Gut Microbiota Metabolites: Unveiling Their Role in Inflammatory Bowel Diseases and Fibrosis
Abstract
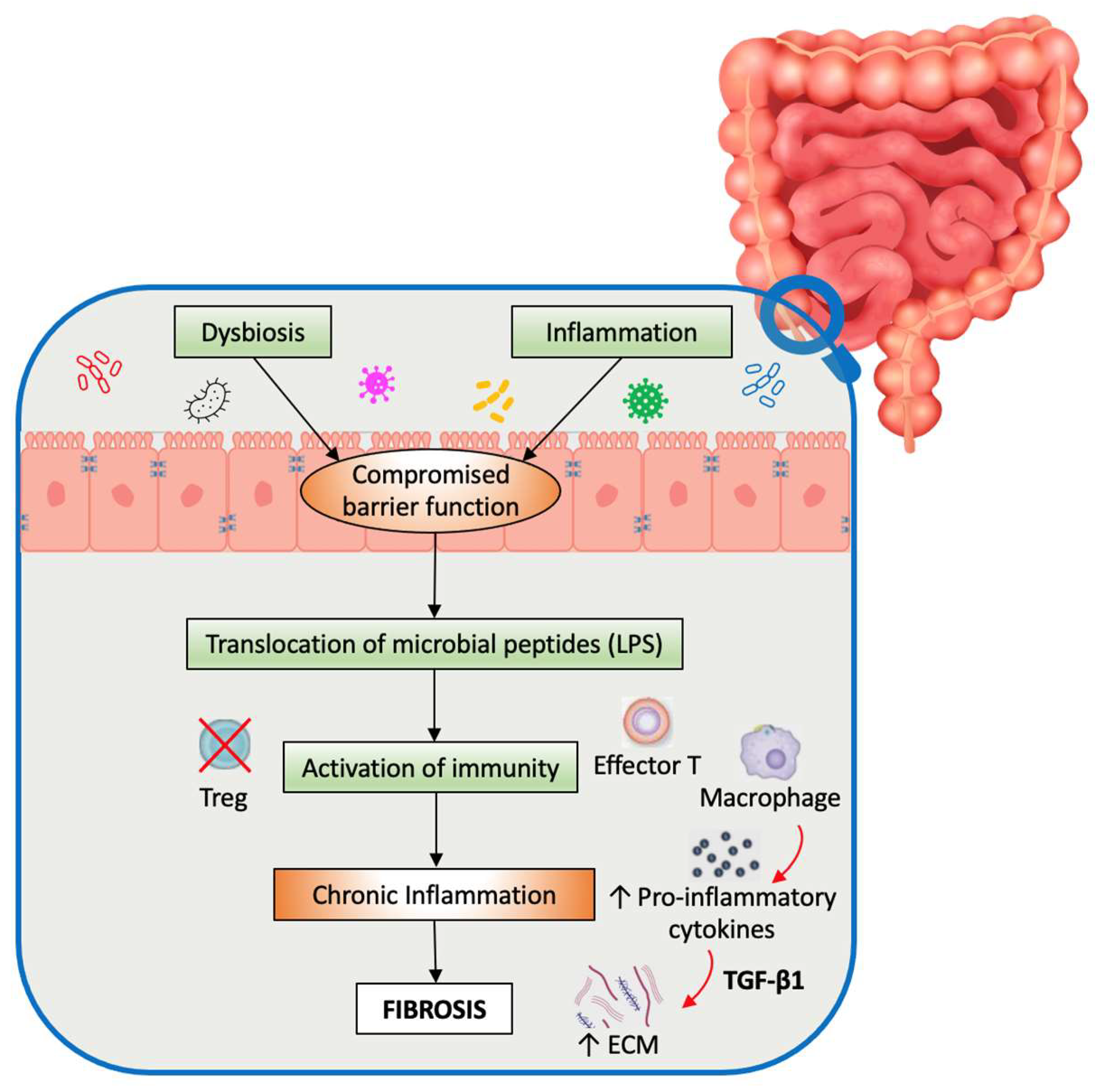
1. Introduction
2. Microbiota Metabolites
2.1. Short-Chain Fatty Acids (SCFAs)
SCFAs and IBD
| SCFAs: Beneficial Effects | SCFAs: Reduction in IBD |
|---|---|
|
2.2. Bile Acids
Bile Acids in IBD
| Bile Acids: Beneficial Effects | Bile Acids in IBD: ↓ Secondary BA, ↑ Primary BA |
|---|---|
|
2.3. Tryptophan
Tryptophan in IBD
| Tryptophan: Beneficial Effects | Tryptophan: Reduction in IBD |
|---|---|
| Development of or aggravating disease activity by: |
3. Microbiota-Induced Fibrosis
4. Treatment and Therapeutic Perspectives
4.1. SCFAs
4.2. Tryptophan
4.3. Bile Acids
4.4. Fibrosis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Park, S.J.; Kim, W.H.; Cheon, J.H. Clinical Characteristics and Treatment of Inflammatory Bowel Disease: A Comparison of Eastern and Western Perspectives. World J. Gastroenterol. WJG 2014, 20, 11525–11537. [Google Scholar] [CrossRef]
- Dong, L.-N.; Wang, M.; Guo, J.; Wang, J.-P. Role of Intestinal Microbiota and Metabolites in Inflammatory Bowel Disease. Chin. Med. J. 2019, 132, 1610–1614. [Google Scholar] [CrossRef]
- Kaplan, G.G.; Ng, S.C. Understanding and Preventing the Global Increase of Inflammatory Bowel Disease. Gastroenterology 2017, 152, 313–321.e2. [Google Scholar] [CrossRef]
- Turpin, W.; Goethel, A.; Bedrani, L.; Croitoru Mdcm, K. Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm. Bowel Dis. 2018, 24, 1133–1148. [Google Scholar] [CrossRef] [PubMed]
- Andersen, V.; Olsen, A.; Carbonnel, F.; Tjønneland, A.; Vogel, U. Diet and Risk of Inflammatory Bowel Disease. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2012, 44, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Parigi, T.L.; Vieujean, S.; Paridaens, K.; Dalgaard, K.; Peyrin-Biroulet, L.; Danese, S. Efficacy, Safety, and Concerns on Microbiota Modulation, Antibiotics, Probiotics, and Fecal Microbial Transplant for Inflammatory Bowel Disease and Other Gastrointestinal Conditions: Results from an International Survey. Microorganisms 2023, 11, 2806. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Gong, X.; Wang, L.; Yu, X.; Dong, Q. Involvement of Reduced Microbial Diversity in Inflammatory Bowel Disease. Gastroenterol. Res. Pract. 2016, 2016, 6951091. [Google Scholar] [CrossRef] [PubMed]
- Postler, T.S.; Ghosh, S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; He, J.; Shen, Y.; Zhang, C.; Wang, J.; Chen, Y. New Frontiers in Genetics, Gut Microbiota, and Immunity: A Rosetta Stone for the Pathogenesis of Inflammatory Bowel Disease. BioMed Res. Int. 2017, 2017, 8201672. [Google Scholar] [CrossRef]
- Alipour, M.; Zaidi, D.; Valcheva, R.; Jovel, J.; Martínez, I.; Sergi, C.; Walter, J.; Mason, A.L.; Wong, G.K.-S.; Dieleman, L.A.; et al. Mucosal Barrier Depletion and Loss of Bacterial Diversity Are Primary Abnormalities in Paediatric Ulcerative Colitis. J. Crohns Colitis 2016, 10, 462–471. [Google Scholar] [CrossRef]
- Rieder, F. The Gut Microbiome in Intestinal Fibrosis: Environmental Protector or Provocateur? Sci. Transl. Med. 2013, 5, 190ps10. [Google Scholar] [CrossRef]
- Friedman, S.L.; Sheppard, D.; Duffield, J.S.; Violette, S. Therapy for Fibrotic Diseases: Nearing the Starting Line. Sci. Transl. Med. 2013, 5, 167sr1. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Fiocchi, C. Intestinal Fibrosis in IBD--a Dynamic, Multifactorial Process. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Zimmermann, E.M.; Remzi, F.H.; Sandborn, W.J. Crohn’s Disease Complicated by Strictures: A Systematic Review. Gut 2013, 62, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Armuzzi, A.; Van Assche, G.; Reinisch, W.; Pineton de Chambrun, G.; Griffiths, A.; Sladek, M.; Preiss, J.C.; Lukas, M.; D’Haens, G. Results of the 2nd Scientific Workshop of the ECCO (IV): Therapeutic Strategies to Enhance Intestinal Healing in Inflammatory Bowel Disease. J. Crohns Colitis 2012, 6, 492–502. [Google Scholar] [CrossRef]
- Peyrin-Biroulet, L.; Oussalah, A.; Williet, N.; Pillot, C.; Bresler, L.; Bigard, M.-A. Impact of Azathioprine and Tumour Necrosis Factor Antagonists on the Need for Surgery in Newly Diagnosed Crohn’s Disease. Gut 2011, 60, 930–936. [Google Scholar] [CrossRef]
