Improved Immunotherapy Outcomes via Cuproptosis Upregulation of HLA-DRA Expression: Promoting the Aggregation of CD4+ and CD8+T Lymphocytes in Clear Cell Renal Cell Carcinoma
Abstract
:1. Introduction
2. Results
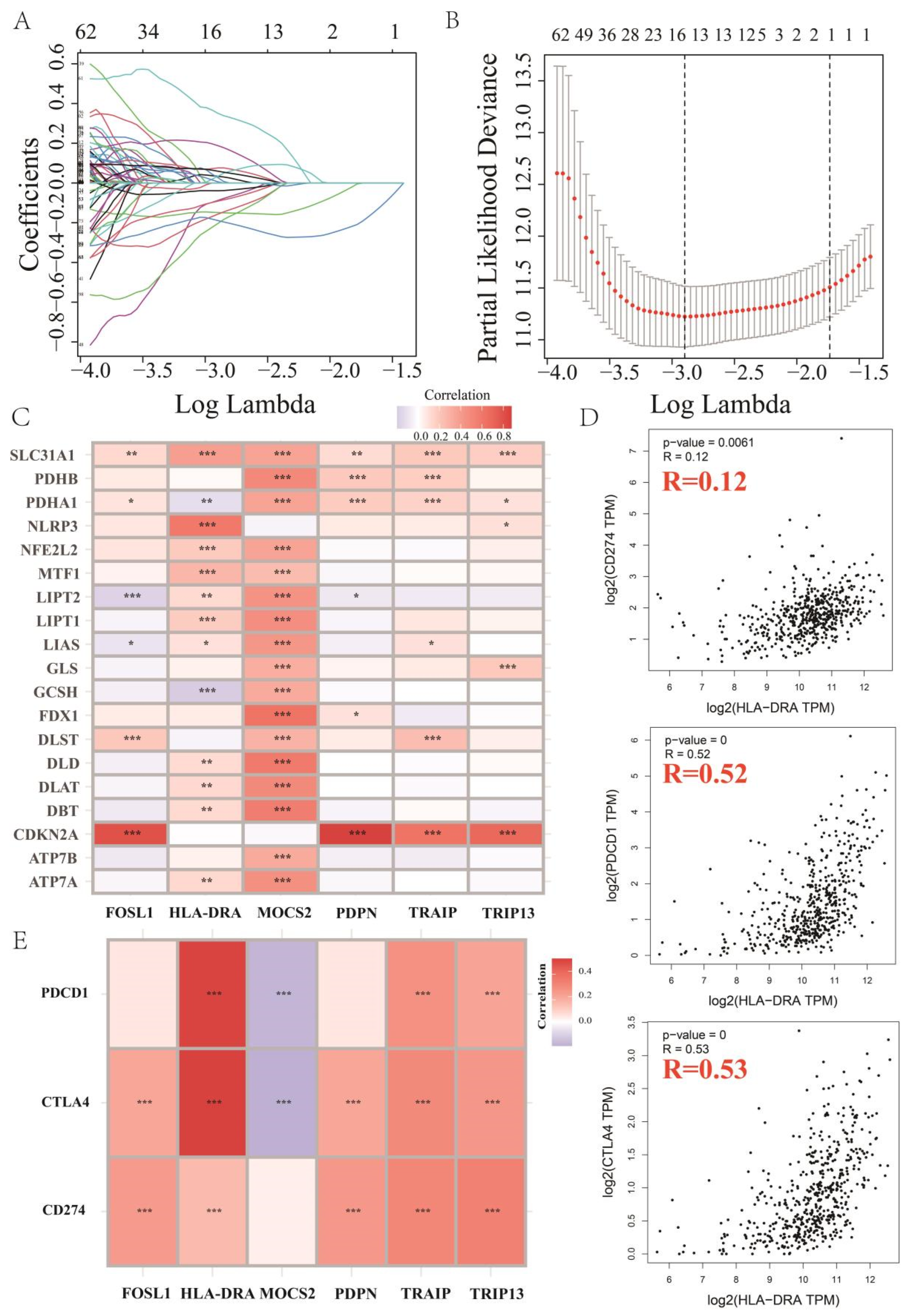
2.1. Key Cuproptosis-Related Gene in ccRCC, HLA-DRA
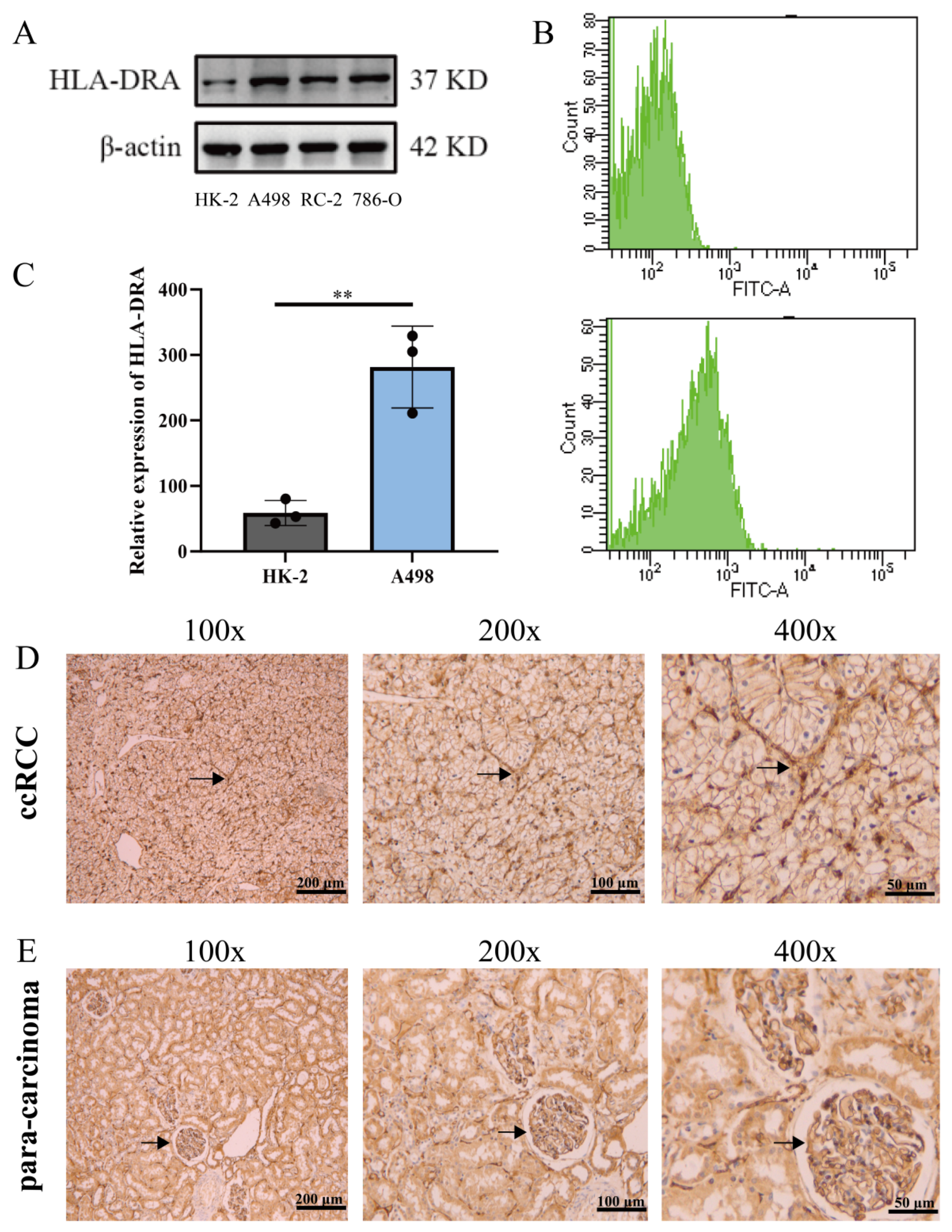
2.2. HLA-DRA Expressed as an MHC-II in ccRCC Cell Lines and Pathological Tissues
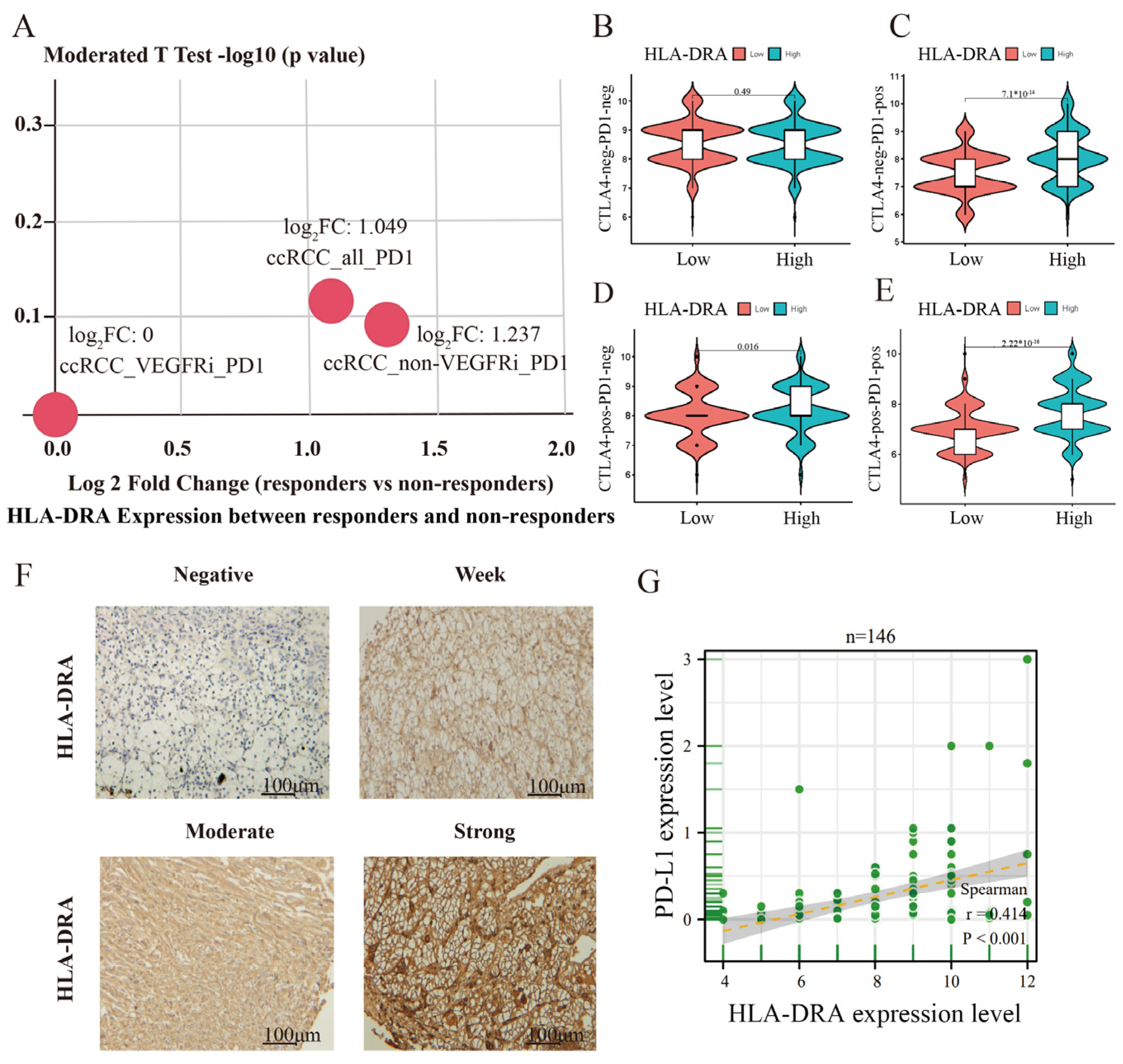
2.3. High HLA-DRA Expression Enhances the Sensitivity of ccRCC to ICI Treatment
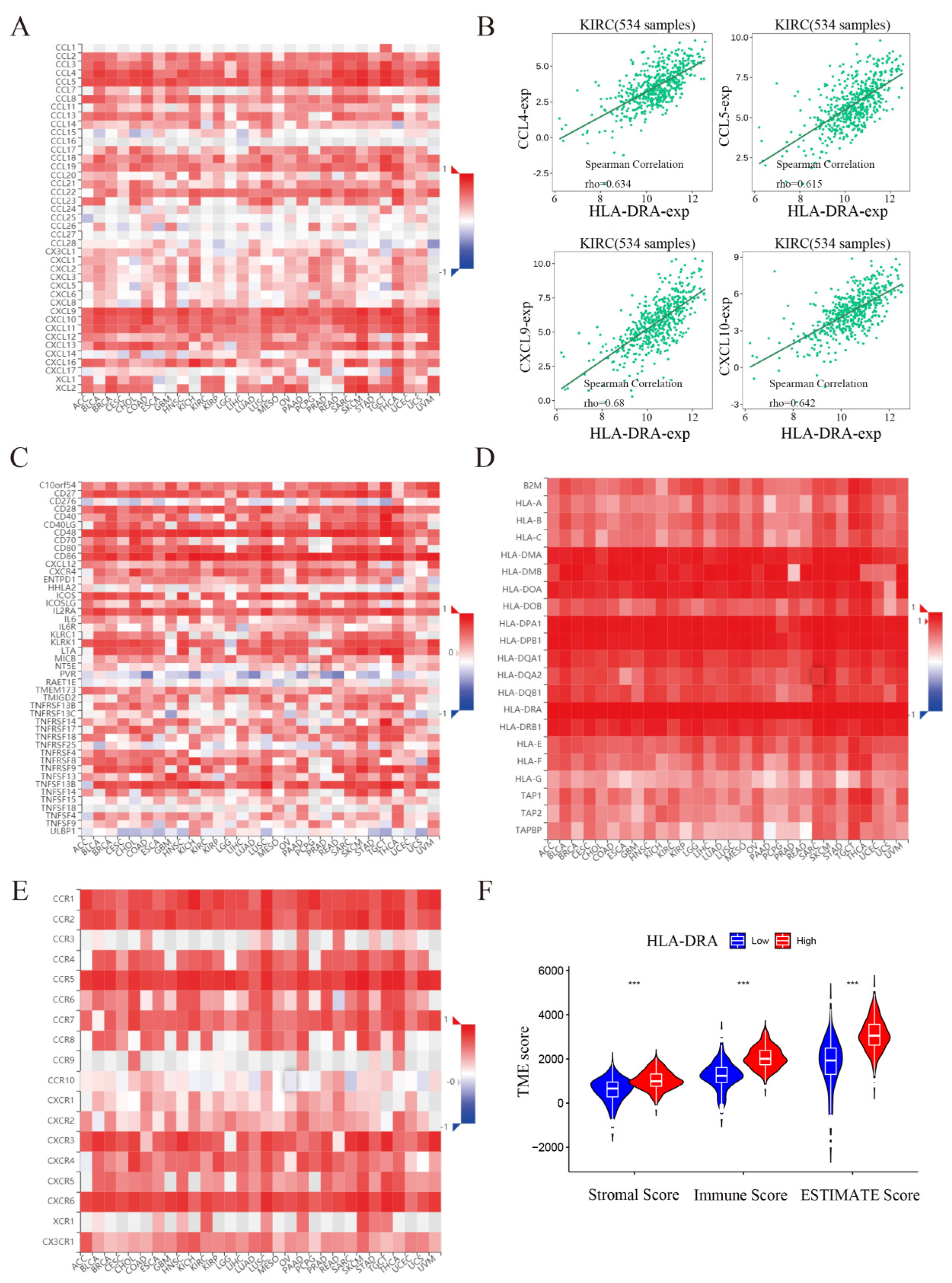
2.4. HLA-DRA Positively Affects the Immune Microenvironment of ccRCC
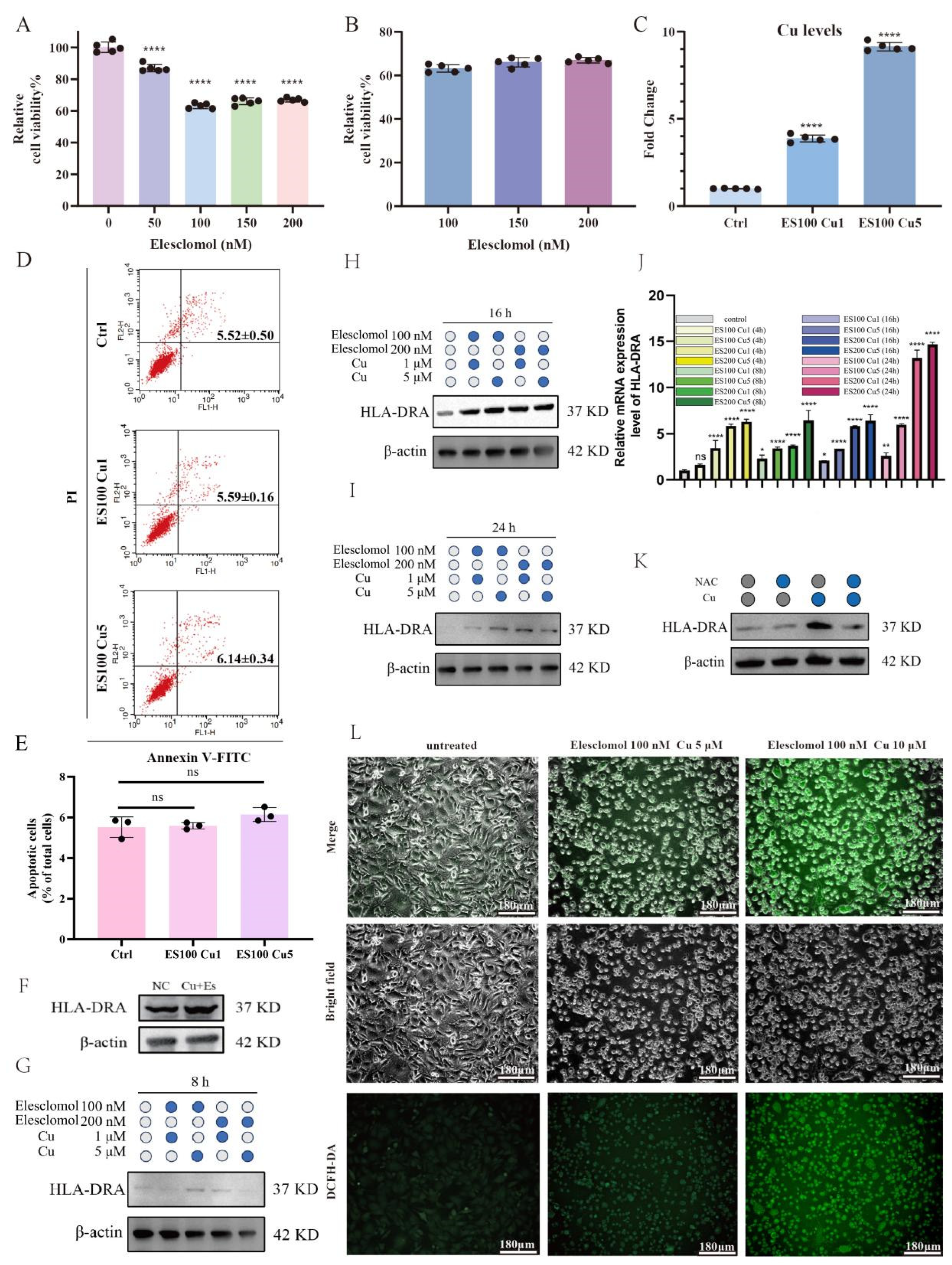
2.5. Cuproptosis Upregulates the Protein Expression of HLA-DRA from the Transcriptional Level in a Dose-Dependent Manner through Reactive Oxygen Species
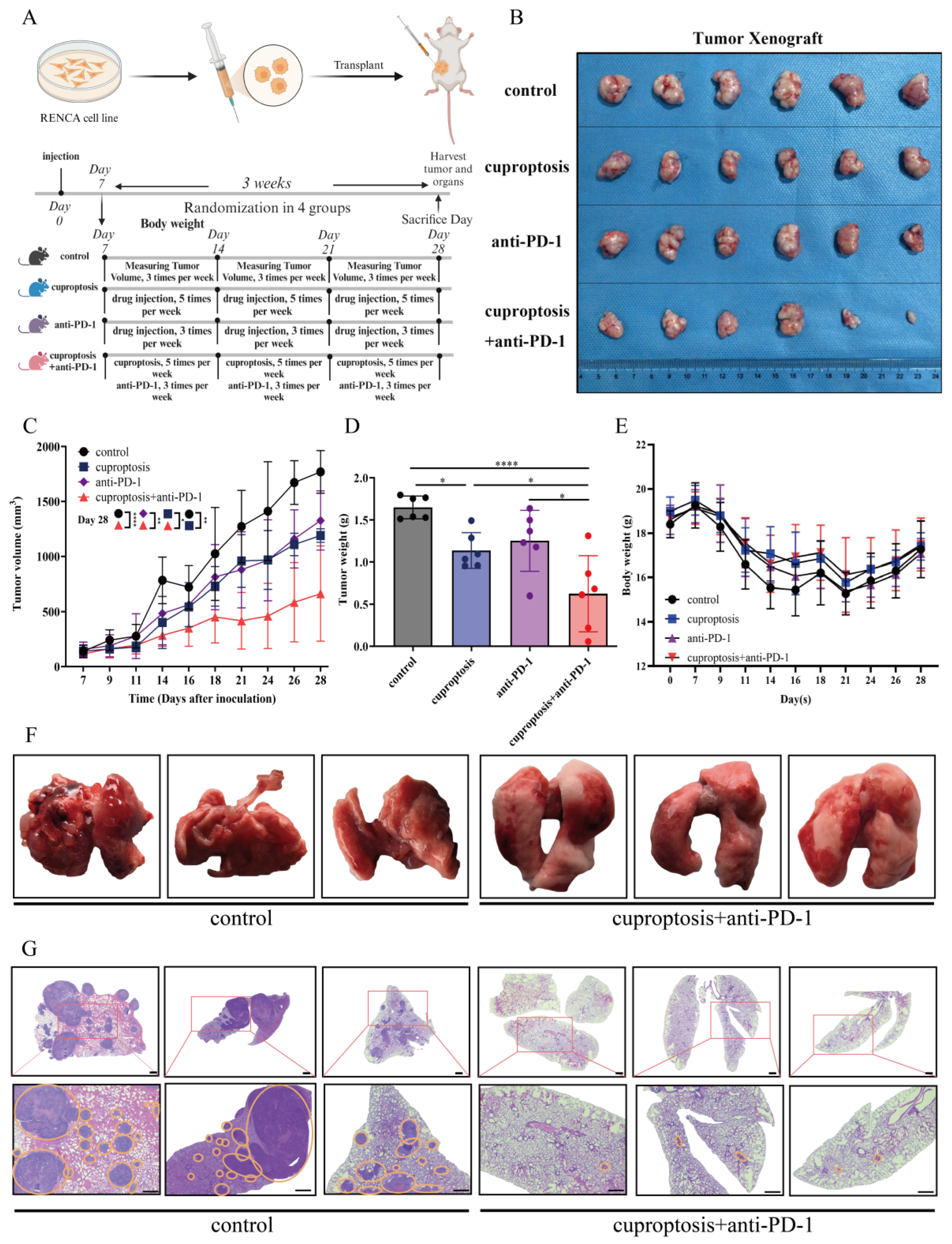
2.6. Cuproptosis Improves the Outcomes of Anti-PD-1 Treatment In Vivo
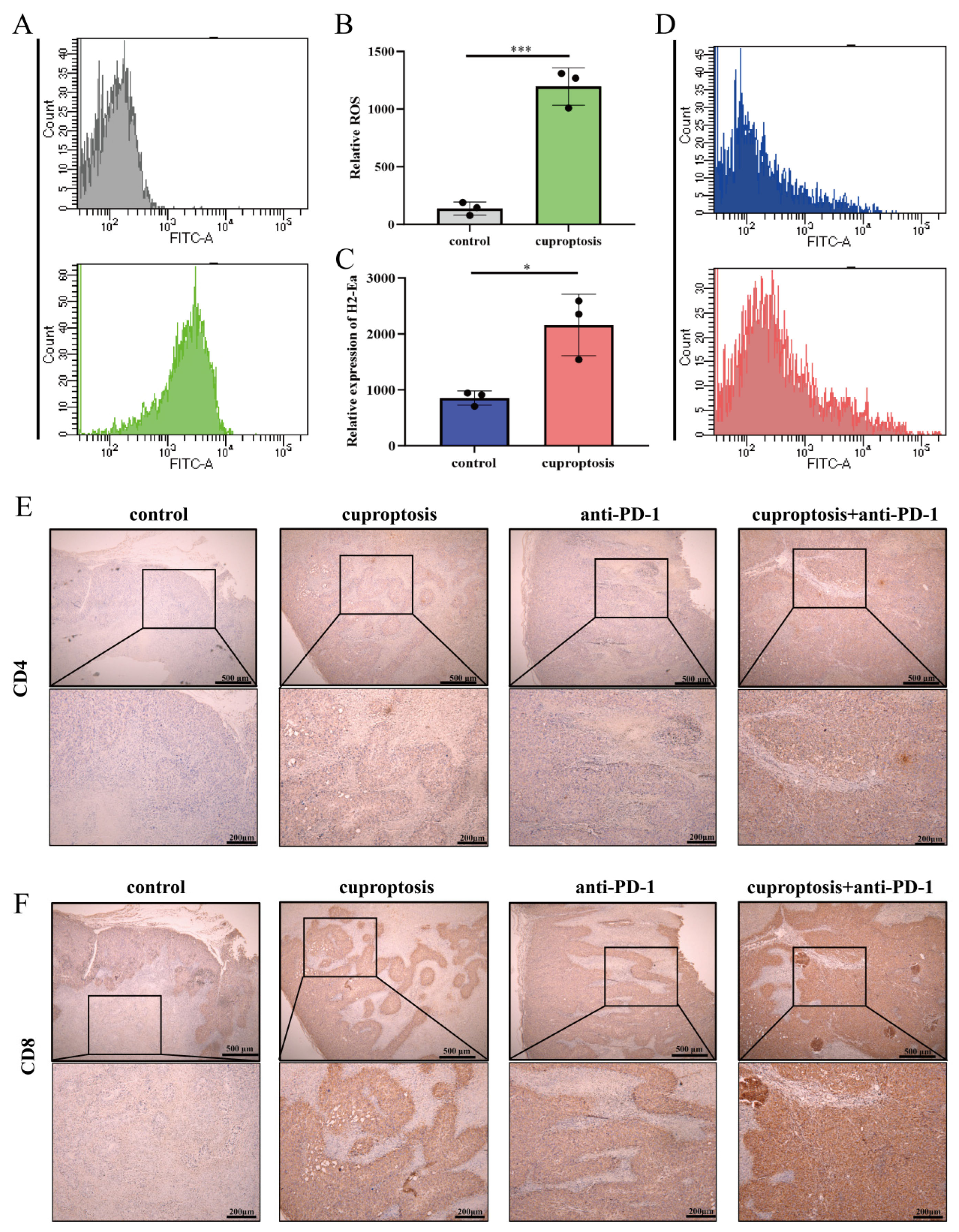
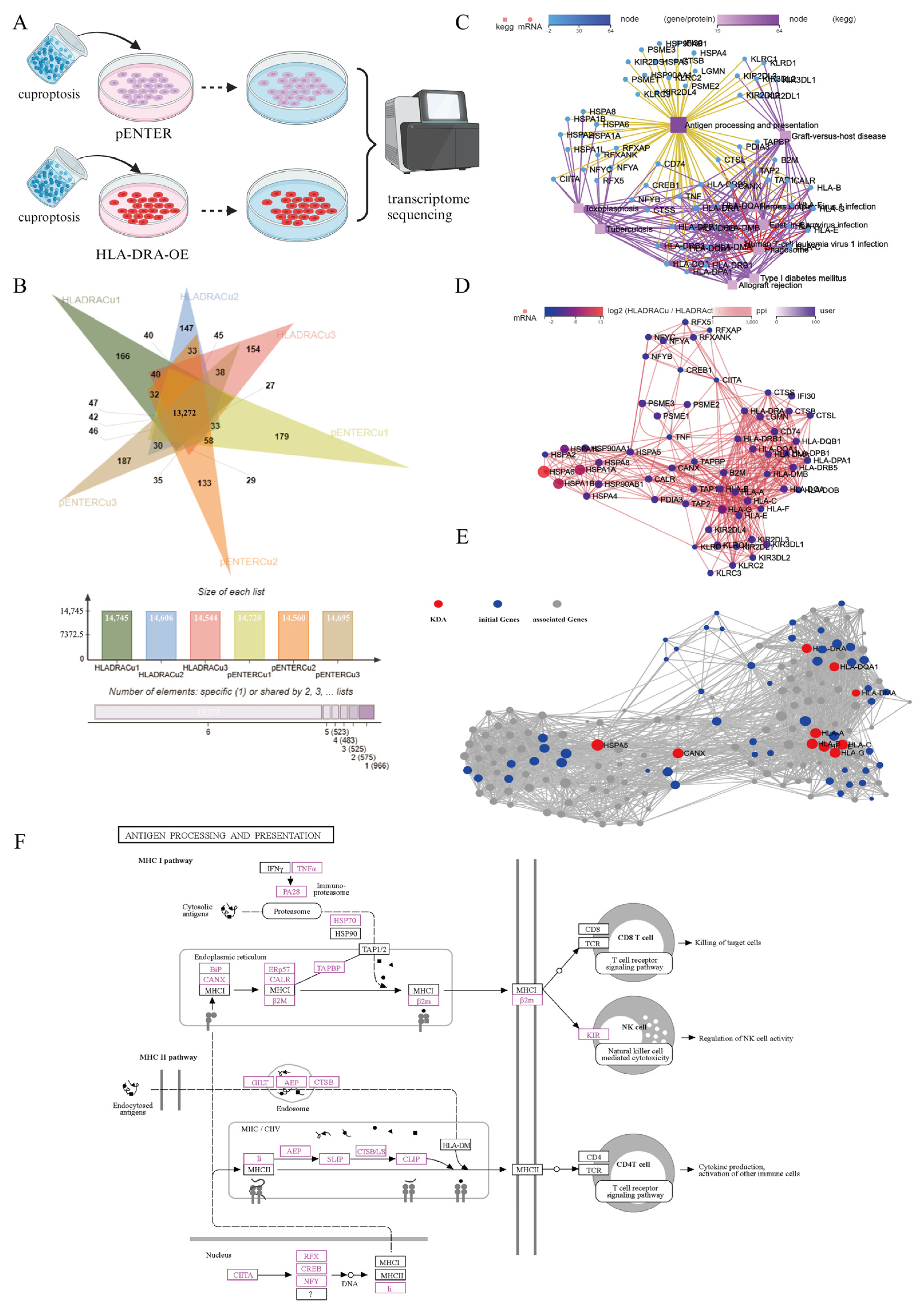
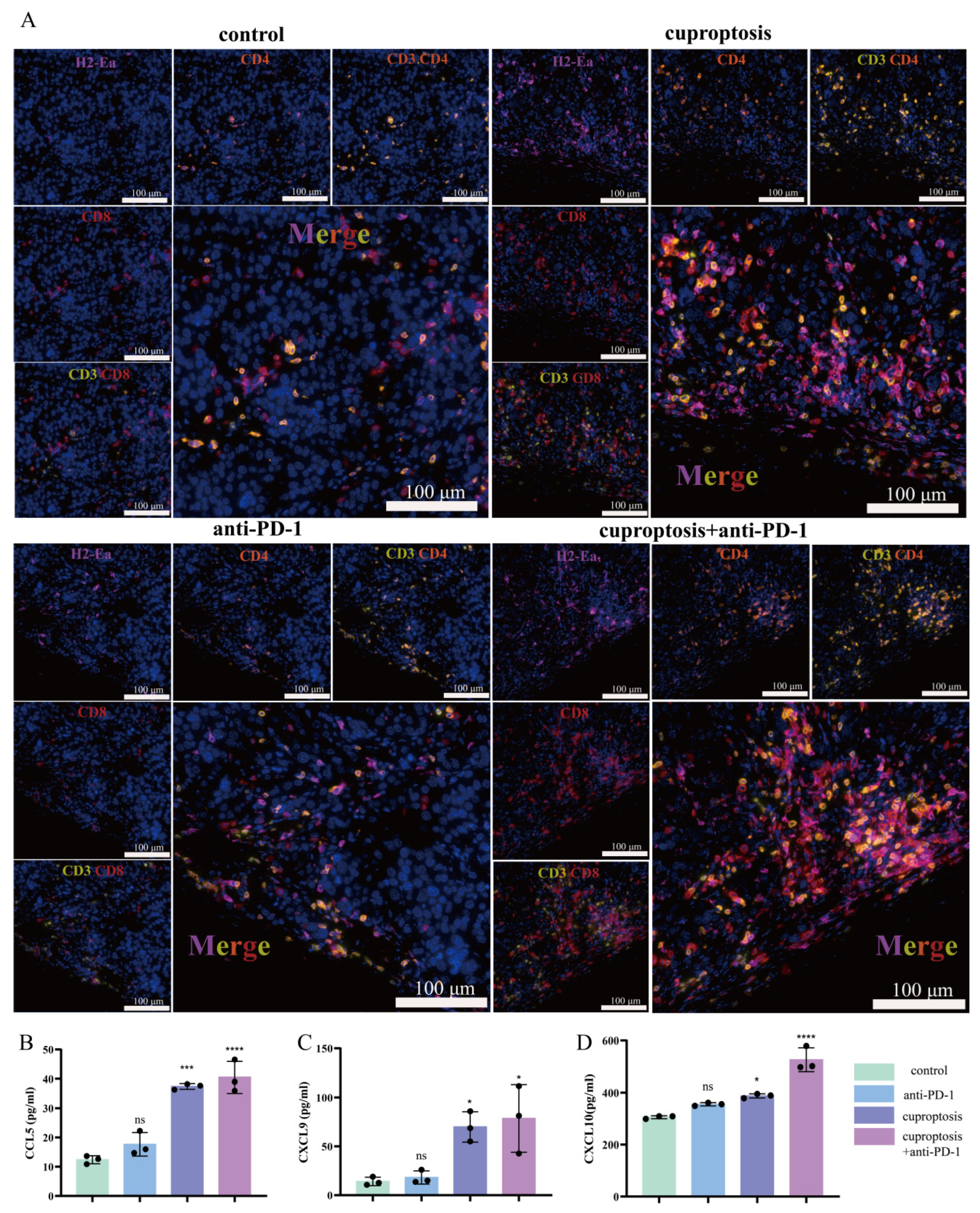
2.7. Upregulated Expression of HLA-DRA via Cuproptosis Synergy with ICI Therapy: Enhanced Aggregation of CD4+ and CD8+ T Cells Associated with Increased Levels of CCL5, CXCL9, and CXCL10
3. Discussion
4. Materials and Methods
4.1. Data Collection and Screening of Cuproptosis-Related Genes (CRGs)
4.2. Functional Enrichment and Drug Sensitivity Analyses
4.3. Analysis of the Immune Microenvironment and Immune Infiltration of the Tumors
4.4. Cell Culture and Cell Lines
4.5. RNA Isolation and Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
4.6. Western Blot Analysis
4.7. Cell Viability Assay
4.8. Cuproptosis Detection
4.9. Transcriptome Sequencing Analysis
4.10. Mouse Syngeneic Tumor Experiment
4.11. Flow Cytometry Detection
4.12. Immunohistochemical (IHC) Staining and Semi-Quantitative Scoring
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Capitanio, U.; Bensalah, K.; Bex, A.; Boorjian, S.A.; Bray, F.; Coleman, J.; Gore, J.L.; Sun, M.; Wood, C.; Russo, P. Epidemiology of Renal Cell Carcinoma. Eur. Urol. 2019, 75, 74–84. [Google Scholar] [CrossRef]
