatum. Z. Naturforschung Sect. C J. Biosci. 2012, 67C, 291–296. [Google Scholar] [CrossRef]
- Mohammed, M.M.D.; Hamdy, A.H.A.; El-Fiky, N.M.; Mettwally, W.S.A.; El-Beih, A.A.; Kobayashi, N. Anti-Influenza A Virus Activity of a New Dihydrochalcone Diglycoside Isolated from the Egyptian Seagrass Thalassodendron Ciliatum (Forsk.) Den Hartog. Nat. Prod. Res. 2014, 28, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Hawas, U.W.; Abou El-Kassem, L.T. Thalassiolin D: A New Flavone O-Glucoside Sulphate from the Seagrass Thalassia Hemprichii. Nat. Prod. Res. 2017, 31, 2369–2374. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Huang, X.Y.; Li, J.; Xin, G.R.; Guo, Y.W. Absolute Configurations of Integracins A, B, and 15′-Dehydroxy-Integracin B. Chirality 2012, 24, 459–462. [Google Scholar] [CrossRef]
- Li, W.; Jiang, Z.; Shen, L.; Pedpradab, P.; Bruhn, T.; Wu, J.; Bringmann, G. Antiviral Limonoids Including Khayanolides from the Trang Mangrove Plant Xylocarpus Moluccensis. J. Nat. Prod. 2015, 78, 1570–1578. [Google Scholar] [CrossRef]
- Tietjen, I.; Williams, D.E.; Read, S.; Kuang, X.T.; Mwimanzi, P.; Wilhelm, E.; Markle, T.; Kinloch, N.N.; Naphen, C.N.; Tenney, K.; et al. Inhibition of NF-ΚB-Dependent HIV-1 Replication by the Marine Natural Product Bengamide A. Antivir. Res. 2018, 152, 94–103. [Google Scholar] [CrossRef]
- Shin, H.J.; Rashid, M.A.; Cartner, L.K.; Bokesch, H.R.; Wilson, J.A.; McMahon, J.B.; Gustafson, K.R. Stellettapeptins A and B, HIV-Inhibitory Cyclic Depsipeptides from the Marine Sponge Stelletta sp. Tetrahedron Lett. 2015, 56, 4215–4219. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Niu, S.; Gao, J.; Zuo, H.; Yuan, J.; Weng, S.; He, J.; Xu, X. A Single WAP Domain (SWD)-Containing Protein with Antiviral Activity from Pacific White Shrimp Litopenaeus Vannamei. Fish. Shellfish. Immunol. 2018, 73, 167–174. [Google Scholar] [CrossRef]
- Tripoteau, L.; Bedoux, G.; Gagnon, J.; Bourgougnon, N. In Vitro Antiviral Activities of Enzymatic Hydrolysates Extracted from Byproducts of the Atlantic Holothurian Cucumaria Frondosa. Process Biochem. 2015, 50, 867–875. [Google Scholar] [CrossRef]
- González-Almela, E.; Sanz, M.A.; García-Moreno, M.; Northcote, P.; Pelletier, J.; Carrasco, L. Differential Action of Pateamine A on Translation of Genomic and Subgenomic MRNAs from Sindbis Virus. Virology 2015, 484, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Guo, J.; Liu, Y.; Lu, A.; Wang, Z.; Li, Y.; Yang, S.; Wang, Q. Marine-Natural-Product Development: First Discovery of Nortopsentin Alkaloids as Novel Antiviral, Anti-Phytopathogenic-Fungus, and Insecticidal Agents. J. Agric. Food Chem. 2018, 66, 4062–4072. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Wang, Z.; Li, G.; Liu, Y.; Xie, Y.; Wang, Q. First Discovery of Polycarpine, Polycarpaurines A and C, and Their Derivatives as Novel Antiviral and Antiphytopathogenic Fungus Agents. J. Agric. Food Chem. 2016, 64, 4264–4272. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.K.; Hsieh, M.K.; Duh, C.Y. New Diterpenoids from Soft Coral Sarcophyton Ehrenbergi. Mar. Drugs 2013, 11, 4318. [Google Scholar] [CrossRef]
- Cheng, S.Y.; Wang, S.K.; Duh, C.Y. Secocrassumol, a Seco-Cembranoid from the Dongsha Atoll Soft Coral Lobophytum Crassum. Mar. Drugs 2014, 12, 6028–6037. [Google Scholar] [CrossRef]
- Cao, F.; Shao, C.L.; Chen, M.; Zhang, M.Q.; Xu, K.X.; Meng, H.; Wang, C.Y. Antiviral C-25 Epimers of 26-Acetoxy Steroids from the South China Sea Gorgonian Echinogorgia Rebekka. J. Nat. Prod. 2014, 77, 1488–1493. [Google Scholar] [CrossRef]
- Gong, K.K.; Tang, X.L.; Zhang, G.; Cheng, C.L.; Zhang, X.W.; Li, P.N.; Li, G.Q. Polyhydroxylated Steroids from the South China Sea Soft Coral Sarcophyton sp. and Their Cytotoxic and Antiviral Activities. Mar. Drugs 2013, 11, 4788–4798. [Google Scholar] [CrossRef]
