Implementation of Modern Therapeutic Drug Monitoring and Lipidomics Approaches in Clinical Practice: A Case Study with Colistin Treatment
Abstract
:1. Introduction
2. Results and Discussion
2.1. Case Presentation
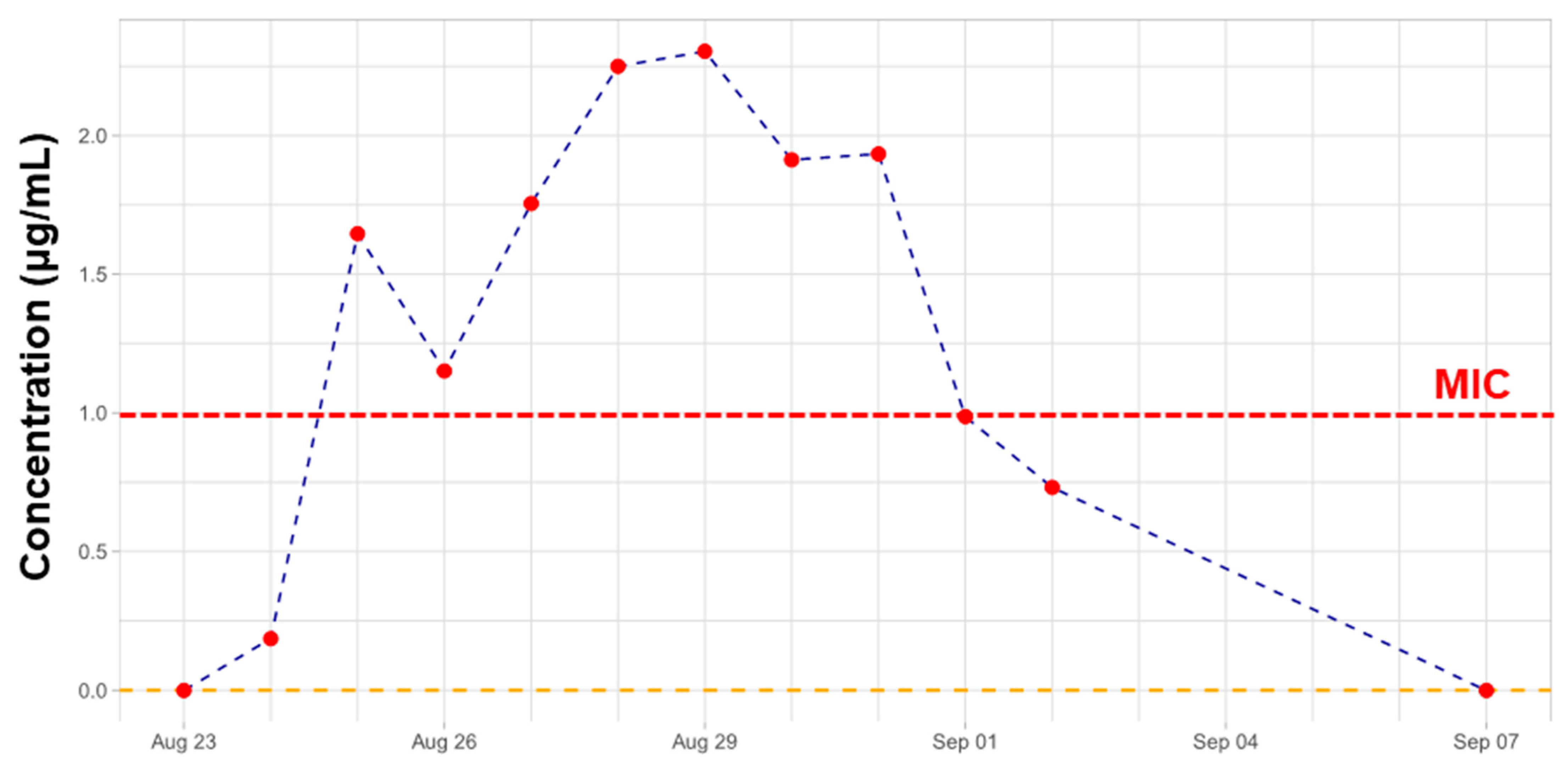
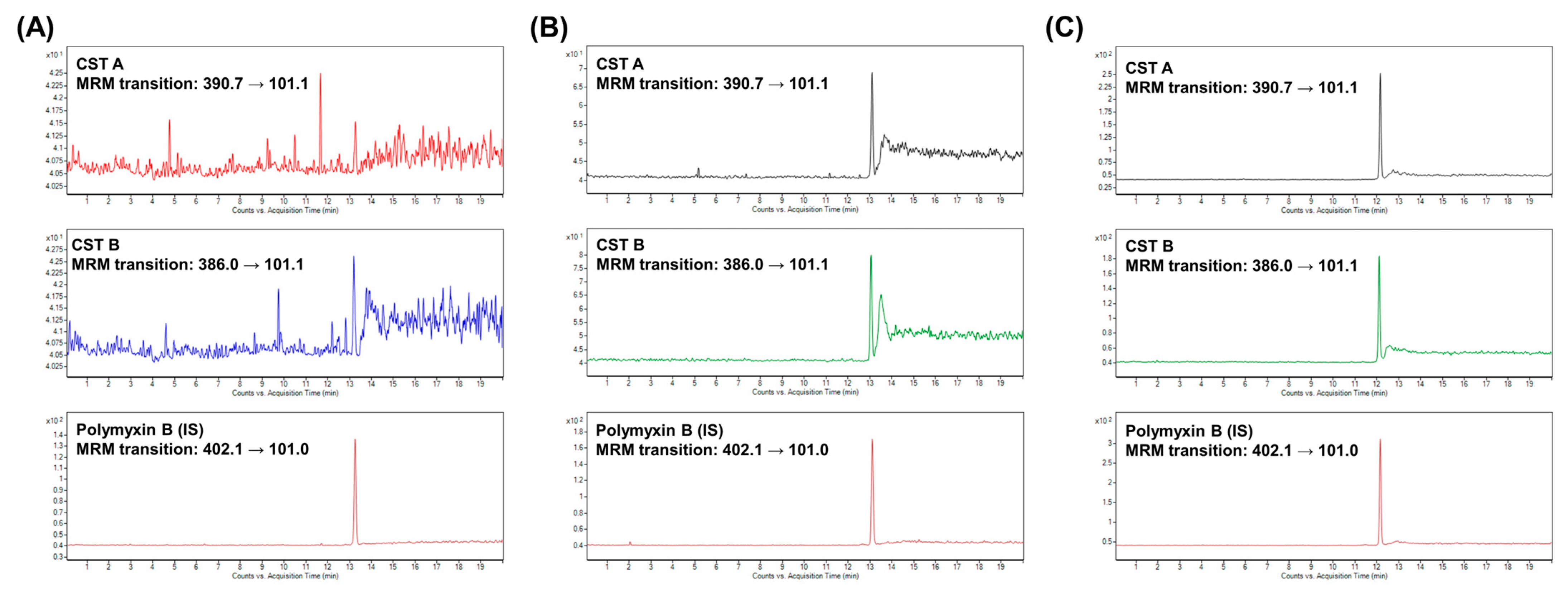
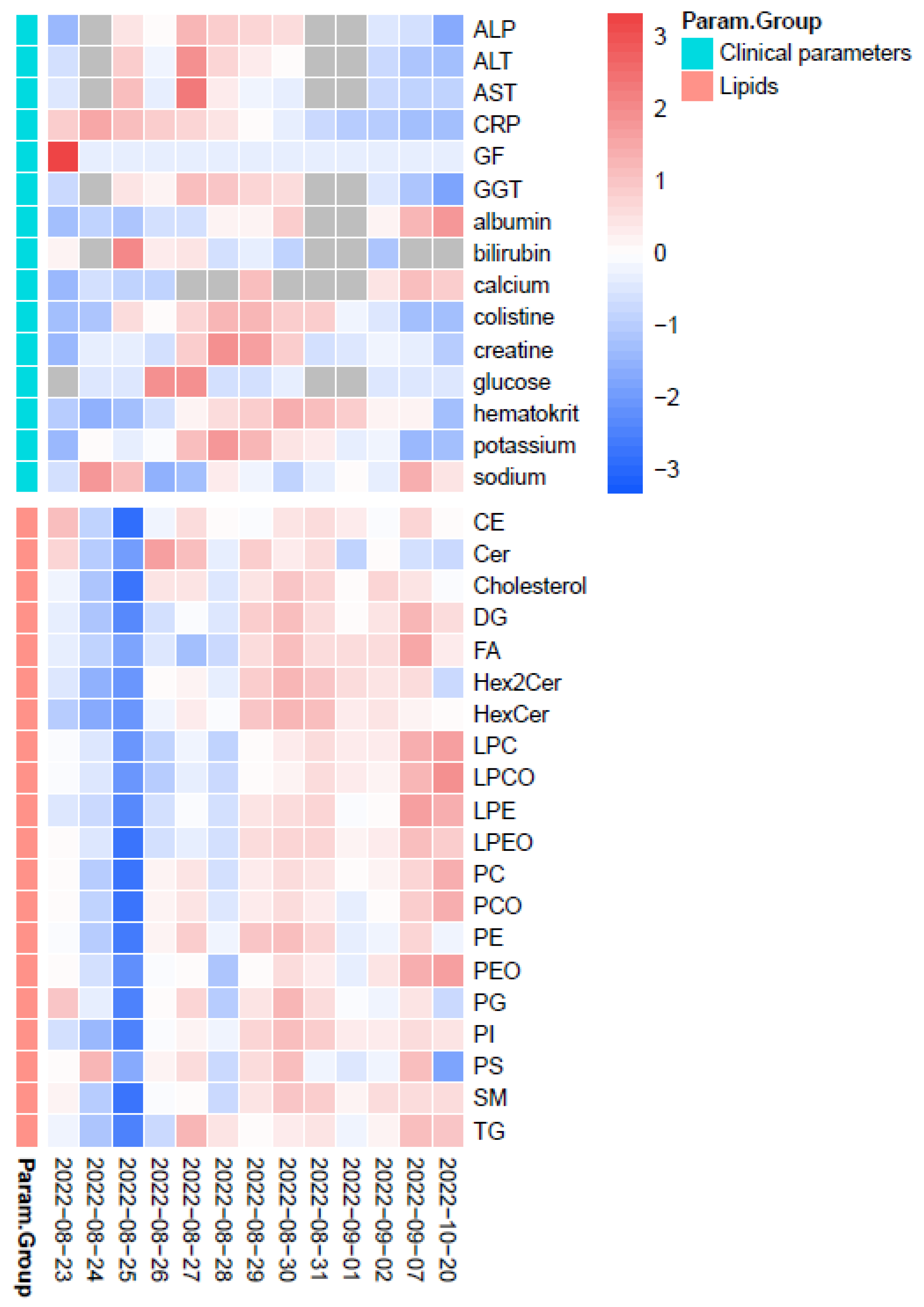
2.2. Therapeutic Drug Monitoring of CST
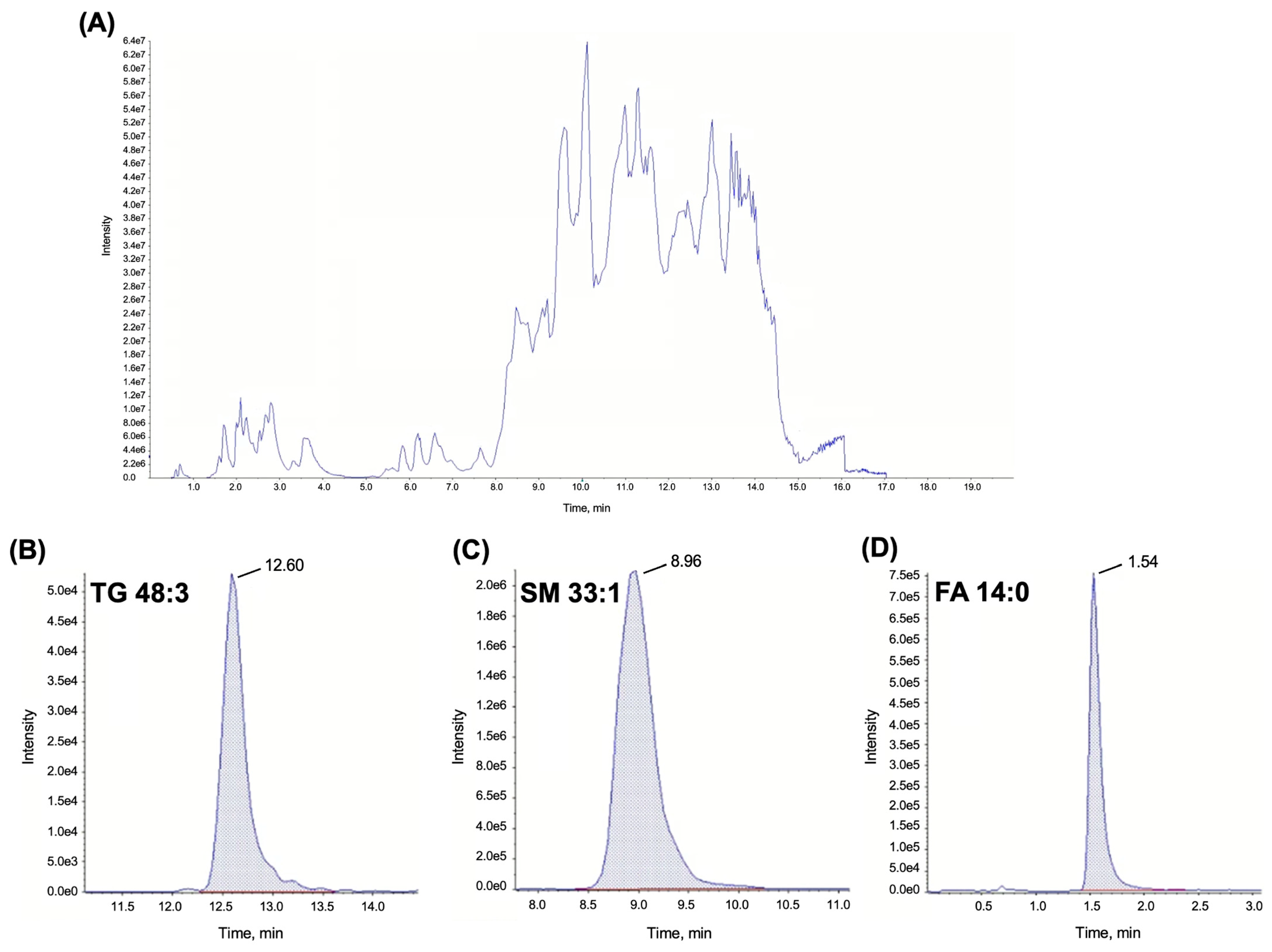
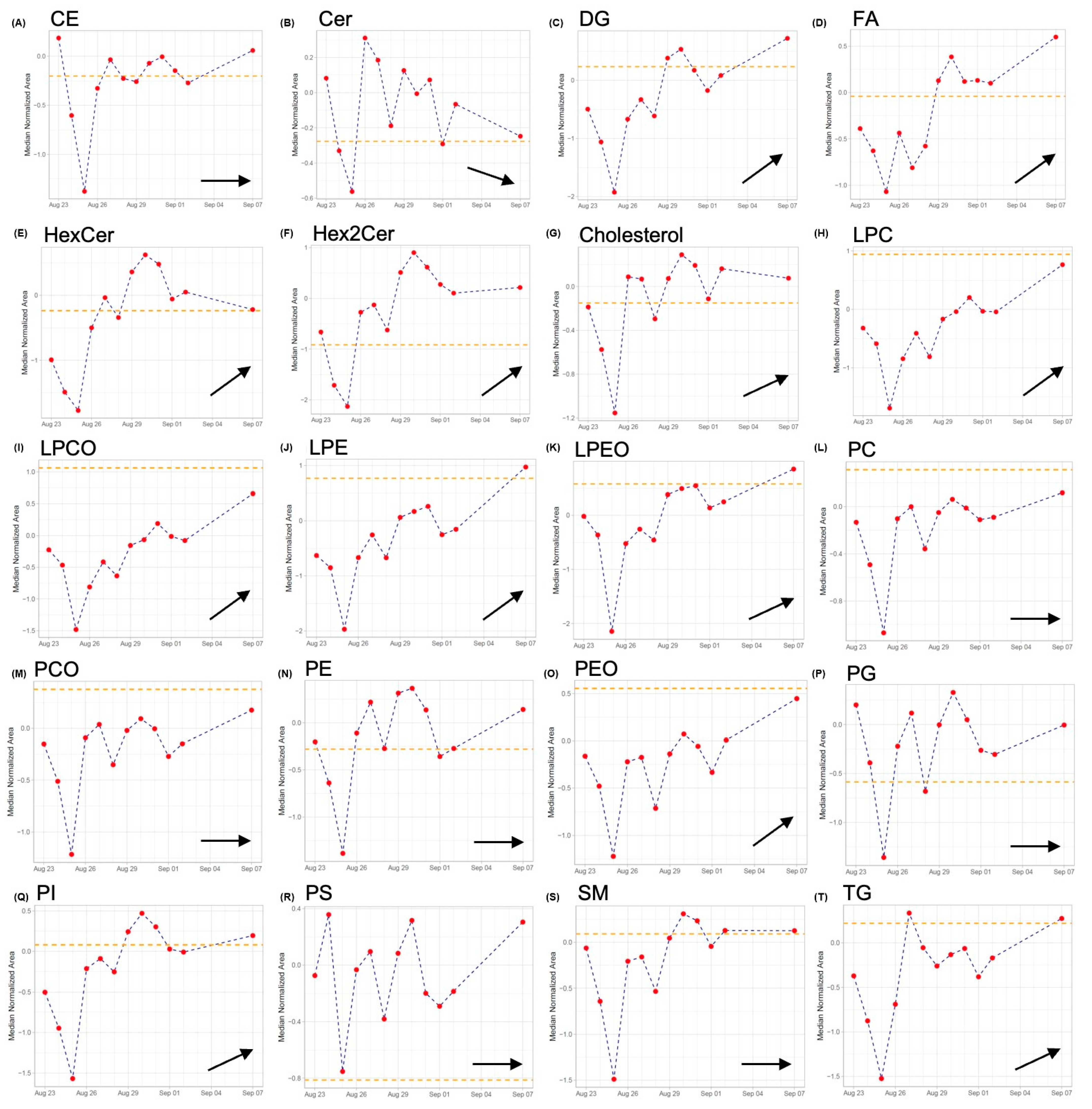
2.3. Lipidomics
2.4. TDM of CST in View of Lipidomics Data
2.5. Drawbacks of the Case Study and Future Perspectives
3. Materials and Methods
3.1. Therapeutic Drug Monitoring of CST
3.1.1. Chemicals and Samples
3.1.2. Instrumentation
3.1.3. Preparation of Standards and Samples
3.2. Lipidomics
3.2.1. Chemicals and Samples
3.2.2. Instrumentation
3.2.3. Preparation of Quality Control (QC) Samples and Clinical Samples
3.2.4. Data and Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Feretzakis, G.; Loupelis, E.; Sakagianni, A.; Skarmoutsou, N.; Michelidou, S.; Velentza, A.; Martsoukou, M.; Valakis, K.; Petropoulou, S.; Koutalas, E. A 2-year single-centre audit on antibiotic resistance of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae strains from intensive care unit and other wards in a general public hospital in Greece. Antibiotics 2019, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial antibiotic resistance: The most critical pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.R.; Balraj, T.A.; Kempegowda, S.N.; Prashant, A. Multidrug-resistant sepsis: A critical healthcare challenge. Antibiotics 2024, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Azad, M.A.K.; Roberts, K.D.; Bergen, P.J. History, Chemistry, and Antibacterial Spectrum. In Polymyxin Antibiotics: From Laboratory Bench to Bedside, 1st ed.; Li, J., Nation, R.L., Kaye, K.S., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 15–36. [Google Scholar]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef] [PubMed]
- Manioglu, S.; Modaresi, S.M.; Ritzmann, N.; Thoma, J.; Overall, S.A.; Harms, A.; Upert, G.; Luther, A.; Barnes, A.B.; Obrecht, D.; et al. Antibiotic polymyxin arranges lipopolysaccharide into crystalline structures to solidify the bacterial membrane. Nat. Commun. 2022, 13, 6195. [Google Scholar] [CrossRef] [PubMed]
- Rychlíčková, J.; Kubíčková, V.; Suk, P.; Urbánek, K. Challenges of colistin use in ICU and therapeutic drug monitoring: A literature review. Antibiotics 2023, 12, 437. [Google Scholar] [CrossRef]
- Meng, L.; Mui, E.; Ha, D.R.; Stave, C.; Deresinski, S.C.; Holubar, M. Comprehensive guidance for antibiotic dosing in obese adults: 2022 update. Pharmacotherapy 2023, 43, 226–246. [Google Scholar] [CrossRef]