- Velázquez, M.; Davies, C.; Marett, R.; Slavin, J.L.; Feirtag, J.M. Effect of Oligosaccharides and Fibre Substitutes on Short-Chain Fatty Acid Production by Human Faecal Microflora. Anaerobe 2000, 6, 87–92. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children from Europe and Rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.W.; de Souza, R.; Kendall, C.W.C.; Emam, A.; Jenkins, D.J.A. Colonic Health: Fermentation and Short Chain Fatty Acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota Metabolite Short Chain Fatty Acids, GPCR, and Inflammatory Bowel Diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short Chain Fatty Acids in Human Large Intestine, Portal, Hepatic and Venous Blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzman, I.R.; Lin, J. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Scheppach, W. Effects of Short Chain Fatty Acids on Gut Morphology and Function. Gut 1994, 35, S35–S38. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.A.; Jackson, J.; Stanton, M.; Rojas-Triana, A.; Bober, L.; Laverty, M.; Yang, X.; Zhu, F.; Liu, J.; Wang, S.; et al. Short-Chain Fatty Acids Act as Antiinflammatory Mediators by Regulating Prostaglandin E2 and Cytokines. World J. Gastroenterol. WJG 2009, 15, 5549–5557. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Babaei, P.; Ji, B.; Nielsen, J. Human Gut Microbiota and Healthy Aging: Recent Developments and Future Prospective. Nutr. Healthy Aging 2016, 4, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St. Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-Phylogenetic Characterization of Microbial Community Imbalances in Human Inflammatory Bowel Diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Martinez-Medina, M.; Aldeguer, X.; Gonzalez-Huix, F.; Acero, D.; Garcia-Gil, L.J. Abnormal Microbiota Composition in the Ileocolonic Mucosa of Crohn’s Disease Patients as Revealed by Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis. Inflamm. Bowel Dis. 2006, 12, 1136–1145. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut Microbiota, Metabolites and Host Immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-Cell Generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Wrzosek, L.; Miquel, S.; Noordine, M.-L.; Bouet, S.; Joncquel Chevalier-Curt, M.; Robert, V.; Philippe, C.; Bridonneau, C.; Cherbuy, C.; Robbe-Masselot, C.; et al. Bacteroides Thetaiotaomicron and Faecalibacterium Prausnitzii Influence the Production of Mucus Glycans and the Development of Goblet Cells in the Colonic Epithelium of a Gnotobiotic Model Rodent. BMC Biol. 2013, 11, 61. [Google Scholar] [CrossRef]
- Willemsen, L.E.M.; Koetsier, M.A.; van Deventer, S.J.H.; van Tol, E.a.F. Short Chain Fatty Acids Stimulate Epithelial Mucin 2 Expression through Differential Effects on Prostaglandin E(1) and E(2) Production by Intestinal Myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Gaudier, E.; Jarry, A.; Blottière, H.M.; de Coppet, P.; Buisine, M.P.; Aubert, J.P.; Laboisse, C.; Cherbut, C.; Hoebler, C. Butyrate Specifically Modulates MUC Gene Expression in Intestinal Epithelial Goblet Cells Deprived of Glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G1168–G1174. [Google Scholar] [CrossRef]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-Sensing Receptors GPR43 and GPR109A Facilitate Dietary Fibre-Induced Gut Homeostasis through Regulation of the Inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria Can Protect from Enteropathogenic Infection through Production of Acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium Prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A Decrease of the Butyrate-Producing Species Roseburia Hominis and Faecalibacterium Prausnitzii Defines Dysbiosis in Patients with Ulcerative Colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Holmes, E.; Khan, F.; Kochhar, S.; Scanlan, P.; Shanahan, F.; Wilson, I.D.; Wang, Y. Rapid and Noninvasive Metabonomic Characterization of Inflammatory Bowel Disease. J. Proteome Res. 2007, 6, 546–551. [Google Scholar] [CrossRef]
- Vernia, P.; Gnaedinger, A.; Hauck, W.; Breuer, R.I. Organic Anions and the Diarrhea of Inflammatory Bowel Disease. Dig. Dis. Sci. 1988, 33, 1353–1358. [Google Scholar] [CrossRef]
- Takaishi, H.; Matsuki, T.; Nakazawa, A.; Takada, T.; Kado, S.; Asahara, T.; Kamada, N.; Sakuraba, A.; Yajima, T.; Higuchi, H.; et al. Imbalance in Intestinal Microflora Constitution Could Be Involved in the Pathogenesis of Inflammatory Bowel Disease. Int. J. Med. Microbiol. 2008, 298, 463–472. [Google Scholar] [CrossRef]