- Pujol, C.A.; Sepúlveda, C.S.; Richmond, V.; Maier, M.S.; Damonte, E.B. Polyhydroxylated Sulfated Steroids Derived from 5α-Cholestanes as Antiviral Agents against Herpes Simplex Virus. Arch. Virol. 2016, 161, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Jiang, L.; Wu, J.; Liu, Z.; Wu, Y. Anti-Tumor and Anti-Virus Activity of Polysaccharides Extracted from Sipunculus Nudus(SNP) on Hepg2.2.15. Int. J. Biol. Macromol. 2016, 87, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Wijanarko, A.; Lischer, K.; Hermansyah, H.; Pratami, D.K.; Sahlan, M. Antiviral Activity of Acanthaster Planci Phospholipase A2 against Human Immunodeficiency Virus. Vet. World 2018, 11, 824. [Google Scholar] [CrossRef]
- Lum, K.Y.; Carroll, A.R.; Ekins, M.G.; Read, S.; Haq, Z.; Tietjen, I.; St John, J.; Davis, R.A. Capillasterin A, a Novel Pyrano[2,3-f]Chromene from the Australian Crinoid Capillaster Multiradiatus. Mar. Drugs 2019, 17, 26. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Fujimoto, Y.; Tamaki, M.; Setiawan, A.; Tanaka, T.; Okuyama-Dobashi, K.; Kasai, H.; Watashi, K.; Wakita, T.; Toyama, M.; et al. Identification of Antiviral Agents Targeting Hepatitis B Virus Promoter from Extracts of Indonesian Marine Organisms by a Novel Cell-Based Screening Assay. Mar. Drugs 2015, 13, 6759–6773. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Tamaki, M.; Kasai, H.; Tanaka, T.; Otoguro, T.; Ryo, A.; Maekawa, S.; Enomoto, N.; de Voogd, N.J.; Tanaka, J.; et al. Inhibitory Effects of Metachromin A on Hepatitis B Virus Production via Impairment of the Viral Promoter Activity. Antivir. Res. 2017, 145, 136–145. [Google Scholar] [CrossRef]
- Salam, K.A.; Furuta, A.; Noda, N.; Tsuneda, S.; Sekiguchi, Y.; Yamashita, A.; Moriishi, K.; Nakakoshi, M.; Tsubuki, M.; Tani, H.; et al. Inhibition of Hepatitis C Virus NS3 Helicase by Manoalide. J. Nat. Prod. 2012, 75, 650–654. [Google Scholar] [CrossRef]
- Salam, K.A.; Furuta, A.; Noda, N.; Tsuneda, S.; Sekiguchi, Y.; Yamashita, A.; Moriishi, K.; Nakakoshi, M.; Tsubuki, M.; Tani, H.; et al. Psammaplin A Inhibits Hepatitis C Virus NS3 Helicase. J. Nat. Med. 2013, 67, 765–772. [Google Scholar] [CrossRef]
- Yu, H.B.; Yang, F.; Sun, F.; Li, J.; Jiao, W.H.; Gan, J.H.; Hu, W.Z.; Lin, H.W. Aaptamine Derivatives with Antifungal and Anti-HIV-1 Activities from the South China Sea Sponge Aaptos Aaptos. Mar. Drugs 2014, 12, 6003. [Google Scholar] [CrossRef]
- Li, G.; Guo, J.; Wang, Z.; Liu, Y.; Song, H.; Wang, Q. Marine Natural Products for Drug Discovery: First Discovery of Kealiinines A-C and Their Derivatives as Novel Antiviral and Antiphytopathogenic Fungus Agents. J. Agric. Food Chem. 2018, 66, 7310–7318. [Google Scholar] [CrossRef]
- Dolashka, P.; Dolashka, P.; Nesterova, N.; Zagorodnya, S.; Dolashki, A.; Baranova, G.; Golovan, A.; Voelter, W. Antiviral Activity of Hemocyanin Rapana Venosa and Its Isoforms Against Epstein-Barr Virus. Glob. J. Pharmacol. 2014, 8, 206–212. [Google Scholar]
- Talaei Zanjani, N.; Miranda-Saksena, M.; Valtchev, P.; Diefenbach, R.J.; Hueston, L.; Diefenbach, E.; Sairi, F.; Gomes, V.G.; Cunningham, A.L.; Dehghani, F. Abalone Hemocyanin Blocks the Entry of Herpes Simplex Virus 1 into Cells: A Potential New Antiviral Strategy. Antimicrob. Agents Chemother. 2015, 60, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Green, T.J.; Robinson, N.; Chataway, T.; Benkendorff, K.; O’Connor, W.; Speck, P. Evidence That the Major Hemolymph Protein of the Pacific Oyster, Crassostrea Gigas, Has Antiviral Activity against Herpesviruses. Antivir. Res. 2014, 110, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Novoa, B.; Romero, A.; Álvarez, Á.L.; Moreira, R.; Pereiro, P.; Costa, M.M.; Dios, S.; Estepa, A.; Parra, F.; Figueras, A. Antiviral Activity of Myticin C Peptide from Mussel: An Ancient Defense against Herpesviruses. J. Virol. 2016, 90, 7692–7702. [Google Scholar] [CrossRef] [PubMed]