- Signoretti, S.; Flaifel, A.; Chen, Y.B.; Reuter, V.E. Renal Cell Carcinoma in the Era of Precision Medicine: From Molecular Pathology to Tissue-Based Biomarkers. J. Clin. Oncol. 2018, 36, JCO2018792259. [Google Scholar] [CrossRef]
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernández-Pello, S.; Giles, R.H.; Hofmann, F.; Hora, M.; Kuczyk, M.A.; et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef]
- Motzer, R.J.; Jonasch, E.; Boyle, S.; Carlo, M.I.; Manley, B.; Agarwal, N.; Alva, A.; Beckermann, K.; Choueiri, T.K.; Costello, B.A.; et al. NCCN Guidelines Insights: Kidney Cancer, Version 1.2021. J. Natl. Compr. Cancer Netw. JNCCN 2020, 18, 1160–1170. [Google Scholar] [CrossRef]
- Cheaib, J.G.; Pierorazio, P.M. How Does Treatment Uncertainty Factor into Decisions to Place Patients on Active Surveillance for Kidney Cancer? Eur. Urol. Focus 2019, 5, 946–948. [Google Scholar] [CrossRef]
- Rathmell, W.K.; Rumble, R.B.; Van Veldhuizen, P.J.; Al-Ahmadie, H.; Emamekhoo, H.; Hauke, R.J.; Louie, A.V.; Milowsky, M.I.; Molina, A.M.; Rose, T.L.; et al. Management of Metastatic Clear Cell Renal Cell Carcinoma: ASCO Guideline. J. Clin. Oncol. 2022, 40, 2957–2995. [Google Scholar] [CrossRef]
- Hagiwara, M.; Fushimi, A.; Matsumoto, K.; Oya, M. The Significance of PARP1 as a biomarker for Predicting the Response to PD-L1 Blockade in Patients with PBRM1-mutated Clear Cell Renal Cell Carcinoma. Eur. Urol. 2022, 81, 145–148. [Google Scholar] [CrossRef]
- Johnson, D.B.; Reynolds, K.L.; Sullivan, R.J.; Balko, J.M.; Patrinely, J.R.; Cappelli, L.C.; Naidoo, J.; Moslehi, J.J. Immune checkpoint inhibitor toxicities: Systems-based approaches to improve patient care and research. Lancet Oncol. 2020, 21, e398–e404. [Google Scholar] [CrossRef]
- Vansteenkiste, J.; Wauters, E.; Reymen, B.; Ackermann, C.J.; Peters, S.; De Ruysscher, D. Current status of immune checkpoint inhibition in early-stage NSCLC. Ann. Oncol. 2019, 30, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Qiao, M.; Zhou, C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell. Mol. Immunol. 2021, 18, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Jiao, D.; Qin, S.; Chu, Q.; Wu, K.; Li, A. Synergistic effect of immune checkpoint blockade and anti-angiogenesis in cancer treatment. Mol. Cancer 2019, 18, 60. [Google Scholar] [CrossRef] [PubMed]