- Louis, P.; Duncan, S.H.; McCrae, S.I.; Millar, J.; Jackson, M.S.; Flint, H.J. Restricted Distribution of the Butyrate Kinase Pathway among Butyrate-Producing Bacteria from the Human Colon. J. Bacteriol. 2004, 186, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Laserna-Mendieta, E.J.; Clooney, A.G.; Carretero-Gomez, J.F.; Moran, C.; Sheehan, D.; Nolan, J.A.; Hill, C.; Gahan, C.G.M.; Joyce, S.A.; Shanahan, F.; et al. Determinants of Reduced Genetic Capacity for Butyrate Synthesis by the Gut Microbiome in Crohn’s Disease and Ulcerative Colitis. J. Crohns Colitis 2018, 12, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut Microbiome Structure and Metabolic Activity in Inflammatory Bowel Disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Le Gall, G.; Noor, S.O.; Ridgway, K.; Scovell, L.; Jamieson, C.; Johnson, I.T.; Colquhoun, I.J.; Kemsley, E.K.; Narbad, A. Metabolomics of Fecal Extracts Detects Altered Metabolic Activity of Gut Microbiota in Ulcerative Colitis and Irritable Bowel Syndrome. J. Proteome Res. 2011, 10, 4208–4218. [Google Scholar] [CrossRef] [PubMed]
- Vernia, P.; Caprilli, R.; Latella, G.; Barbetti, F.; Magliocca, F.M.; Cittadini, M. Fecal Lactate and Ulcerative Colitis. Gastroenterology 1988, 95, 1564–1568. [Google Scholar] [CrossRef] [PubMed]
- Hove, H.; Mortensen, P.B. Influence of Intestinal Inflammation (IBD) and Small and Large Bowel Length on Fecal Short-Chain Fatty Acids and Lactate. Dig. Dis. Sci. 1995, 40, 1372–1380. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of Inflammatory Responses by Gut Microbiota and Chemoattractant Receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Tye, H.; Yu, C.-H.; Simms, L.A.; de Zoete, M.R.; Kim, M.L.; Zakrzewski, M.; Penington, J.S.; Harapas, C.R.; Souza-Fonseca-Guimaraes, F.; Wockner, L.F.; et al. NLRP1 Restricts Butyrate Producing Commensals to Exacerbate Inflammatory Bowel Disease. Nat. Commun. 2018, 9, 3728. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal Microbe-Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the Gut Microbiota in Disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L. Bile Acids: Regulation of Synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.W. The Enzymes, Regulation, and Genetics of Bile Acid Synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef]
- Schaap, F.G.; Trauner, M.; Jansen, P.L.M. Bile Acid Receptors as Targets for Drug Development. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 55–67. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Boney-Montoya, J.; Choi, M.; He, T.; Sunny, N.E.; Satapati, S.; Suino-Powell, K.; Xu, H.E.; Gerard, R.D.; Finck, B.N.; et al. FGF15/19 Regulates Hepatic Glucose Metabolism by Inhibiting the CREB-PGC-1α Pathway. Cell Metab. 2011, 13, 729–738. [Google Scholar] [CrossRef]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El Alam, G.; Bou Sleiman, M.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile Acids Signal via TGR5 to Activate Intestinal Stem Cells and Epithelial Regeneration. Gastroenterology 2020, 159, 956–968.e8. [Google Scholar] [CrossRef]
- Wang, Y.-D.; Chen, W.-D.; Yu, D.; Forman, B.M.; Huang, W. The G-Protein-Coupled Bile Acid Receptor, Gpbar1 (TGR5), Negatively Regulates Hepatic Inflammatory Response through Antagonizing Nuclear Factor κ Light-Chain Enhancer of Activated B Cells (NF-κB) in Mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef]
- Keitel, V.; Donner, M.; Winandy, S.; Kubitz, R.; Häussinger, D. Expression and Function of the Bile Acid Receptor TGR5 in Kupffer Cells. Biochem. Biophys. Res. Commun. 2008, 372, 78–84. [Google Scholar] [CrossRef]
- Huang, W.; Ma, K.; Zhang, J.; Qatanani, M.; Cuvillier, J.; Liu, J.; Dong, B.; Huang, X.; Moore, D.D. Nuclear Receptor-Dependent Bile Acid Signaling Is Required for Normal Liver Regeneration. Science 2006, 312, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Jain, U.; Lai, C.-W.; Xiong, S.; Goodwin, V.M.; Lu, Q.; Muegge, B.D.; Christophi, G.P.; VanDussen, K.L.; Cummings, B.P.; Young, E.; et al. Temporal Regulation of the Bacterial Metabolite Deoxycholate during Colonic Repair Is Critical for Crypt Regeneration. Cell Host Microbe 2018, 24, 353–363.e5. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.M.; Marchesi, J.R. Functional and Comparative Metagenomic Analysis of Bile Salt Hydrolase Activity in the Human Gut Microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [PubMed]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G.M. Regulation of Host Weight Gain and Lipid Metabolism by Bacterial Bile Acid Modification in the Gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef]