- Lillsunde, K.E.; Festa, C.; Adel, H.; De Marino, S.; Lombardi, V.; Tilvi, S.; Nawrot, D.A.; Zampella, A.; D’Souza, L.; D’Auria, M.V.; et al. Bioactive Cembrane Derivatives from the Indian Ocean Soft Coral, Sinularia Kavarattiensis. Mar. Drugs 2014, 12, 4045–4068. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.T.; Wang, S.K.; Dai, C.F.; Duh, C.Y. Briacavatolides A-C, New Briaranes from the Taiwanese Octocoral Briareum Excavatum. Mar. Drugs 2012, 10, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.K.; Yeh, T.T.; Duh, C.Y. Briacavatolides D-F, New Briaranes from the Taiwanese Octocoral Briareum Excavatum. Mar. Drugs 2012, 10, 2103–2110. [Google Scholar] [CrossRef]
- Ahmed, S.; Ibrahim, A.; Arafa, A.S. Anti-H5N1 Virus Metabolites from the Red Sea Soft Coral, Sinularia Candidula. Tetrahedron Lett. 2013, 54, 2377–2381. [Google Scholar] [CrossRef]
- Du, Z.-Q.; Ren, Q.; Huang, A.M.; Fang, W.H.; Zhou, J.F.; Gao, L.J.; Li, X.C. A Novel Peroxinectin Involved in Antiviral and Antibacterial Immunity of Mud Crab, Scylla Paramamosain. Mol. Biol. Rep. 2013, 40, 6873–6881. [Google Scholar] [CrossRef]
- Peng, H.; Liu, H.P.; Chen, B.; Hao, H.; Wang, K.J. Optimized Production of Scygonadin in Pichia Pastoris and Analysis of Its Antimicrobial and Antiviral Activities. Protein Expr. Purif. 2012, 82, 37–44. [Google Scholar] [CrossRef]
- Liu, H.P.; Chen, R.Y.; Zhang, Q.X.; Wang, Q.Y.; Li, C.R.; Peng, H.; Cai, L.; Zheng, C.Q.; Wang, K.J. Characterization of Two Isoforms of Antiliopolysacchride Factors (Sp-ALFs) from the Mud Crab Scylla Paramamosain. Fish. Shellfish. Immunol. 2012, 33, 1–10. [Google Scholar] [CrossRef]
- Zhan, S.; Aweya, J.J.; Wang, F.; Yao, D.; Zhong, M.; Chen, J.; Li, S.; Zhang, Y. Litopenaeus Vannamei Attenuates White Spot Syndrome Virus Replication by Specific Antiviral Peptides Generated from Hemocyanin. Dev. Comp. Immunol. 2019, 91, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.-Q.; Wang, Y.; Ma, H.Y.; Shen, X.L.; Wang, K.; Du, J.; Yu, X.D.; Fang, W.H.; Li, X.C. A New Crustin Homologue (SpCrus6) Involved in the Antimicrobial and Antiviral Innate Immunity in Mud Crab, Scylla Paramamosain. Fish. Shellfish. Immunol. 2019, 84, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Khong, H.Y.; Mao, W.; Chen, X.; Bao, L.; Wen, X.; Xu, Y. Tunicates as Sources of High-Quality Nutrients and Bioactive Compounds for Food/Feed and Pharmaceutical Applications: A Review. Foods 2023, 12, 3684. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Harper, M.K.; Pond, C.D.; Barrows, L.R.; Ireland, C.M.; Van Wagoner, R.M. Thiazoline Peptides and a Tris-Phenethyl Urea from Didemnum Molle with Anti-HIV Activity. J. Nat. Prod. 2012, 75, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Smitha, D.; Kumar, M.M.K.; Ramana, H.; Rao, D.V. Rubrolide R: A New Furanone Metabolite from the Ascidian Synoicum of the Indian Ocean. Nat. Prod. Res. 2014, 28, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.E.; Pond, C.D.; Pierce, E.; Harmer, Z.P.; Kwan, J.; Zachariah, M.M.; Harper, M.K.; Wyche, T.P.; Matainaho, T.K.; Bugni, T.S.; et al. Accessing Chemical Diversity from the Uncultivated Symbionts of Small Marine Animals. Nat. Chem. Biol. 2018, 14, 179–185. [Google Scholar] [CrossRef]
- Migliolo, L.; Silva, O.N.; Silva, P.A.; Costa, M.P.; Costa, C.R.; Nolasco, D.O.; Barbosa, J.A.R.G.; Silva, M.R.R.; Bemquerer, M.P.; Lima, L.M.P.; et al. Structural and Functional Characterization of a Multifunctional Alanine-Rich Peptide Analogue from Pleuronectes Americanus. PLoS ONE 2012, 7, e47047. [Google Scholar] [CrossRef]