- Lavacchi, D.; Pellegrini, E.; Palmieri, V.E.; Doni, L.; Mela, M.M.; Di Maida, F.; Amedei, A.; Pillozzi, S.; Carini, M.; Antonuzzo, L. Immune Checkpoint Inhibitors in the Treatment of Renal Cancer: Current State and Future Perspective. Int. J. Mol. Sci. 2020, 21, 4691. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Min, J.; Wang, F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct. Target. Ther. 2022, 7, 378. [Google Scholar] [CrossRef] [PubMed]
- Hadian, K.; Stockwell, B.R. The therapeutic potential of targeting regulated non-apoptotic cell death. Nat. Rev. Drug Discov. 2023, 22, 723–742. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yang, Y.; Gao, Y.; He, J. Cuproptosis: Mechanisms and links with cancers. Mol. Cancer 2023, 22, 46. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Tang, R.; Xiao, M.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Targeting cell death pathways for cancer therapy: Recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J. Hematol. Oncol. 2022, 15, 174. [Google Scholar] [CrossRef]
- Li, Y. Copper homeostasis: Emerging target for cancer treatment. IUBMB Life 2020, 72, 1900–1908. [Google Scholar] [CrossRef]
- Michniewicz, F.; Saletta, F.; Rouaen, J.R.C.; Hewavisenti, R.V.; Mercatelli, D.; Cirillo, G.; Giorgi, F.M.; Trahair, T.; Ziegler, D.; Vittorio, O. Copper: An Intracellular Achilles’ Heel Allowing the Targeting of Epigenetics, Kinase Pathways, and Cell Metabolism in Cancer Therapeutics. ChemMedChem 2021, 16, 2315–2329. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Heesters, B.A.; van Megesen, K.; Tomris, I.; de Vries, R.P.; Magri, G.; Spits, H. Characterization of human FDCs reveals regulation of T cells and antigen presentation to B cells. J. Exp. Med. 2021, 218, e20210790. [Google Scholar] [CrossRef]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef] [PubMed]
- Borst, J.; Ahrends, T.; Bąbała, N.; Melief, C.J.M.; Kastenmüller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Kreiter, S.; Vormehr, M.; van de Roemer, N.; Diken, M.; Löwer, M.; Diekmann, J.; Boegel, S.; Schrörs, B.; Vascotto, F.; Castle, J.C.; et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015, 520, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Forero, A.; Li, Y.; Chen, D.; Grizzle, W.E.; Updike, K.L.; Merz, N.D.; Downs-Kelly, E.; Burwell, T.C.; Vaklavas, C.; Buchsbaum, D.J.; et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol. Res. 2016, 4, 390–399. [Google Scholar] [CrossRef]