- Koch, B.C.P.; Muller, A.E.; Hunfeld, N.G.M.; de Winter, B.C.M.; Ewoldt, T.M.J.; Abdulla, A.; Endeman, H. Therapeutic drug monitoring of antibiotics in critically ill patients: Current practice and future perspectives with focus on clinical outcome. Ther. Drug Monit. 2022, 44, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A. Introduction to therapeutic drug monitoring: Frequently and less frequently monitored drugs. In Therapeutic Drug Monitoring, 1st ed.; Dasgupta, A., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 1–29. [Google Scholar]
- Roberts, J.A.; Ross, N.; Peterson, D.L.; Martin, J.H. Therapeutic drug monitoring of antimicrobials. Br. J. Clin. Pharmacol. 2012, 73, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Mann, K.; Hiemke, C.; Schmidt, L.G.; Bates, D.W. Appropriateness of therapeutic drug monitoring for antidepressants in routine psychiatric inpatient care. Ther. Drug Monit. 2006, 28, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.S. Best practice in therapeutic drug monitoring. Br. J. Clin. Pharmacol. 2001, 52, 5S–10S. [Google Scholar] [CrossRef] [PubMed]
- Hammet-Stabler, C.A.; Dasgupta, A. Therapeutic Drug Monitoring Data: A Concise Guide, 3rd ed.; AACC Press: Washington, DC, USA, 2007; 241p. [Google Scholar]
- Thormann, W. Progress of capillary electrophoresis in therapeutic drug monitoring and clinical and forensic toxicology. Ther. Drug Monit. 2002, 24, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Ates, H.C.; Roberts, J.A.; Lipman, J.; Cass, A.E.G.; Urban, G.A.; Dincer, C. On-site therapeutic drug monitoring. Trends Biotechnol. 2020, 38, 1262–1277. [Google Scholar] [CrossRef] [PubMed]
- Shihabi, Z.K. Capillary electrophoresis for the determination of drugs in biological fluids. In Handbook of Analytical Separations, 1st ed.; Hempel, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2004; Volume 5, pp. 77–94. [Google Scholar]
- Le Brun, P.P.H.; de Graaf, A.I.; Vinks, A.A.T.M.M. High-performance liquid chromatographic method for the determination of colistin in serum. Ther. Drug Monit. 2000, 22, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Milne, R.W.; Nation, R.L.; Turnidge, J.D.; Coulthard, K.; Johnson, D.W. A simple method for the assay of colistin in human plasma, using pre-column derivatization with 9-fluorenylmethyl chloroformate in solid-phase extraction cartridges and reversed-phase high-performance liquid chromatography. J. Chromatogr. B 2001, 761, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.D.; Stern, R.C.; O’Riordan, M.A.; Blumer, J.L. The pharmacokinetics of colistin in patients with cystic fibrosis. J. Clin. Pharmacol. 2001, 41, 645–654. [Google Scholar] [CrossRef]
- Chepyala, D.; Tsai, I.-L.; Sun, H.-Y.; Lin, S.-W.; Kuo, C.-H. Development and validation of a high-performance liquid chromatography-fluorescence detection method for the accurate quantification of colistin in human plasma. J. Chromatogr. B 2015, 980, 48–54. [Google Scholar] [CrossRef]