- Vilas Boas, L.C.P.; de Lima, L.M.P.; Migliolo, L.; Mendes, G.d.S.; de Jesus, M.G.; Franco, O.L.; Silva, P.A. Linear Antimicrobial Peptides with Activity against Herpes Simplex Virus 1 and Aichi Virus. Biopolymers 2017, 108, e22871. [Google Scholar] [CrossRef]
- Sommeng, A.N.; Muhammad Yusuf Arya, R.; Ginting, M.J.; Pratami, D.K.; Hermansyah, H.; Sahlan, M.; Wijanarko, A. Antiretroviral Activity of Pterois Volitans (Red Lionfish) Venom in the Early Development of Human Immunodeficiency Virus/Acquired Immunodeficiency Syndrome Antiretroviral Alternative Source. Vet. World 2019, 12, 309–315. [Google Scholar] [CrossRef]
- Xin, Y.; Li, W.; Lu, L.; Zhou, L.; Victor, D.W.; Xuan, S. Antiviral Effects of Stichopus Japonicus Acid Mucopolysaccharide on Hepatitis B Virus Transgenic Mice. J. Ocean. Univ. China 2016, 15, 719–725. [Google Scholar] [CrossRef]
- Gentile, D.; Patamia, V.; Scala, A.; Sciortino, M.T.; Piperno, A.; Rescifina, A. Putative Inhibitors of SARS-CoV-2 Main Protease from a Library of Marine Natural Products: A Virtual Screening and Molecular Modeling Study. Mar. Drugs 2020, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Antonio, A.D.S.; Wiedemann, L.S.M.; Veiga-Junior, V.F. Natural Products’ Role against COVID-19. RSC Adv. 2020, 10, 23379–23393. [Google Scholar] [CrossRef]
- Chojnacka, K.; Witek-Krowiak, A.; Skrzypczak, D.; Mikula, K.; Młynarz, P. Phytochemicals Containing Biologically Active Polyphenols as an Effective Agent against COVID-19-Inducing Coronavirus. J. Funct. Foods 2020, 73, 104146. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Ghosh, S.; Garg, S.; Ghosh, S.; Jana, A.; Samat, R.; Mukherjee, N.; Roy, R.; Ghosh, S. An Overview of Key Potential Therapeutic Strategies for Combat in the COVID-19 Battle. RSC Adv. 2020, 10, 28243–28266. [Google Scholar] [CrossRef]
- Twomey, J.D.; Luo, S.; Dean, A.Q.; Bozza, W.P.; Nalli, A.; Zhang, B. COVID-19 Update: The Race to Therapeutic Development. Drug Resist. Updates 2020, 53, 100733. [Google Scholar] [CrossRef] [PubMed]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; MacKens-Kiani, T.; Cheng, J.; et al. Structural Basis for Translational Shutdown and Immune Evasion by the Nsp1 Protein of SARS-CoV-2. Science 2020, 369, 1249–1256. [Google Scholar] [CrossRef]
- Emrani, J.; Ahmed, M.; Jeffers-Francis, L.; Teleha, J.C.; Mowa, N.; Newman, R.H.; Thomas, M.D. SARS-CoV-2, Infection, Transmission, Transcription, Translation, Proteins, and Treatment: A Review. Int. J. Biol. Macromol. 2021, 193, 1249–1273. [Google Scholar] [CrossRef]
- Xu, X.; Chen, P.; Wang, J.; Feng, J.; Zhou, H.; Li, X.; Zhong, W.; Hao, P. Evolution of the Novel Coronavirus from the Ongoing Wuhan Outbreak and Modeling of Its Spike Protein for Risk of Human Transmission. Sci. China Life Sci. 2020, 63, 457–460. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Physiological and Molecular Triggers for SARS-CoV Membrane Fusion and Entry into Host Cells. Virology 2018, 517, 3–8. [Google Scholar] [CrossRef]
- Mengist, H.M.; Dilnessa, T.; Jin, T. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front. Chem. 2021, 9, 622898. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, D.; Pandav, K.; Patel, M.; Riva-Moscoso, A.; Singh, B.M.; Patel, A.; Min, Z.C.; Singh-Makkar, S.; Sana, M.K.; Sanchez-Dopazo, R.; et al. Omega 3 Fatty Acids and COVID-19: A Comprehensive Review. Infect. Chemother. 2020, 52, 478–495. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- De Lira, S.P.; Seleghim, M.H.R.; Williams, D.E.; Marion, F.; Hamill, P.; Jean, F.; Andersen, R.J.; Hajdu, E.; Berlinck, R.G.S. A SARS-Coronovirus 3CL Protease Inhibitor Isolated from the Marine Sponge Axinella Cf. Corrugata: Structure Elucidation and Synthesis. J. Braz. Chem. Soc. 2007, 18, 440–443. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Abdelrheem, D.A.; El-Mageed, H.R.A.; Mohamed, H.S.; Rahman, A.A.; Elsayed, K.N.M.; Ahmed, S.A. Destabilizing the Structural Integrity of COVID-19 by Caulerpin and Its Derivatives along with Some Antiviral Drugs: An in Silico Approaches for a Combination Therapy. Struct. Chem. 2020, 31, 2391–2412. [Google Scholar] [CrossRef]