- Park, I.A.; Hwang, S.-H.; Song, I.H.; Heo, S.-H.; Kim, Y.-A.; Bang, W.S.; Park, H.S.; Lee, M.; Gong, G.; Lee, H.J. Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS ONE 2017, 12, e0182786. [Google Scholar] [CrossRef]
- Rodig, S.J.; Gusenleitner, D.; Jackson, D.G.; Gjini, E.; Giobbie-Hurder, A.; Jin, C.; Chang, H.; Lovitch, S.B.; Horak, C.; Weber, J.S.; et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 2018, 10, eaar3342. [Google Scholar] [CrossRef] [PubMed]
- Roemer, M.G.M.; Redd, R.A.; Cader, F.Z.; Pak, C.J.; Abdelrahman, S.; Ouyang, J.; Sasse, S.; Younes, A.; Fanale, M.; Santoro, A.; et al. Major Histocompatibility Complex Class II and Programmed Death Ligand 1 Expression Predict Outcome After Programmed Death 1 Blockade in Classic Hodgkin Lymphoma. J. Clin. Oncol. 2018, 36, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Tannir, N.M.; Papadopoulos, K.P.; Wong, D.J.; Aljumaily, R.; Hung, A.; Afable, M.; Kim, J.S.; Ferry, D.; Drakaki, A.; Bendell, J.; et al. Pegilodecakin as monotherapy or in combination with anti-PD-1 or tyrosine kinase inhibitor in heavily pretreated patients with advanced renal cell carcinoma: Final results of cohorts A, G, H and I of IVY Phase I study. Int. J. Cancer 2021, 149, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Dengjel, J.; Nastke, M.D.; Gouttefangeas, C.; Gitsioudis, G.; Schoor, O.; Altenberend, F.; Müller, M.; Krämer, B.; Missiou, A.; Sauter, M.; et al. Unexpected abundance of HLA class II presented peptides in primary renal cell carcinomas. Clin. Cancer Res. 2006, 12 Pt 1, 4163–4170. [Google Scholar] [CrossRef]
- Armstrong, T.D.; Clements, V.K.; Martin, B.K.; Ting, J.P.-Y.; Ostrand-Rosenberg, S. Major histocompatibility complex class II-transfected tumor cells present endogenous antigen and are potent inducers of tumor-specific immunity. Proc. Natl. Acad. Sci. USA 1997, 94, 6886–6891. [Google Scholar] [CrossRef]
- Lee, C.H.; Shah, A.Y.; Rasco, D.; Rao, A.; Taylor, M.H.; Di Simone, C.; Hsieh, J.J.; Pinto, A.; Shaffer, D.R.; Girones Sarrio, R.; et al. Lenvatinib plus pembrolizumab in patients with either treatment-naive or previously treated metastatic renal cell carcinoma (Study 111/KEYNOTE-146): A phase 1b/2 study. Lancet Oncol. 2021, 22, 946–958. [Google Scholar] [CrossRef]