- Inagaki, T.; Moschetta, A.; Lee, Y.-K.; Peng, L.; Zhao, G.; Downes, M.; Yu, R.T.; Shelton, J.M.; Richardson, J.A.; Repa, J.J.; et al. Regulation of Antibacterial Defense in the Small Intestine by the Nuclear Bile Acid Receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 3920–3925. [Google Scholar] [CrossRef]
- Lorenzo-Zúñiga, V.; Bartolí, R.; Planas, R.; Hofmann, A.F.; Viñado, B.; Hagey, L.R.; Hernández, J.M.; Mañé, J.; Alvarez, M.A.; Ausina, V.; et al. Oral Bile Acids Reduce Bacterial Overgrowth, Bacterial Translocation, and Endotoxemia in Cirrhotic Rats. Hepatology 2003, 37, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Kurdi, P.; Kawanishi, K.; Mizutani, K.; Yokota, A. Mechanism of Growth Inhibition by Free Bile Acids in Lactobacilli and Bifidobacteria. J. Bacteriol. 2006, 188, 1979–1986. [Google Scholar] [CrossRef]
- D’Aldebert, E.; Biyeyeme Bi Mve, M.-J.; Mergey, M.; Wendum, D.; Firrincieli, D.; Coilly, A.; Fouassier, L.; Corpechot, C.; Poupon, R.; Housset, C.; et al. Bile Salts Control the Antimicrobial Peptide Cathelicidin through Nuclear Receptors in the Human Biliary Epithelium. Gastroenterology 2009, 136, 1435–1443. [Google Scholar] [CrossRef]
- Termén, S.; Tollin, M.; Rodriguez, E.; Sveinsdóttir, S.H.; Jóhannesson, B.; Cederlund, A.; Sjövall, J.; Agerberth, B.; Gudmundsson, G.H. PU. 1 and Bacterial Metabolites Regulate the Human Gene CAMP Encoding Antimicrobial Peptide LL-37 in Colon Epithelial Cells. Mol. Immunol. 2008, 45, 3947–3955. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.L.; Renooij, W.; Murzilli, S.; Klomp, L.W.J.; Siersema, P.D.; Schipper, M.E.I.; Danese, S.; et al. Farnesoid X Receptor Activation Inhibits Inflammation and Preserves the Intestinal Barrier in Inflammatory Bowel Disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.B.M.S.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile Acid Is a Host Factor That Regulates the Composition of the Cecal Microbiota in Rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; et al. Precision Microbiome Reconstitution Restores Bile Acid Mediated Resistance to Clostridium Difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting Dysbiosis, Bile-Acid Dysmetabolism and Gut Inflammation in Inflammatory Bowel Diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Battat, R.; Scherl, E.J.; Lukin, D.; Charilaou, P.; Mahtani, P.; Gerber, J.; Gandara, J.A.; Dündar, F.; Zumbo, P.; Betel, D.; et al. Increased Primary Bile Acids with Ileocolonic Resection Impact Ileal Inflammation and Gut Microbiota in Inflammatory Bowel Disease. J. Crohns Colitis 2022, 17, 795–803. [Google Scholar] [CrossRef] [PubMed]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The Bile Acid Receptor FXR Is a Modulator of Intestinal Innate Immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef]
- Biagioli, M.; Carino, A.; Cipriani, S.; Francisci, D.; Marchianò, S.; Scarpelli, P.; Sorcini, D.; Zampella, A.; Fiorucci, S. The Bile Acid Receptor GPBAR1 Regulates the M1/M2 Phenotype of Intestinal Macrophages and Activation of GPBAR1 Rescues Mice from Murine Colitis. J. Immunol. 2017, 199, 718–733. [Google Scholar] [CrossRef]
- Badawy, A.A.-B. Kynurenine Pathway of Tryptophan Metabolism: Regulatory and Functional Aspects. Int. J. Tryptophan Res. 2017, 10, 1178646917691938. [Google Scholar] [CrossRef]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s Metabolites in Exercise, Inflammation, and Mental Health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef]
- Côté, F.; Thévenot, E.; Fligny, C.; Fromes, Y.; Darmon, M.; Ripoche, M.-A.; Bayard, E.; Hanoun, N.; Saurini, F.; Lechat, P.; et al. Disruption of the Nonneuronal Tph1 Gene Demonstrates the Importance of Peripheral Serotonin in Cardiac Function. Proc. Natl. Acad. Sci. USA 2003, 100, 13525–13530. [Google Scholar] [CrossRef]
- Qiu, J.; Heller, J.J.; Guo, X.; Chen, Z.E.; Fish, K.; Fu, Y.-X.; Zhou, L. The Aryl Hydrocarbon Receptor Regulates Gut Immunity through Modulation of Innate Lymphoid Cells. Immunity 2012, 36, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Zenewicz, L.A.; Yancopoulos, G.D.; Valenzuela, D.M.; Murphy, A.J.; Stevens, S.; Flavell, R.A. Innate and Adaptive Interleukin-22 Protects Mice from Inflammatory Bowel Disease. Immunity 2008, 29, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl Hydrocarbon Receptor-Induced Signals up-Regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterology 2011, 141, 237–248, 248.e1. [Google Scholar] [CrossRef] [PubMed]