- Jansson, B.; Karvanen, M.; Cars, O.; Plachouras, D.; Friberg, L.E. Quantitative analysis of colistin A and colistin B in plasma and culture medium using a simple precipitation step followed by LC/MS/MS. J. Pharm. Biomed. Anal. 2009, 49, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, J.; Gerber, J.P.; Milne, R.W. Determination of colistin in human plasma, urine and other biological samples using LC–MS/MS. J. Chromatogr. B 2008, 862, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Gobin, P.; Lemaître, F.; Marchand, S.; Couet, W.; Olivier, J.-C. Assay of colistin and colistin methanesulfonate in plasma and urine by liquid chromatography-tandem mass spectrometry. Antimicrob. Agents Chemother. 2010, 54, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Dotsikas, Y.; Markopoulou, C.K.; Koundourellis, J.E.; Loukas, Y.L. Validation of a novel LC-MS/MS method for the quantitation of colistin A and B in human plasma. J. Sep. Sci. 2011, 34, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Gikas, E.; Bazoti, F.N.; Katsimardou, M.; Anagnostopoulos, D.; Papanikolaou, K.; Inglezos, I.; Skoutelis, A.; Daikos, G.L.; Tsarbopoulos, A. Determination of colistin A and colistin B in human plasma by UPLC–ESI high resolution tandem MS: Application to a pharmacokinetic study. J. Pharm. Biomed. Anal. 2013, 83, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.-L.; Sun, H.-Y.; Chen, G.-Y.; Lin, S.-W.; Kuo, C.-H. Simultaneous quantification of antimicrobial agents for multidrug-resistant bacterial infections in human plasma by ultra-high-pressure liquid chromatography–tandem mass spectrometry. Talanta 2013, 116, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Leporati, M.; Bua, R.O.; Mariano, F.; Carignano, P.; Stella, M.; Biancone, L.; Vincenti, M. Determination by LC–MS/MS of colistins A and B in plasma and ultrafiltrate from critically ill patients undergoing continuous venovenous hemodiafiltration. Ther. Drug Monit. 2014, 36, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Mercier, T.; Tissot, F.; Gardiol, C.; Corti, N.; Wehrli, S.; Guidi, M.; Csajka, C.; Buclin, T.; Couet, W.; Marchetti, O.; et al. High-throughput hydrophilic interaction chromatography coupled to tandem mass spectrometry for the optimized quantification of the anti-Gram-negatives antibiotic colistin A/B and its pro-drug colistimethate. J. Chromatogr. A 2014, 1369, 52–63. [Google Scholar] [CrossRef]
- Zhao, M.; Wu, X.-J.; Fan, Y.-X.; Guo, B.-N.; Zhang, J. Development and validation of a UHPLC–MS/MS assay for colistin methanesulphonate (CMS) and colistin in human plasma and urine using weak-cation exchange solid-phase extraction. J. Pharm. Biomed. Anal. 2016, 124, 303–308. [Google Scholar] [CrossRef]
- Bihan, K.; Lu, Q.; Enjalbert, M.; Apparuit, M.; Langeron, O.; Rouby, J.-J.; Funck-Brentano, C.; Zahr, N. Determination of colistin and colistimethate levels in human plasma and urine by high-performance liquid chromatography–tandem mass spectrometry. Ther. Drug Monit. 2016, 38, 796–803. [Google Scholar] [CrossRef]