- Graham, J.; Wells, J.C.; Dudani, S.; Gan, C.L.; Donskov, F.; Lee, J.L.; Kollmannsberger, C.K.; Meza, L.; Beuselinck, B.; Hansen, A.; et al. Outcomes of patients with advanced non-clear cell renal cell carcinoma treated with first-line immune checkpoint inhibitor therapy. Eur. J. Cancer 2022, 171, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Zhang, J.; Yang, Y.; Fleishman, J.S.; Wang, Y.; Wang, J.; Chen, J.; Li, Y.; Wang, H. Cuproptosis: A novel therapeutic target for overcoming cancer drug resistance. Drug Resist. Updat. 2023, 72, 101018. [Google Scholar] [CrossRef] [PubMed]
- Lévy, J.; Cacheux, W.; Bara, M.A.; L’hermitte, A.; Lepage, P.; Fraudeau, M.; Trentesaux, C.; Lemarchand, J.; Durand, A.; Crain, A.-M.; et al. Intestinal inhibition of Atg7 prevents tumour initiation through a microbiome-influenced immune response and suppresses tumour growth. Nat. Cell Biol. 2015, 17, 1062–1073. [Google Scholar] [CrossRef]
- Yang, A.; Herter-Sprie, G.; Zhang, H.; Lin, E.Y.; Biancur, D.; Wang, X.; Deng, J.; Hai, J.; Yang, S.; Wong, K.-K.; et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018, 8, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Ding, J.; Wang, C.; Zhou, X.; Gao, W.; Huang, H.; Shao, F.; Liu, Z. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature 2020, 579, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Lang, X.; Green, M.D.; Wang, W.; Yu, J.; Choi, J.E.; Jiang, L.; Liao, P.; Zhou, J.; Zhang, Q.; Dow, A.; et al. Radiotherapy and immunotherapy promote tumoral lipid oxidation and ferroptosis via synergistic repression of SLC7A11. Cancer Discov. 2019, 9, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Xia, S.; Kong, Q.; Li, S.; Liu, X.; Junqueira, C.; Meza-Sosa, K.F.; Mok, T.M.Y.; Ansara, J.; et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature 2020, 579, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701. [Google Scholar] [CrossRef]
- Wang, W.; Green, M.; Choi, J.E.; Gijón, M.; Kennedy, P.D.; Johnson, J.K.; Liao, P.; Lang, X.; Kryczek, I.; Sell, A.; et al. CD8 T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019, 569, 270–274. [Google Scholar] [CrossRef]