- Al Adem, K.; Shanti, A.; Stefanini, C.; Lee, S. Inhibition of SARS-CoV-2 Entry into Host Cells Using Small Molecules. Pharmaceuticals 2020, 13, 447. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.; Arora, P.; Prajapati, S.K. Can Algal Derived Bioactive Metabolites Serve as Potential Therapeutics for the Treatment of SARS-CoV-2 Like Viral Infection? Front. Microbiol. 2020, 11, 596374. [Google Scholar] [CrossRef]
- Pendyala, B.; Patras, A.; Dash, C. Phycobilins as Potent Food Bioactive Broad-Spectrum Inhibitors Against Proteases of SARS-CoV-2 and Other Coronaviruses: A Preliminary Study. Front. Microbiol. 2021, 12, 645713. [Google Scholar] [CrossRef]
- Gyebi, G.A.; Ogunro, O.B.; Adegunloye, A.P.; Ogunyemi, O.M.; Afolabi, S.O. Potential Inhibitors of Coronavirus 3-Chymotrypsin-like Protease (3CL pro): An in Silico Screening of Alkaloids and Terpenoids from African Medicinal Plants. J. Biomol. Struct. Dyn. 2020, 39, 3396–3408. [Google Scholar] [CrossRef]
- Erlina, L.; Paramita, R.I.; Kusuma, W.A.; Fadilah, F.; Tedjo, A.; Pratomo, I.P.; Ramadhanti, N.S.; Nasution, A.K.; Surado, F.K.; Fitriawan, A.; et al. Virtual Screening of Indonesian Herbal Compounds as COVID-19 Supportive Therapy: Machine Learning and Pharmacophore Modeling Approaches. BMC Complement. Med. Ther. 2022, 22, 207. [Google Scholar] [CrossRef]
- Jiao, X.; Jin, X.; Ma, Y.; Yang, Y.; Li, J.; Liang, L.; Liu, R.; Li, Z. A Comprehensive Application: Molecular Docking and Network Pharmacology for the Prediction of Bioactive Constituents and Elucidation of Mechanisms of Action in Component-Based Chinese Medicine. Comput. Biol. Chem. 2021, 90, 107402. [Google Scholar] [CrossRef] [PubMed]
- Surti, M.; Patel, M.; Adnan, M.; Moin, A.; Ashraf, S.A.; Siddiqui, A.J.; Snoussi, M.; Deshpande, S.; Reddy, M.N. Ilimaquinone (Marine Sponge Metabolite) as a Novel Inhibitor of SARS-CoV-2 Key Target Proteins in Comparison with Suggested COVID-19 Drugs: Designing, Docking and Molecular Dynamics Simulation Study. RSC Adv. 2020, 10, 37707–37720. [Google Scholar] [CrossRef]
- Vivar-Sierra, A.; Araiza-Macías, M.J.; Hernández-Contreras, J.P.; Vergara-Castañeda, A.; Ramírez-Vélez, G.; Pinto-Almazán, R.; Salazar, J.R.; Loza-Mejía, M.A. In Silico Study of Polyunsaturated Fatty Acids as Potential SARS-CoV-2 Spike Protein Closed Conformation Stabilizers: Epidemiological and Computational Approaches. Molecules 2021, 26, 711. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhao, L.; Wen, H.; Li, Y. Herbal Combinations against COVID-19: A Network Pharmacology, Molecular Docking and Dynamics Study. J. Integr. Med. 2023, 21, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, Y.; Li, H.; Li, W.; Wu, X.; Li, Z. Application of Network Pharmacology in the Prevention and Treatment of COVID-19 by Traditional Chinese Medicine. Tradit. Med. Res. 2022, 7, 21. [Google Scholar] [CrossRef]
- Zaporozhets, T.S.; Besednova, N.N. Biologically Active Compounds from Marine Organisms in the Strategies for Combating Coronaviruses. AIMS Microbiol. 2020, 6, 470–494. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Chauhan, N.; Kuddus, M. Exploring the Therapeutic Potential of Marine-Derived Bioactive Compounds against COVID-19. Environ. Sci. Pollut. Res. 2021, 28, 52798–52809. [Google Scholar] [CrossRef]
- Arunkumar, M.; Gunaseelan, S.; Kubendran Aravind, M.; Mohankumar, V.; Anupam, P.; Harikrishnan, M.; Siva, A.; Ashokkumar, B.; Varalakshmi, P. Marine Algal Antagonists Targeting 3CL Protease and Spike Glycoprotein of SARS-CoV-2: A Computational Approach for Anti-COVID-19 Drug Discovery. J. Biomol. Struct. Dyn. 2021, 40, 8961–8988. [Google Scholar] [CrossRef]