- Cangemi, G.; Barco, S.; Castagnola, E.; Tripodi, G.; Favata, F.; D’Avolio, A. Development and validation of UHPLC–MS/MS methods for the quantification of colistin in plasma and dried plasma spots. J. Pharm. Biomed. Anal. 2016, 129, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Binhashim, N.H.; Alvi, S.N.; Hammami, M.M. LC-MS/MS method for determination of colistin in human plasma: Validation and stability studies. IJAMSC 2021, 9, 1–11. [Google Scholar] [CrossRef]
- Matar, K.M.; Al-Refai, B. Quantification of colistin in plasma by liquid chromatography-tandem mass spectrometry: Application to a pharmacokinetic study. Sci. Rep. 2020, 10, 8198. [Google Scholar] [CrossRef] [PubMed]
- Qi, B.; Gijsen, M.; Van Brantegem, P.; De Vocht, T.; Deferm, N.; Abza, G.B.; Nauwelaerts, N.; Wauters, J.; Spriet, I.; Annaert, P. Quantitative determination of colistin A/B and colistin methanesulfonate in biological samples using hydrophilic interaction chromatography tandem mass spectrometry. Drug Test. Anal. 2020, 12, 1183–1195. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yu, S.; Chai, G.; Liu, J.; Zhou, Q.T. An LC-MS/MS method for simultaneous analysis of the cystic fibrosis therapeutic drugs colistin, ivacaftor and ciprofloxacin. J. Pharm. Anal. 2021, 11, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, Q.; Li, X.; Wang, G.; Xing, B.; Li, Z. Novel method for determination of colistin sulfate in human plasma by high-performance liquid chromatography-tandem mass spectrometry and its clinical applications in critically ill patients. J. Pharmacol. Toxicol. Methods 2024, 127, 107502. [Google Scholar] [CrossRef] [PubMed]
- Neef, S.K.; Hinderer, A.D.; Arbash, W.; Kinzig, M.; Sörgel, F.; Wunder, C.; Schwab, M.; Hofmann, U. A high performance liquid chromatography-tandem mass spectrometry assay for therapeutic drug monitoring of 10 drug compounds commonly used for antimicrobial therapy in plasma and serum of critically ill patients: Method optimization, validation, cross-validation and clinical application. Clin. Chim. Acta 2024, 559, 119690. [Google Scholar] [CrossRef] [PubMed]
- Cizmarova, I.; Parrak, V.; Secnik, P., Jr.; Secnik, P.; Sopko, L.; Nemergutova, K.; Kovac, A.; Mikus, P.; Piestansky, J. A simple and green capillary electrophoresis-mass spectrometry method for therapeutic drug monitoring of colistin in clinical plasma samples. Helyion 2023, 9, e23111. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Mun, S.; Lee, J.; Kang, H.-G. Discovery and validation of protein biomarkers for monitoring the effectiveness of drug treatment for major depressive disorder. J. Psychiatr. Res. 2024, 169, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and valuable tools towards diagnosis, prognosis and treatment of COVID-19 and other diseases. Helyion 2023, 9, e13323. [Google Scholar] [CrossRef]