- Veatch, J.R.; Lee, S.M.; Fitzgibbon, M.; Chow, I.-T.; Jesernig, B.; Schmitt, T.; Kong, Y.Y.; Kargl, J.; Houghton, A.M.; Thompson, J.A.; et al. Tumor-infiltrating BRAFV600E-specific CD4+ T cells correlated with complete clinical response in melanoma. J. Clin. Investig. 2018, 128, 1563–1568. [Google Scholar] [CrossRef] [PubMed]
- Marty, R.; Thompson, W.K.; Salem, R.M.; Zanetti, M.; Carter, H. Evolutionary Pressure against MHC Class II Binding Cancer Mutations. Cell 2018, 175, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Bou Nasser Eddine, F.; Forlani, G.; Lombardo, L.; Tedeschi, A.; Tosi, G.; Accolla, R.S. CIITA-driven MHC class II expressing tumor cells can efficiently prime naive CD4+ TH cells in vivo and vaccinate the host against parental MHC-II-negative tumor cells. Oncoimmunology 2017, 6, e1261777. [Google Scholar] [CrossRef] [PubMed]
- Linnemann, C.; van Buuren, M.M.; Bies, L.; Verdegaal, E.M.E.; Schotte, R.; Calis, J.J.A.; Behjati, S.; Velds, A.; Hilkmann, H.; el Atmioui, D.; et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat. Med. 2015, 21, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Estrada, M.V.; Salgado, R.; Sanchez, V.; Doxie, D.B.; Opalenik, S.R.; Vilgelm, A.E.; Feld, E.; Johnson, A.S.; Greenplate, A.R.; et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 2016, 7, 10582. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, T.; Kamma, H.; Fujiwara, M.; Matsui, M.; Horiguchi, H.; Satoh, H.; Fujimoto, M.; Yokoyama, K.; Ogata, T. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J. Pathol. 1999, 187, 191–199. [Google Scholar] [CrossRef]
- Holling, T.M.; Bergevoet, M.W.T.; Wilson, L.; Van Eggermond, M.C.J.A.; Schooten, E.; Steenbergen, R.D.M.; Snijders, P.J.F.; Jager, M.J.; Van den Elsen, P.J. A role for EZH2 in silencing of IFN-gamma inducible MHC2TA transcription in uveal melanoma. J. Immunol. 2007, 179, 5317–5325. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, S.Y.; Zeng, L.; Ma, H.; Zhang, Y.; Yang, H.; Liu, Y.; Fang, S.; Zhao, J.; Xu, Y.; et al. An Enzyme-Engineered Nonporous Copper(I) Coordination Polymer Nanoplatform for Cuproptosis-Based Synergistic Cancer Therapy. Adv. Mater. 2022, 34, e2204733, Erratum in Adv. Mater. 2023, 35, e2300773. [Google Scholar] [CrossRef]