- Kwon, P.S.; Oh, H.; Kwon, S.-J.; Jin, W.; Zhang, F.; Fraser, K.; Hong, J.J.; Linhardt, R.J.; Dordick, J.S. Sulfated Polysaccharides Effectively Inhibit SARS-CoV-2 in Vitro. Cell Discov. 2020, 6, 50. [Google Scholar] [CrossRef]
- Morokutti-Kurz, M.; Fröba, M.; Graf, P.; Große, M.; Grassauer, A.; Auth, J.; Schubert, U.; Prieschl-Grassauer, E. Iota-Carrageenan Neutralizes SARS-CoV-2 and Inhibits Viral Replication in Vitro. PLoS ONE 2021, 16, e0237480. [Google Scholar] [CrossRef]
- Reynolds, D.; Huesemann, M.; Edmundson, S.; Sims, A.; Hurst, B.; Cady, S.; Beirne, N.; Freeman, J.; Berger, A.; Gao, S. Viral Inhibitors Derived from Macroalgae, Microalgae, and Cyanobacteria: A Review of Antiviral Potential throughout Pathogenesis. Algal Res. 2021, 57, 102331. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Peng, H.; Wang, Q.; Liu, Z.; Dong, X.; Wen, C.; Ai, C.; Zhang, Y.; Wang, Z.; Zhu, B. Inhibitory Activities of Marine Sulfated Polysaccharides against SARS-CoV-2. Food Funct. 2020, 11, 7415–7420. [Google Scholar] [CrossRef] [PubMed]
- Panggabean, J.A.; Adiguna, S.P.; Rahmawati, S.I.; Ahmadi, P.; Zainuddin, E.N.; Bayu, A.; Putra, M.Y. Antiviral Activities of Algal-Based Sulfated Polysaccharides. Molecules 2022, 27, 1178. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Shin, H.; Lee, M.K.; Kwon, O.S.; Shin, J.S.; Kim, Y.I.; Kim, C.W.; Lee, H.R.; Kim, M. Antiviral Activity of Lambda-Carrageenan against Influenza Viruses and Severe Acute Respiratory Syndrome Coronavirus 2. Sci. Rep. 2021, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.K.; Kim, K.; Kim, I.; Chun, S.H.; Oh, T.H.; Kim, J.U.; Kim, J.; Jung, W.; Moon, H.; Ku, B.; et al. Inhibition of SARS-CoV-2 Virus Entry by the Crude Polysaccharides of Seaweeds and Abalone Viscera in Vitro. Mar. Drugs 2021, 19, 219. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Aroca, Á.; Ferrandis-Montesinos, M.; Wang, R. Antiviral Properties of Alginate-Based Biomaterials: Promising Antiviral Agents against SARS-CoV-2. ACS Appl. Bio. Mater. 2021, 4, 5897–5907. [Google Scholar] [CrossRef] [PubMed]
- Cano-Vicent, A.; Hashimoto, R.; Takayama, K.; Serrano-Aroca, Á. Biocompatible Films of Calcium Alginate Inactivate Enveloped Viruses Such as SARS-CoV-2. Polymers 2022, 14, 1483. [Google Scholar] [CrossRef]
- You, Y.; Song, H.; Wang, L.; Peng, H.; Sun, Y.; Ai, C.; Wen, C.; Zhu, B.; Song, S. Structural Characterization and SARS-CoV-2 Inhibitory Activity of a Sulfated Polysaccharide from Caulerpa Lentillifera. Carbohydr. Polym. 2022, 280, 119006. [Google Scholar] [CrossRef]
- Yao, Y.; Luo, Z.; Zhang, X. In Silico Evaluation of Marine Fish Proteins as Nutritional Supplements for COVID-19 Patients. Food Funct. 2020, 11, 5565–5572. [Google Scholar] [CrossRef]
- Riyadi, P.H.; Tanod, W.A.; Wahyudi, D.; Susanto, E.; Fahmi, A.S.; Aisiah, S. Potential of Tilapia (Oreochromis Niloticus) Viscera Bioactive Peptides as Antiviral for SARS-CoV-2 (COVID 19). IOP Conf. Ser. Earth Environ. Sci. 2020, 584, 012004. [Google Scholar] [CrossRef]
- White, K.M.; Rosales, R.; Yildiz, S.; Kehrer, T.; Miorin, L.; Moreno, E.; Jangra, S.; Uccellini, M.B.; Rathnasinghe, R.; Coughlan, L.; et al. Plitidepsin Has Potent Preclinical Efficacy against SARS-CoV-2 by Targeting the Host Protein EEF1A. Science 2021, 371, 926–931. [Google Scholar] [CrossRef]
- Ding, Y.; Zhu, X.; Hao, L.; Zhao, M.; Hua, Q.; An, F. Bioactive Indolyl Diketopiperazines from the Marine Derived Endophytic Aspergillus Versicolor DY180635. Mar. Drugs 2020, 18, 338. [Google Scholar] [CrossRef] [PubMed]
- Sahin, S.; Calapoğlu, F.; Ozmen, I. Didemnins Inhibit COVID-19 Main Protease (MPRO). Biointerface Res. Appl. Chem. 2021, 11, 8204–8209. [Google Scholar] [CrossRef]