- Aronson, J.K.; Ferner, R.E. Biomarkers—A general review. Curr. Protoc. Pharmacol. 2017, 76, 9.23.1–9.23.17. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, L.; Khakpoor-Koosheh, M.; Mirzaei, H.; Mirzaei, H.R. Biomarkers for predicting the outcome of various cancer immunotherapies. Crit. Rev. Oncol. Hematol. 2021, 157, 103161. [Google Scholar] [CrossRef] [PubMed]
- Astarita, G.; Kelly, R.S.; Lasky-Su, J. Metabolomics and lipidomics strategies in modern drug discovery and development. Drug Discov. Today 2023, 28, 103751. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Cheng, Y.; Zhu, H.; Zha, L. Metabolomics and lipidomics in non-small cell lung cancer. Clin. Chim. Acta 2024, 555, 117823. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, M.; Cheng, X.; Liu, X.; Sun, H.; Guo, Z.; Li, J.; Tang, X.; Wang, Z.; Sun, W.; et al. LC-MS-Based Plasma Metabolomics and Lipidomics Analyses for Differential Diagnosis of Bladder Cancer and Renal Cell Carcinoma. Front. Oncol. 2020, 10, 717. [Google Scholar] [CrossRef] [PubMed]
- Koen, N.; van Breda, S.V.; Loots, D.T. Metabolomics of colistin methanesulfonate treated Mycobacterium tuberculosis. Tuberculosis 2018, 111, 154–160. [Google Scholar] [CrossRef]
- Carfrae, L.A.; Rachwalski, K.; French, S.; Gorzevich, R.; Seidel, L.; Tsai, C.N.; Tu, M.M.; MacNair, C.R.; Ovchinnikova, O.G.; Clarke, B.R.; et al. Inhibiting fatty acid synthesis overcomes colistin resistance. Nat. Microbiol. 2023, 8, 1026–1038. [Google Scholar] [CrossRef] [PubMed]
- Meikle, T.G.; Huynh, K.; Giles, C.; Meikle, P.J. Clinical lipidomics: Realizing the potential of lipid profiling. J. Lipid Res. 2021, 62, 100127. [Google Scholar] [CrossRef] [PubMed]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from Sample Preparation to Data Analysis: A Primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef]
- Xu, T.; Hu, C.; Xuan, Q.; Xu, G. Recent Advances in Analytical Strategies for Mass Spectrometry-Based Lipidomics. Anal. Chim. Acta 2020, 1137, 156–169. [Google Scholar] [CrossRef]
- Kao, Y.C.; Ho, P.C.; Tu, Y.K.; Jou, I.M.; Tsai, K.J. Lipids and Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 1505. [Google Scholar] [CrossRef] [PubMed]
- Ulmer, C.Z.; Koelmel, J.P.; Jones, C.M.; Garrett, T.J.; Aristizabal-Henao, J.J.; Vesper, H.W.; Bowden, J.A. A Review of Efforts to Improve Lipid Stability during Sample Preparation and Standardization Efforts to Ensure Accuracy in the Reporting of Lipid Measurements. Lipids 2021, 56, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.E.; Zhu, S.; Liu, H. Recent Progresses of Derivatization Approaches in the Targeted Lipidomics Analysis by Mass Spectrometry. J. Sep. Sci. 2020, 43, 1838–1846. [Google Scholar] [CrossRef] [PubMed]