- Metidji, A.; Omenetti, S.; Crotta, S.; Li, Y.; Nye, E.; Ross, E.; Li, V.; Maradana, M.R.; Schiering, C.; Stockinger, B. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity 2018, 49, 353–362.e5. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Innocentin, S.; Withers, D.R.; Roberts, N.A.; Gallagher, A.R.; Grigorieva, E.F.; Wilhelm, C.; Veldhoen, M. Exogenous Stimuli Maintain Intraepithelial Lymphocytes via Aryl Hydrocarbon Receptor Activation. Cell 2011, 147, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- Desbonnet, L.; Clarke, G.; Traplin, A.; O’Sullivan, O.; Crispie, F.; Moloney, R.D.; Cotter, P.D.; Dinan, T.G.; Cryan, J.F. Gut Microbiota Depletion from Early Adolescence in Mice: Implications for Brain and Behaviour. Brain. Behav. Immun. 2015, 48, 165–173. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F.; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut Microbes Promote Colonic Serotonin Production through an Effect of Short-Chain Fatty Acids on Enterochromaffin Cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 1395–1403. [Google Scholar] [CrossRef]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Häsler, R.; et al. Increased Tryptophan Metabolism Is Associated With Activity of Inflammatory Bowel Diseases. Gastroenterology 2017, 153, 1504–1516.e2. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 Impacts Colitis by Altering Gut Microbiota Metabolism of Tryptophan into Aryl Hydrocarbon Receptor Ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 Links Amino Acid Malnutrition to Microbial Ecology and Intestinal Inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Takamura, T.; Harama, D.; Fukumoto, S.; Nakamura, Y.; Shimokawa, N.; Ishimaru, K.; Ikegami, S.; Makino, S.; Kitamura, M.; Nakao, A. Lactobacillus Bulgaricus OLL1181 Activates the Aryl Hydrocarbon Receptor Pathway and Inhibits Colitis. Immunol. Cell Biol. 2011, 89, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef] [PubMed]
- Karakan, T.; Karatas, A.; Cindoruk, M.; Gulbahar, O. P306 Serum Tryptophan Metabolites as a Biomarker for Disease Severity in Patients with IBD. J. Crohns Colitis 2023, 17, i450. [Google Scholar] [CrossRef]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function via the Xenobiotic Sensor PXR and Toll-like Receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of Fibrosis: Therapeutic Translation for Fibrotic Disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef]
- Wynn, T.A. Cellular and Molecular Mechanisms of Fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef]
- Latella, G.; Rogler, G.; Bamias, G.; Breynaert, C.; Florholmen, J.; Pellino, G.; Reif, S.; Speca, S.; Lawrance, I.C. Results of the 4th Scientific Workshop of the ECCO (I): Pathophysiology of Intestinal Fibrosis in IBD. J. Crohns Colitis 2014, 8, 1147–1165. [Google Scholar] [CrossRef]
- Fiocchi, C.; Lund, P.K. Themes in Fibrosis and Gastrointestinal Inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G677–G683. [Google Scholar] [CrossRef]
- Burke, J.P.; Mulsow, J.J.; O’Keane, C.; Docherty, N.G.; Watson, R.W.G.; O’Connell, P.R. Fibrogenesis in Crohn’s Disease. Am. J. Gastroenterol. 2007, 102, 439–448. [Google Scholar] [CrossRef]
- Mourelle, M.; Salas, A.; Guarner, F.; Crespo, E.; García-Lafuente, A.; Malagelada, J.R. Stimulation of Transforming Growth Factor Beta1 by Enteric Bacteria in the Pathogenesis of Rat Intestinal Fibrosis. Gastroenterology 1998, 114, 519–526. [Google Scholar] [CrossRef]
- Mow, W.S.; Vasiliauskas, E.A.; Lin, Y.-C.; Fleshner, P.R.; Papadakis, K.A.; Taylor, K.D.; Landers, C.J.; Abreu-Martin, M.T.; Rotter, J.I.; Yang, H.; et al. Association of Antibody Responses to Microbial Antigens and Complications of Small Bowel Crohn’s Disease. Gastroenterology 2004, 126, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Liew, F.Y.; Xu, D.; Brint, E.K.; O’Neill, L.A.J. Negative Regulation of Toll-like Receptor-Mediated Immune Responses. Nat. Rev. Immunol. 2005, 5, 446–458. [Google Scholar] [CrossRef]