- Ashhurst, A.S.; Tang, A.H.; Fajtová, P.; Yoon, M.C.; Aggarwal, A.; Bedding, M.J.; Stoye, A.; Beretta, L.; Pwee, D.; Drelich, A.; et al. Potent Anti-SARS-CoV-2 Activity by the Natural Product Gallinamide A and Analogues via Inhibition of Cathepsin L. J. Med. Chem. 2022, 65, 2956–2970. [Google Scholar] [CrossRef] [PubMed]
- Ahmmed, M.K.; Bhowmik, S.; Giteru, S.G.; Zilani, M.N.H.; Adadi, P.; Islam, S.S.; Kanwugu, O.N.; Haq, M.; Ahmmed, F.; Ng, C.C.W.; et al. An Update of Lectins from Marine Organisms: Characterization, Extraction Methodology, and Potential Biofunctional Applications. Mar. Drugs 2022, 20, 430. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, R.P.; Kumar, I.; Yadav, P.; Singh, S.K.; Kaushalendra; Singh, P.K.; Gupta, R.K.; Singh, S.M.; Kesawat, M.S.; et al. Algal Metabolites Can Be an Immune Booster against COVID-19 Pandemic. Antioxidants 2022, 11, 452. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Xu, W.; Gu, C.; Cai, X.; Qu, D.; Lu, L.; Xie, Y.; Jiang, S. Griffithsin with A Broad-Spectrum Antiviral Activity by Binding Glycans in Viral Glycoprotein Exhibits Strong Synergistic Effect in Combination with A Pan-Coronavirus Fusion Inhibitor Targeting SARS-CoV-2 Spike S2 Subunit. Virol. Sin. 2020, 35, 857–860. [Google Scholar] [CrossRef]
- Pendyala, B.; Patras, A. In Silico Screening of Food Bioactive Compounds to Predict Potential Inhibitors of COVID-19 Main Protease (Mpro) and RNA-Dependent RNA Polymerase (RdRp). ChemRxiv 2020. [Google Scholar] [CrossRef]
- Petit, L.; Vernès, L.; Cadoret, J.P. Docking and in Silico Toxicity Assessment of Arthrospira Compounds as Potential Antiviral Agents against SARS-CoV-2. J. Appl. Phycol. 2021, 33, 1579–1602. [Google Scholar] [CrossRef]
- Hirata, T.; Tanaka, M.; Ooike, M.; Tsunomura, T.; Sakaguchi, M. Antioxidant Activities of Phycocyanobilin Prepared from Spirulina Platensis. J. Appl. Phycol. 2000, 12, 435–439. [Google Scholar] [CrossRef]
- Raj, K.T.; Ranjithkumar, R.; Kanthesh, B.M.; Gopenath, T.S. Algal Metabolites Can Be an Immune Booster against COVID-19 Pandemic. Int. J. Pharm. Sci. Res. 2020, 11, 4271–4278. [Google Scholar] [CrossRef]
- Toelzer, C.; Gupta, K.; Yadav, S.K.N.; Borucu, U.; Davidson, A.D.; Williamson, M.K.; Shoemark, D.K.; Garzoni, F.; Staufer, O.; Milligan, R.; et al. Free Fatty Acid Binding Pocket in the Locked Structure of SARS-CoV-2 Spike Protein. Science 2020, 370, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Zahran, E.M.; Albohy, A.; Khalil, A.; Ibrahim, A.H.; Ahmed, H.A.; El-Hossary, E.M.; Bringmann, G.; Abdelmohsen, U.R. Bioactivity Potential of Marine Natural Products from Scleractinia-Associated Microbes and In Silico Anti-SARS-CoV-2 Evaluation. Mar. Drugs 2020, 18, 645. [Google Scholar] [CrossRef] [PubMed]
- Tassakka, A.C.M.A.R.; Sumule, O.; Massi, M.N.; Sulfahri; Manggau, M.; Iskandar, I.W.; Alam, J.F.; Permana, A.D.; Liao, L.M. Potential Bioactive Compounds as SARS-CoV-2 Inhibitors from Extracts of the Marine Red Alga Halymenia Durvillei (Rhodophyta)—A Computational Study. Arab. J. Chem. 2021, 14, 103393. [Google Scholar] [CrossRef] [PubMed]
- Aatif, M.; Muteeb, G.; Alsultan, A.; Alshoaibi, A.; Khelif, B.Y. Dieckol and Its Derivatives as Potential Inhibitors of SARS-CoV-2 Spike Protein (Uk Strain: VUI 202012/01): A Computational Study. Mar. Drugs 2021, 19, 242. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.T.; Ali, A.; Wang, Q.; Irfan, M.; Khan, A.; Zeb, M.T.; Zhang, Y.-J.; Chinnasamy, S.; Wei, D.-Q. Marine Natural Compounds as Potents Inhibitors against the Main Protease of SARS-CoV-2—A Molecular Dynamic Study. J. Biomol. Struct. Dyn. 2020, 39, 3627–3637. [Google Scholar] [CrossRef] [PubMed]
- Youssef, F.S.; Alshammari, E.; Ashour, M.L. Bioactive Alkaloids from Genus Aspergillus: Mechanistic Interpretation of Their Antimicrobial and Potential SARS-CoV-2 Inhibitory Activity Using Molecular Modelling. Int. J. Mol. Sci. 2021, 22, 1866. [Google Scholar] [CrossRef]