- Kostidis, S.; Sánchez-López, E.; Giera, M. Lipidomics analysis in drug discovery and development. Curr. Opin. Chem. Biol. 2023, 72, 102256. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.E.A.; Ontiveros, E.N.; Arreola, R.; González, A.M.E.; Bores, A.M.G.; Urrutia, R.E.L.; Castro, I.P.; Correa, M.S.S.; Parra, E.A.E. Lipidomics as a tool in the diagnosis and clinical therapy. In Fatty Acids–From Biosynthesis to Human Health, 1st ed.; Froyen, E., Ed.; InTech Open: Rijeka, Croatia, 2022; pp. 1–23. [Google Scholar] [CrossRef]
- Sulaiman, D.; Wu, D.; Black, L.P.; Williams, K.J.; Graim, K.; Datta, S.; Reddy, S.T.; Guirgis, F.W. Lipidomic changes in a novel sepsis outcome-based analysis reveals potent pro-inflammatory and pro-resolving signaling lipids. Clin. Transl. Sci. 2024, 17, e13745. [Google Scholar] [CrossRef] [PubMed]
- Mecatti, G.C.; Messias, M.C.F.; de Oliveira Carvalho, P. Lipidomic profile and candidate biomarkers in septic patients. Lipids Health Dis. 2020, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Mosevoll, K.A.; Hansen, B.A.; Gundersen, I.M.; Reikvam, H.; Bruserud, Ø.; Bruserud, Ø.; Wendelbo, Ø. Patients with Bacterial Sepsis Are Heterogeneous with Regard to Their Systemic Lipidomic Profiles. Metabolites 2023, 13, 52. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, J.; Zhang, J.; Rong, J.; Wang, X.; Zhao, C.; Zhang, H.; Shi, H.; Wu, W. U HPLC-MS/MS-based untargeted lipidomics analysis of septic patients. Clin. Chim. Acta 2023, 544, 117336. [Google Scholar] [CrossRef] [PubMed]
- Mecatti, G.C.; Fernandes Messias, M.C.; Sant’Anna Paiola, R.M.; Figueiredo Angolini, C.F.; da Silva Cunha, I.B.; Eberlin, M.N.; de Oliveira Carvalho, P. Lipidomic Profiling of Plasma and Erythrocytes From Septic Patients Reveals Potential Biomarker Candidates. Biomark. Insights 2018, 26, 13. [Google Scholar] [CrossRef]
- Mansell, T.; Saffery, R.; Burugupalli, S.; Ponsonby, A.-L.; Tang, M.L.K.; O’Hely, M.; Bekkering, S.; Smith, A.A.T.; Rowland, R.; Barwon Infant Study Investigator Group; et al. Early life infection and proinflammatory, atherogenic metabolomic and lipidomic profiles in infancy: A population-based cohort study. eLife 2022, 11, e75170. [Google Scholar] [CrossRef]
- Chen, H.; Chen, J.-B.; Du, L.-N.; Yuan, H.-X.; Shan, J.-J.; Wang, S.-C.; Ye, J.; Lin, L.-L. Integration of lipidomics and metabolomics reveals plasma and urinary profiles associated with pediatric Mycoplasma pneumoniae infections and its severity. Biomed. Chromatogr. 2024, 38, e5817. [Google Scholar] [CrossRef] [PubMed]
- Anh, N.K.; Phat, N.K.; Yen, N.T.H.; Jayanti, R.P.; Thu, V.T.A.; Park, Y.J.; Cho, Y.-S.; Shin, J.-G.; Kim, D.-H.; Oh, J.Y.; et al. Comprehensive lipid profiles investigation reveals host metabolic and immune alterations during anti-tuberculosis treatment: Implications for therapeutic monitoring. Biomed. Pharmacother. 2023, 158, 114187. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.J.; Ellis, S.R.; Hofstaedter, C.E.; Heeren, R.M.A.; Ernst, R.K. Spatial lipidomics reveals biased phospholipid remodeling in acute Pseudomonas lung infection. iScience 2023, 26, 107700. [Google Scholar] [CrossRef] [PubMed]