- Guo, B.; Yang, F.; Zhang, L.; Zhao, Q.; Wang, W.; Yin, L.; Chen, D.; Wang, M.; Han, S.; Xiao, H.; et al. Cuproptosis Induced by ROS Responsive Nanoparticles with Elesclomol and Copper Combined with αPD-L1 for Enhanced Cancer Immunotherapy. Adv. Mater. 2023, 35, e2212267. [Google Scholar] [CrossRef]
- de Miguel, M.; Calvo, E. Clinical Challenges of Immune Checkpoint Inhibitors. Cancer Cell 2020, 38, 326–333. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Hellmann, M.D. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell 2020, 37, 443–455. [Google Scholar] [CrossRef]
- Barata, P.C.; Rini, B.I. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J. Clin. 2017, 67, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.N.; Rathmell, J.C. Metabolic programming and immune suppression in the tumor microenvironment. Cancer Cell 2023, 41, 421–433. [Google Scholar] [CrossRef] [PubMed]
- White, L.G.; Goy, H.E.; Rose, A.J.; McLellan, A.D. Controlling Cell Trafficking: Addressing Failures in CAR T and NK Cell Therapy of Solid Tumours. Cancers 2022, 14, 978. [Google Scholar] [CrossRef] [PubMed]
- Dangaj, D.; Bruand, M.; Grimm, A.J.; Ronet, C.; Barras, D.; Duttagupta, P.A.; Lanitis, E.; Duraiswamy, J.; Tanyi, J.L.; Benencia, F.; et al. Cooperation between Constitutive and Inducible Chemokines Enables T Cell Engraftment and Immune Attack in Solid Tumors. Cancer Cell 2019, 35, 885–900.e10. [Google Scholar] [CrossRef] [PubMed]
- Limagne, E.; Nuttin, L.; Thibaudin, M.; Jacquin, E.; Aucagne, R.; Bon, M.; Revy, S.; Barnestein, R.; Ballot, E.; Truntzer, C.; et al. MEK inhibition overcomes chemoimmunotherapy resistance by inducing CXCL10 in cancer cells. Cancer Cell 2022, 40, 136–152.e12. [Google Scholar] [CrossRef]
- Polishchuk, E.V.; Merolla, A.; Lichtmannegger, J.; Romano, A.; Indrieri, A.; Ilyechova, E.Y.; Concilli, M.; De Cegli, R.; Crispino, R.; Mariniello, M.; et al. Activation of Autophagy, Observed in Liver Tissues from Patients with Wilson Disease and from ATP7B-Deficient Animals, Protects Hepatocytes from Copper-Induced Apoptosis. Gastroenterology 2019, 156, 1173–1189.e5. [Google Scholar] [CrossRef]