- Dauphinee, S.M.; Karsan, A. Lipopolysaccharide Signaling in Endothelial Cells. Lab. Investig. J. Tech. Methods Pathol. 2006, 86, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.P.; Cunningham, M.F.; Watson, R.W.G.; Docherty, N.G.; Coffey, J.C.; O’Connell, P.R. Bacterial Lipopolysaccharide Promotes Profibrotic Activation of Intestinal Fibroblasts. Br. J. Surg. 2010, 97, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Sazuka, S.; Katsuno, T.; Nakagawa, T.; Saito, M.; Saito, K.; Maruoka, D.; Matsumura, T.; Arai, M.; Miyauchi, H.; Matsubara, H.; et al. Fibrocytes Are Involved in Inflammation as Well as Fibrosis in the Pathogenesis of Crohn’s Disease. Dig. Dis. Sci. 2014, 59, 760–768. [Google Scholar] [CrossRef] [PubMed]
- van Tol, E.A.; Holt, L.; Li, F.L.; Kong, F.M.; Rippe, R.; Yamauchi, M.; Pucilowska, J.; Lund, P.K.; Sartor, R.B. Bacterial Cell Wall Polymers Promote Intestinal Fibrosis by Direct Stimulation of Myofibroblasts. Am. J. Physiol. 1999, 277, G245–G255. [Google Scholar] [CrossRef] [PubMed]
- Jacob, N.; Jacobs, J.P.; Kumagai, K.; Ha, C.W.Y.; Kanazawa, Y.; Lagishetty, V.; Altmayer, K.; Hamill, A.M.; Von Arx, A.; Sartor, R.B.; et al. Inflammation-Independent TL1A-Mediated Intestinal Fibrosis Is Dependent on the Gut Microbiome. Mucosal Immunol. 2018, 11, 1466–1476. [Google Scholar] [CrossRef] [PubMed]
- Grassl, G.A.; Valdez, Y.; Bergstrom, K.S.B.; Vallance, B.A.; Finlay, B.B. Chronic Enteric Salmonella Infection in Mice Leads to Severe and Persistent Intestinal Fibrosis. Gastroenterology 2008, 134, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Small, C.-L.N.; Reid-Yu, S.A.; McPhee, J.B.; Coombes, B.K. Persistent Infection with Crohn’s Disease-Associated Adherent-Invasive Escherichia Coli Leads to Chronic Inflammation and Intestinal Fibrosis. Nat. Commun. 2013, 4, 1957. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A Frameshift Mutation in NOD2 Associated with Susceptibility to Crohn’s Disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 Leucine-Rich Repeat Variants with Susceptibility to Crohn’s Disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 Is a General Sensor of Peptidoglycan through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Kamada, N. Contribution of the Gut Microbiota to Intestinal Fibrosis in Crohn’s Disease. Front. Med. 2022, 9, 826240. [Google Scholar] [CrossRef] [PubMed]
- Büning, C.; Genschel, J.; Bühner, S.; Krüger, S.; Kling, K.; Dignass, A.; Baier, P.; Bochow, B.; Ockenga, J.; Schmidt, H.H.-J.; et al. Mutations in the NOD2/CARD15 Gene in Crohn’s Disease Are Associated with Ileocecal Resection and Are a Risk Factor for Reoperation. Aliment. Pharmacol. Ther. 2004, 19, 1073–1078. [Google Scholar] [CrossRef]
- Adler, J.; Rangwalla, S.C.; Dwamena, B.A.; Higgins, P.D.R. The Prognostic Power of the NOD2 Genotype for Complicated Crohn’s Disease: A Meta-Analysis. Am. J. Gastroenterol. 2011, 106, 699–712. [Google Scholar] [CrossRef]
- McKaig, B.C.; Hughes, K.; Tighe, P.J.; Mahida, Y.R. Differential Expression of TGF-Beta Isoforms by Normal and Inflammatory Bowel Disease Intestinal Myofibroblasts. Am. J. Physiol. Cell Physiol. 2002, 282, C172–C182. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg Induction by a Rationally Selected Mixture of Clostridia Strains from the Human Microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Oka Akihiko|Researcher Information|J-GLOBAL. Available online: https://jglobal.jst.go.jp/en/detail?JGLOBAL_ID=201301093268253326 (accessed on 5 January 2024).
- Henn, M.R.; O’Brien, E.J.; Diao, L.; Feagan, B.G.; Sandborn, W.J.; Huttenhower, C.; Wortman, J.R.; McGovern, B.H.; Wang-Weigand, S.; Lichter, D.I.; et al. A Phase 1b Safety Study of SER-287, a Spore-Based Microbiome Therapeutic, for Active Mild to Moderate Ulcerative Colitis. Gastroenterology 2021, 160, 115–127.e30. [Google Scholar] [CrossRef]
- Seres Therapeutics Announces Topline Results for SER-287 Phase 2b Study in Mild-to-Moderate Ulcerative Colitis|Seres Therapeutics. Available online: https://ir.serestherapeutics.com/news-releases/news-release-details/seres-therapeutics-announces-topline-results-ser-287-phase-2b (accessed on 5 January 2024).
- Martinez, A.; O’Brien, E.J.; Balasubramanian, D.; Chafee, M.; Pina, A.; Narendar, P.; Kieser, K.J.; Vulic, M.; Diao, L.; Weiner, B.; et al. 681 ser-301, an investigational, rationally-designed bacterial consortium for mild-to-moderate ulcerative colitis, recapitulates the effects of ser-287, a consortium of firmicute spores, on remission associated microbial metabolites and host gene expression. Gastroenterology 2021, 160, S-135. [Google Scholar] [CrossRef]
- ANZCTR-Registration. Available online: https://anzctr.org.au/Trial/Registration/TrialReview.aspx?ACTRN=12620001103954 (accessed on 5 January 2024).