- Goetz, L.H.; Schork, N.J. Personalized medicine. Motivation, challenges, and progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Qiu, S.; Ji, L.; Lu, H.; Wu, S.; Chen, Q.; Zou, X.; Hu, Q.; Feng, T.; Chen, S.; et al. Pathogenic characterization of a Micrococcus luteus strain isolated from an infant. Front. Pediatr. 2023, 11, 1303040. [Google Scholar] [CrossRef]
- Haseeb, A.; Faidah, H.S.; Alghamdi, S.; Alotaibi, A.F.; Elrggal, M.E.; Mahrous, A.J.; Abuhussain, S.S.A.; Obaid, N.A.; Algethamy, M.; AlQarni, A.; et al. Dose optimization of colistin: A systematic review. Antibiotics 2021, 10, 1454. [Google Scholar] [CrossRef] [PubMed]
- Nation, R.L.; Rigatto, M.H.; Falci, D.R.; Zavascki, A.P. Polymyxin acute kidney injury: Dosing and other strategies to reduce toxicity. Antibiotics 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Regenthal, R.; Krueger, M.; Koeppel, C.; Preiss, R. Drug levels: Therapeutic and toxic serum/plasma concentrations of common drugs. J. Clin. Monit. Comput. 1999, 15, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Sedlák, F.; Kvasnička, A.; Marešová, B.; Brumarová, R.; Dobešová, D.; Dostálová, K.; Šrámková, K.; Pehr, M.; Šácha, P.; Friedecký, D.; et al. Parallel Metabolomics and Lipidomics of a PSMA/GCPII Deficient Mouse Model Reveal Alteration of NAAG Levels and Brain Lipid Composition. ACS Chem. Neurosci. 2024, 15, 1342–1355. [Google Scholar] [CrossRef]
- Doß, S.; Blessing, C.; Haller, K.; Richter, G.; Sauer, M. Influence of Antibiotics on Functionality and Viability of Liver Cells In Vitro. Curr. Issues Mol. Biol. 2022, 44, 4639–4657. [Google Scholar] [CrossRef]
- de Pablo, M.A.; Angeles Puertollano, M.; Alvarez de Cienfuegos, G. Immune cell functions, lipids and host natural resistance. FEMS Immunol. Med. Microbiol. 2000, 4, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Andersen, C.J.; Blesso, C.N. The Role of Lipids in the Regulation of Immune Responses. Nutrients 2023, 15, 3899. [Google Scholar] [CrossRef] [PubMed]
- Sorlí, L.; Luque, S.; Grau, S.; Berenguer, N.; Segura, C.; Montero, M.M.; Álvarez-Lerma, F.; Knobel, H.; Benito, N.; Horcada, J.P. Trough colistin plasma level is an independent risk factor for nephrotoxicity: A prospective observational cohort study. BMC Infect. Dis. 2013, 13, 380. [Google Scholar] [CrossRef] [PubMed]
- Olešová, D.; Dobešová, D.; Majerová, P.; Brumarová, R.; Kvasnička, A.; Kouřil, Š.; Stevens, E.; Hanes, J.; Fialová, Ľ.; Michalicová, A.; et al. Changes in lipid metabolism track with the progression of neurofibrillary pathology in tauopathies. J. Neuroinflammation 2024, 21, 78. [Google Scholar] [CrossRef]