- Muteeb, G.; Alshoaibi, A.; Aatif, M.; Rehman, M.T.; Qayyum, M.Z. Screening Marine Algae Metabolites as High-Affinity Inhibitors of SARS-CoV-2 Main Protease (3CLpro): An in Silico Analysis to Identify Novel Drug Candidates to Combat COVID-19 Pandemic. Appl. Biol. Chem. 2020, 63, 79. [Google Scholar] [CrossRef]
- Abdelrheem, D.A.; Ahmed, S.A.; Abd El-Mageed, H.R.; Mohamed, H.S.; Rahman, A.A.; Elsayed, K.N.M.; Ahmed, S.A. The Inhibitory Effect of Some Natural Bioactive Compounds against SARS-CoV-2 Main Protease: Insights from Molecular Docking Analysis and Molecular Dynamic Simulation. J. Environ. Sci. Health Part. A 2020, 55, 1373–1386. [Google Scholar] [CrossRef]
- Gaudêncio, S.P.; Pereira, F. A Computer-Aided Drug Design Approach to Predict Marine Drug-Like Leads for SARS-CoV-2 Main Protease Inhibition. Mar. Drugs 2020, 18, 633. [Google Scholar] [CrossRef]
- Kumar, V.; Parate, S.; Yoon, S.; Lee, G.; Lee, K.W. Computational Simulations Identified Marine-Derived Natural Bioactive Compounds as Replication Inhibitors of SARS-CoV-2. Front. Microbiol. 2021, 12, 647295. [Google Scholar] [CrossRef] [PubMed]
- Sepay, N.; Sekar, A.; Halder, U.C.; Alarifi, A.; Afzal, M. Anti-COVID-19 Terpenoid from Marine Sources: A Docking, Admet and Molecular Dynamics Study. J. Mol. Struct. 2021, 1228, 129433. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N.; Basharat, Z.; Yousuf, M.; Castaldo, G.; Rastrelli, L.; Khan, H. Virtual Screening of Natural Products against Type II Transmembrane Serine Protease (TMPRSS2), the Priming Agent of Coronavirus 2 (SARS-CoV-2). Molecules 2020, 25, 2271. [Google Scholar] [CrossRef]
- Bharathi, M.; Sivamaruthi, B.S.; Kesika, P.; Thangaleela, S.; Chaiyasut, C. In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer. Mar. Drugs 2022, 20, 148. [Google Scholar] [CrossRef] [PubMed]
- Ambarwati, R.; Rahayu, D.A.; Khaleyla, F.; Wisanti; Putri, E.K. Angiotensin-Converting Enzyme 2 (ACE2) of Marine Biota: A Preliminary Study of Potential Therapy for SARS-CoV-2 Infectiontle. J. Phys. Conf. Ser. 2020; in press. [Google Scholar] [CrossRef]
- Fayed, M.A.A.; El-Behairy, M.F.; Abdallah, I.A.; Abdel-Bar, H.M.; Elimam, H.; Mostafa, A.; Moatasim, Y.; Abouzid, K.A.M.; Elshaier, Y.A.M.M. Structure- and Ligand-Based in Silico Studies towards the Repurposing of Marine Bioactive Compounds to Target SARS-CoV-2. Arab. J. Chem. 2021, 14, 103092. [Google Scholar] [CrossRef] [PubMed]
- Srivastav, A.K.; Jaiswal, J.; Kumar, U. In Silico Bioprospecting of Antiviral Compounds from Marine Fungi and Mushroom for Rapid Development of Nutraceuticals against SARS-CoV-2. J. Biomol. Struct. Dyn. 2021, 41, 1574–1585. [Google Scholar] [CrossRef]
- Neufurth, M.; Wang, X.; Tolba, E.; Lieberwirth, I.; Wang, S.; Schröder, H.C.; Müller, W.E.G. The Inorganic Polymer, Polyphosphate, Blocks Binding of SARS-CoV-2 Spike Protein to ACE2 Receptor at Physiological Concentrations. Biochem. Pharmacol. 2020, 182, 114215. [Google Scholar] [CrossRef]
- Geahchan, S.; Ehrlich, H.; Rahman, M.A. The Anti-Viral Applications of Marine Resources for COVID-19 Treatment: An Overview. Mar. Drugs 2021, 19, 409. [Google Scholar] [CrossRef]
- Müller, W.E.G.; Neufurth, M.; Wang, S.; Tan, R.; Schröder, H.C.; Wang, X. Morphogenetic (Mucin Expression) as Well as Potential Anti-Corona Viral Activity of the Marine Secondary Metabolite Polyphosphate on A549 Cells. Mar. Drugs 2020, 18, 639. [Google Scholar] [CrossRef]
- Neufurth, M.; Wang, X.; Wang, S.; Schröder, H.C.; Müller, W.E.G. Caged Dexamethasone/Quercetin Nanoparticles, Formed of the Morphogenetic Active Inorganic Polyphosphate, Are Strong Inducers of MUC5AC. Mar. Drugs 2021, 19, 64. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).