- Aubert, L.; Nandagopal, N.; Steinhart, Z.; Lavoie, G.; Nourreddine, S.; Berman, J.; Saba-El-Leil, M.K.; Papadopoli, D.; Lin, S.; Hart, T.; et al. Copper bioavailability is a KRAS-specific vulnerability in colorectal cancer. Nat. Commun. 2020, 11, 3701. [Google Scholar] [CrossRef]
- Kahlson, M.A.; Dixon, S.J. Copper-induced cell death. Science 2022, 375, 1231–1232. [Google Scholar] [CrossRef]
- Dong, J.; Wang, X.; Xu, C.; Gao, M.; Wang, S.; Zhang, J.; Tong, H.; Wang, L.; Han, Y.; Cheng, N.; et al. Inhibiting NLRP3 inflammasome activation prevents copper-induced neuropathology in a murine model of Wilson’s disease. Cell Death Dis. 2021, 12, 87. [Google Scholar] [CrossRef]
- Ren, X.; Li, Y.; Zhou, Y.; Hu, W.; Yang, C.; Jing, Q.; Zhou, C.; Wang, X.; Hu, J.; Wang, L.; et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol. 2021, 46, 102122. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, C.; Han, F.; Lin, X.; Cao, L.; Liu, C.; Ji, Q.; Cui, J.; Yao, Y.; Wang, B.; et al. The regulation loop of MARVELD1 interacting with PARP1 in DNA damage response maintains genome stability and promotes therapy resistance of cancer cells. Cell Death Differ. 2023, 30, 922–937. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Liu, Y.; Xiong, F.; Wang, C. Improved Immunotherapy Outcomes via Cuproptosis Upregulation of HLA-DRA Expression: Promoting the Aggregation of CD4+ and CD8+T Lymphocytes in Clear Cell Renal Cell Carcinoma. Pharmaceuticals 2024, 17, 678. https://doi.org/10.3390/ph17060678
Wang B, Liu Y, Xiong F, Wang C. Improved Immunotherapy Outcomes via Cuproptosis Upregulation of HLA-DRA Expression: Promoting the Aggregation of CD4+ and CD8+T Lymphocytes in Clear Cell Renal Cell Carcinoma. Pharmaceuticals. 2024; 17(6):678. https://doi.org/10.3390/ph17060678
Chicago/Turabian StyleWang, Bowen, Yiwen Liu, Feng Xiong, and Chunyang Wang. 2024. "Improved Immunotherapy Outcomes via Cuproptosis Upregulation of HLA-DRA Expression: Promoting the Aggregation of CD4+ and CD8+T Lymphocytes in Clear Cell Renal Cell Carcinoma" Pharmaceuticals 17, no. 6: 678. https://doi.org/10.3390/ph17060678
APA StyleWang, B., Liu, Y., Xiong, F., & Wang, C. (2024). Improved Immunotherapy Outcomes via Cuproptosis Upregulation of HLA-DRA Expression: Promoting the Aggregation of CD4+ and CD8+T Lymphocytes in Clear Cell Renal Cell Carcinoma. Pharmaceuticals, 17(6), 678. https://doi.org/10.3390/ph17060678