| Clinical Parameter | Day 1 | Day 2 *1 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 *2 | Day 9 | Day 10 | Day 11 | Day 12 | Follow Up *3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serum creatinine (μmol/L) | 70.6 | 89.9 | 89.7 | 83.9 | 109.7 | 129 | 125 | 111 | 84 | 85.7 | 92.3 | 89.5 | 76.2 |
| GF (mL/s) | 1.89 | 1.56 | 1.56 | 1.69 | 1.22 | 1.00 | 1.04 | 1.20 | 1.69 | 1.65 | 1.51 | 1.56 | 1.83 |
| ALT (μkat/L) | 1.27 | n.d. | 3.46 | 2.00 | 5.00 | 3.24 | 2.66 | 2.37 | n.d. | n.d. | 1.12 | 0.53 | 0.35 |
| AST (μkat/L) | 1.46 | n.d. | 5.27 | 1.87 | 8.34 | 3.40 | 2.21 | 1.83 | n.d. | n.d. | 0.78 | 0.57 | 0.43 |
| GGT (μkat/L) | 3.79 | n.d. | 7.10 | 6.31 | 9.11 | 8.66 | 8.04 | 7.61 | n.d. | n.d. | 4.47 | 2.78 | 0.77 |
| ALP (μkat/L) | 2.28 | n.d. | 5.14 | 4.37 | 6.32 | 5.60 | 5.55 | 5.30 | n.d. | n.d. | 3.66 | 3.56 | 1.89 |
| Albumin (g/L) | 26.6 | 28.6 | 27.0 | 29.5 | 29.6 | 33.0 | 33.0 | 35.9 | n.d. | n.d. | 33.0 | 38.0 | 40.7 |
| CRP (mg/L) | 335 | 442 | 364.8 | 338 | 313.4 | 263.5 | 201.0 | 153.4 | 80.2 | 49.5 | 37.26 | 8.35 | 5.37 |
| Glucose (mmol/L) | n.d. | 5.88 | 5.77 | 16.19 | 16.4 | 4.9 | 5.18 | 6.11 | n.d. | n.d. | 5.32 | 5.82 | 5.58 |
| Bilirubin (mmol/L) | 8.4 | n.d. | 11.8 | 8.6 | 8.8 | 6.9 | 7.4 | 6.5 | n.d. | n.d. | 6.1 | n.d. | n.d. |
| Na (mmol/L) | 134 | 145 | 142 | 130 | 131 | 138 | 136 | 133 | 135 | 137 | 135 | 143 | 139 |
| K (mmol/L) | 3.41 | 3.94 | 3.80 | 3.92 | 4.38 | 4.60 | 4.40 | 4.10 | 4.06 | 3.80 | 3.88 | 3.40 | 3.48 |
| Hematocrit (%) | 26.4 | 24.4 | 25.1 | 27.3 | 29.6 | 30.7 | 32.3 | 34 | 32.8 | 32.4 | 30 | 30 | 25.3 |
| Time (min) | MP A [%] | MP B [%] |
|---|---|---|
| 0.00 | 68 | 32 |
| 1.50 | 68 | 32 |
| 15.50 | 15 | 85 |
| 15.60 | 3 | 97 |
| 18.00 | 3 | 97 |
| 18.01 | 68 | 32 |
| 20.00 | 68 | 32 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerhardtova, I.; Cizmarova, I.; Jankech, T.; Olesova, D.; Jampilek, J.; Parrak, V.; Nemergutova, K.; Sopko, L.; Piestansky, J.; Kovac, A. Implementation of Modern Therapeutic Drug Monitoring and Lipidomics Approaches in Clinical Practice: A Case Study with Colistin Treatment. Pharmaceuticals 2024, 17, 753. https://doi.org/10.3390/ph17060753
Gerhardtova I, Cizmarova I, Jankech T, Olesova D, Jampilek J, Parrak V, Nemergutova K, Sopko L, Piestansky J, Kovac A. Implementation of Modern Therapeutic Drug Monitoring and Lipidomics Approaches in Clinical Practice: A Case Study with Colistin Treatment. Pharmaceuticals. 2024; 17(6):753. https://doi.org/10.3390/ph17060753
Chicago/Turabian StyleGerhardtova, Ivana, Ivana Cizmarova, Timotej Jankech, Dominika Olesova, Josef Jampilek, Vojtech Parrak, Kristina Nemergutova, Ladislav Sopko, Juraj Piestansky, and Andrej Kovac. 2024. "Implementation of Modern Therapeutic Drug Monitoring and Lipidomics Approaches in Clinical Practice: A Case Study with Colistin Treatment" Pharmaceuticals 17, no. 6: 753. https://doi.org/10.3390/ph17060753