- Sugimoto, S.; Naganuma, M.; Kanai, T. Indole Compounds May Be Promising Medicines for Ulcerative Colitis. J. Gastroenterol. 2016, 51, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Mori, Y.; Matsui, S.; Takigami, H.; Fujino, J.; Kitagawa, H.; Miller, C.A.; Kato, T.; Saeki, K.; Matsuda, T. Indirubin and Indigo Are Potent Aryl Hydrocarbon Receptor Ligands Present in Human Urine. J. Biol. Chem. 2001, 276, 31475–31478. [Google Scholar] [CrossRef]
- Kawai, S.; Iijima, H.; Shinzaki, S.; Hiyama, S.; Yamaguchi, T.; Araki, M.; Iwatani, S.; Shiraishi, E.; Mukai, A.; Inoue, T.; et al. Indigo Naturalis Ameliorates Murine Dextran Sodium Sulfate-Induced Colitis via Aryl Hydrocarbon Receptor Activation. J. Gastroenterol. 2017, 52, 904–919. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.-Y.; Ma, L.; Zhang, C.; Lin, J.-Z.; Han, L.; He, Y.-N.; Xie, C.-G. Exploring the Mechanism of Indigo Naturalis in the Treatment of Ulcerative Colitis Based on TLR4/MyD88/NF-κB Signaling Pathway and Gut Microbiota. Front. Pharmacol. 2021, 12, 674416. [Google Scholar] [CrossRef]
- Kim, C.J.; Kovacs-Nolan, J.A.; Yang, C.; Archbold, T.; Fan, M.Z.; Mine, Y. L-Tryptophan Exhibits Therapeutic Function in a Porcine Model of Dextran Sodium Sulfate (DSS)-Induced Colitis. J. Nutr. Biochem. 2010, 21, 468–475. [Google Scholar] [CrossRef]
- Bettenworth, D.; Nowacki, T.M.; Ross, M.; Kyme, P.; Schwammbach, D.; Kerstiens, L.; Thoennissen, G.B.; Bokemeyer, C.; Hengst, K.; Berdel, W.E.; et al. Nicotinamide Treatment Ameliorates the Course of Experimental Colitis Mediated by Enhanced Neutrophil-Specific Antibacterial Clearance. Mol. Nutr. Food Res. 2014, 58, 1474–1490. [Google Scholar] [CrossRef]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef]
- Wilson, A.; Almousa, A.; Teft, W.A.; Kim, R.B. Attenuation of Bile Acid-Mediated FXR and PXR Activation in Patients with Crohn’s Disease. Sci. Rep. 2020, 10, 1866. [Google Scholar] [CrossRef]
- Cipriani, S.; Mencarelli, A.; Chini, M.G.; Distrutti, E.; Renga, B.; Bifulco, G.; Baldelli, F.; Donini, A.; Fiorucci, S. The Bile Acid Receptor GPBAR-1 (TGR5) Modulates Integrity of Intestinal Barrier and Immune Response to Experimental Colitis. PLoS ONE 2011, 6, e25637. [Google Scholar] [CrossRef]
- Van den Bossche, L.; Hindryckx, P.; Devisscher, L.; Devriese, S.; Van Welden, S.; Holvoet, T.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Vanden Bussche, J.; et al. Ursodeoxycholic Acid and Its Taurine- or Glycine-Conjugated Species Reduce Colitogenic Dysbiosis and Equally Suppress Experimental Colitis in Mice. Appl. Environ. Microbiol. 2017, 83, e02766-16. [Google Scholar] [CrossRef] [PubMed]
- Alemi, F.; Poole, D.P.; Chiu, J.; Schoonjans, K.; Cattaruzza, F.; Grider, J.R.; Bunnett, N.W.; Corvera, C.U. The Receptor TGR5 Mediates the Prokinetic Actions of Intestinal Bile Acids and Is Required for Normal Defecation in Mice. Gastroenterology 2013, 144, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Carino, A.; Biagioli, M.; Marchianò, S.; Fiorucci, C.; Bordoni, M.; Roselli, R.; Di Giorgio, C.; Baldoni, M.; Ricci, P.; Monti, M.C.; et al. Opposite Effects of the FXR Agonist Obeticholic Acid on Mafg and Nrf2 Mediate the Development of Acute Liver Injury in Rodent Models of Cholestasis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158733. [Google Scholar] [CrossRef] [PubMed]
- Van den Bossche, L.; Borsboom, D.; Devriese, S.; Van Welden, S.; Holvoet, T.; Devisscher, L.; Hindryckx, P.; De Vos, M.; Laukens, D. Tauroursodeoxycholic Acid Protects Bile Acid Homeostasis under Inflammatory Conditions and Dampens Crohn’s Disease-like Ileitis. Lab. Investig. J. Tech. Methods Pathol. 2017, 97, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, J.; Suo, Y.; Zheng, Z.; Wang, J.; Lv, L.; Huo, C.; Wang, Z.; Li, J.; Sun, W.; et al. Tauroursodeoxycholate Improves 2,4,6-Trinitrobenzenesulfonic Acid-Induced Experimental Acute Ulcerative Colitis in Mice. Int. Immunopharmacol. 2016, 36, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Withers, D.R.; Hepworth, M.R.; Wang, X.; Mackley, E.C.; Halford, E.E.; Dutton, E.E.; Marriott, C.L.; Brucklacher-Waldert, V.; Veldhoen, M.; Kelsen, J.; et al. Transient Inhibition of ROR-Γt Therapeutically Limits Intestinal Inflammation by Reducing TH17 Cells and Preserving Group 3 Innate Lymphoid Cells. Nat. Med. 2016, 22, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between Gut Bacteria and Bile in Health and Disease. Mol. Aspects Med. 2017, 56, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Barrangou, R.; Theriot, C.M. Bile Salt Hydrolases: Gatekeepers of Bile Acid Metabolism and Host-Microbiome Crosstalk in the Gastrointestinal Tract. PLoS Pathog. 2019, 15, e1007581. [Google Scholar] [CrossRef]
- Joyce, S.A.; Gahan, C.G.M. Bile Acid Modifications at the Microbe-Host Interface: Potential for Nutraceutical and Pharmaceutical Interventions in Host Health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, L.A.; Jones, B.V. Dysbiosis Modulates Capacity for Bile Acid Modification in the Gut Microbiomes of Patients with Inflammatory Bowel Disease: A Mechanism and Marker of Disease? Gut 2012, 61, 1642–1643. [Google Scholar] [CrossRef] [PubMed]
- Danese, S.; Sans, M.; Spencer, D.M.; Beck, I.; Doñate, F.; Plunkett, M.L.; de la Motte, C.; Redline, R.; Shaw, D.E.; Levine, A.D.; et al. Angiogenesis Blockade as a New Therapeutic Approach to Experimental Colitis. Gut 2007, 56, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.T.; Taylor, K.D.; Lin, Y.-C.; Hang, T.; Gaiennie, J.; Landers, C.J.; Vasiliauskas, E.A.; Kam, L.Y.; Rojany, M.; Papadakis, K.A.; et al. Mutations in NOD2 Are Associated with Fibrostenosing Disease in Patients with Crohn’s Disease. Gastroenterology 2002, 123, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Nakagome, K.; Dohi, M.; Okunishi, K.; Tanaka, R.; Miyazaki, J.; Yamamoto, K. In Vivo IL-10 Gene Delivery Attenuates Bleomycin Induced Pulmonary Fibrosis by Inhibiting the Production and Activation of TGF-Beta in the Lung. Thorax 2006, 61, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Louis, H.; Van Laethem, J.L.; Wu, W.; Quertinmont, E.; Degraef, C.; Van den Berg, K.; Demols, A.; Goldman, M.; Le Moine, O.; Geerts, A.; et al. Interleukin-10 Controls Neutrophilic Infiltration, Hepatocyte Proliferation, and Liver Fibrosis Induced by Carbon Tetrachloride in Mice. Hepatology 1998, 28, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Kashima, S.; Fujiya, M.; Konishi, H.; Ueno, N.; Inaba, Y.; Moriichi, K.; Tanabe, H.; Ikuta, K.; Ohtake, T.; Kohgo, Y. Polyphosphate, an Active Molecule Derived from Probiotic Lactobacillus Brevis, Improves the Fibrosis in Murine Colitis. Transl. Res. J. Lab. Clin. Med. 2015, 166, 163–175. [Google Scholar] [CrossRef]
- Park, J.-S.; Choi, J.; Kwon, J.Y.; Jung, K.-A.; Yang, C.W.; Park, S.-H.; Cho, M.-L. A Probiotic Complex, Rosavin, Zinc, and Prebiotics Ameliorate Intestinal Inflammation in an Acute Colitis Mouse Model. J. Transl. Med. 2018, 16, 37. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, X.; Hao, Y.; Ding, J.; Shen, J.; Xue, Z.; Qi, W.; Li, Z.; Song, Y.; Zhang, T.; et al. Protective Effects of a Novel Probiotic Strain, Lactococcus Lactis ML2018, in Colitis: In Vivo and in Vitro Evidence. Food Funct. 2019, 10, 1132–1145. [Google Scholar] [CrossRef]
- Deng, X.; Zheng, C.; Wang, S.; Yang, R.; Liu, Z.; Chen, T. Treatment with a Probiotic Combination Reduces Abdominal Adhesion in Rats by Decreasing Intestinal Inflammation and Restoring Microbial Composition. Oncol. Rep. 2020, 43, 986–998. [Google Scholar] [CrossRef]
- Lombardi, F.; Augello, F.R.; Palumbo, P.; Mollsi, E.; Giuliani, M.; Cimini, A.M.; Cifone, M.G.; Cinque, B. Soluble Fraction from Lysate of a High Concentration Multi-Strain Probiotic Formulation Inhibits TGF-Β1-Induced Intestinal Fibrosis on CCD-18Co Cells. Nutrients 2021, 13, 882. [Google Scholar] [CrossRef] [PubMed]
| Agent | Mechanism | Model | Reference |
|---|---|---|---|
| Polyphosphate | ↓ IL-1β, TNF-α | Cellular models: DSS- and TNBS-induced colitis | [150] |
| 12 probiotics, prebiotics, rosavin and zinc | ↓ IL-6, IL-1β, IL-17 ↑ IL-10 | Mouse models: DSS-induced colitis | [151] |
| Lactococcus lactis ML2018 | ↓ NF-κB and MAPK signaling ↑ SCFAs | Mouse models: DSS-induced colitis | [152] |
| 4 probiotics | ↓ IL-1β, TNF-α ↓ TLR4/NF-κB and TGF-β1/Smad signaling ↑ microbial balance | Rat models with abdominal adhesions | [153] |
| Multi-Strain Probiotic Formulation (Vivomixx) | ↓ TGF-β1 | Cellular models: CCD-18Co cells cultured with TGF-β1 | [154] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardi, F.; D’Amico, F.; Bencardino, S.; Faggiani, I.; Fanizza, J.; Zilli, A.; Parigi, T.L.; Allocca, M.; Danese, S.; Furfaro, F. Gut Microbiota Metabolites: Unveiling Their Role in Inflammatory Bowel Diseases and Fibrosis. Pharmaceuticals 2024, 17, 347. https://doi.org/10.3390/ph17030347
Bernardi F, D’Amico F, Bencardino S, Faggiani I, Fanizza J, Zilli A, Parigi TL, Allocca M, Danese S, Furfaro F. Gut Microbiota Metabolites: Unveiling Their Role in Inflammatory Bowel Diseases and Fibrosis. Pharmaceuticals. 2024; 17(3):347. https://doi.org/10.3390/ph17030347
Chicago/Turabian StyleBernardi, Francesca, Ferdinando D’Amico, Sarah Bencardino, Ilaria Faggiani, Jacopo Fanizza, Alessandra Zilli, Tommaso Lorenzo Parigi, Mariangela Allocca, Silvio Danese, and Federica Furfaro. 2024. "Gut Microbiota Metabolites: Unveiling Their Role in Inflammatory Bowel Diseases and Fibrosis" Pharmaceuticals 17, no. 3: 347. https://doi.org/10.3390/ph17030347
APA StyleBernardi, F., D’Amico, F., Bencardino, S., Faggiani, I., Fanizza, J., Zilli, A., Parigi, T. L., Allocca, M., Danese, S., & Furfaro, F. (2024). Gut Microbiota Metabolites: Unveiling Their Role in Inflammatory Bowel Diseases and Fibrosis. Pharmaceuticals, 17(3), 347. https://doi.org/10.3390/ph17030347






