Unveiling the Bioactive Efficacy of Cupressus sempervirens ‘Stricta’ Essential Oil: Composition, In Vitro Activities, and In Silico Analyses
Abstract
1. Introduction
2. Results
2.1. Chemical Composition of CSSLEO
2.2. In Vitro Assessments
2.2.1. Cytotoxic Potential and Selectivity of CSSLEO
2.2.2. Antidiabetic Activity
α-Glucosidase Inhibition Activity
α-Amylase Inhibition Activity
2.2.3. Antioxidant Activity
2.2.4. Antimicrobial Activity
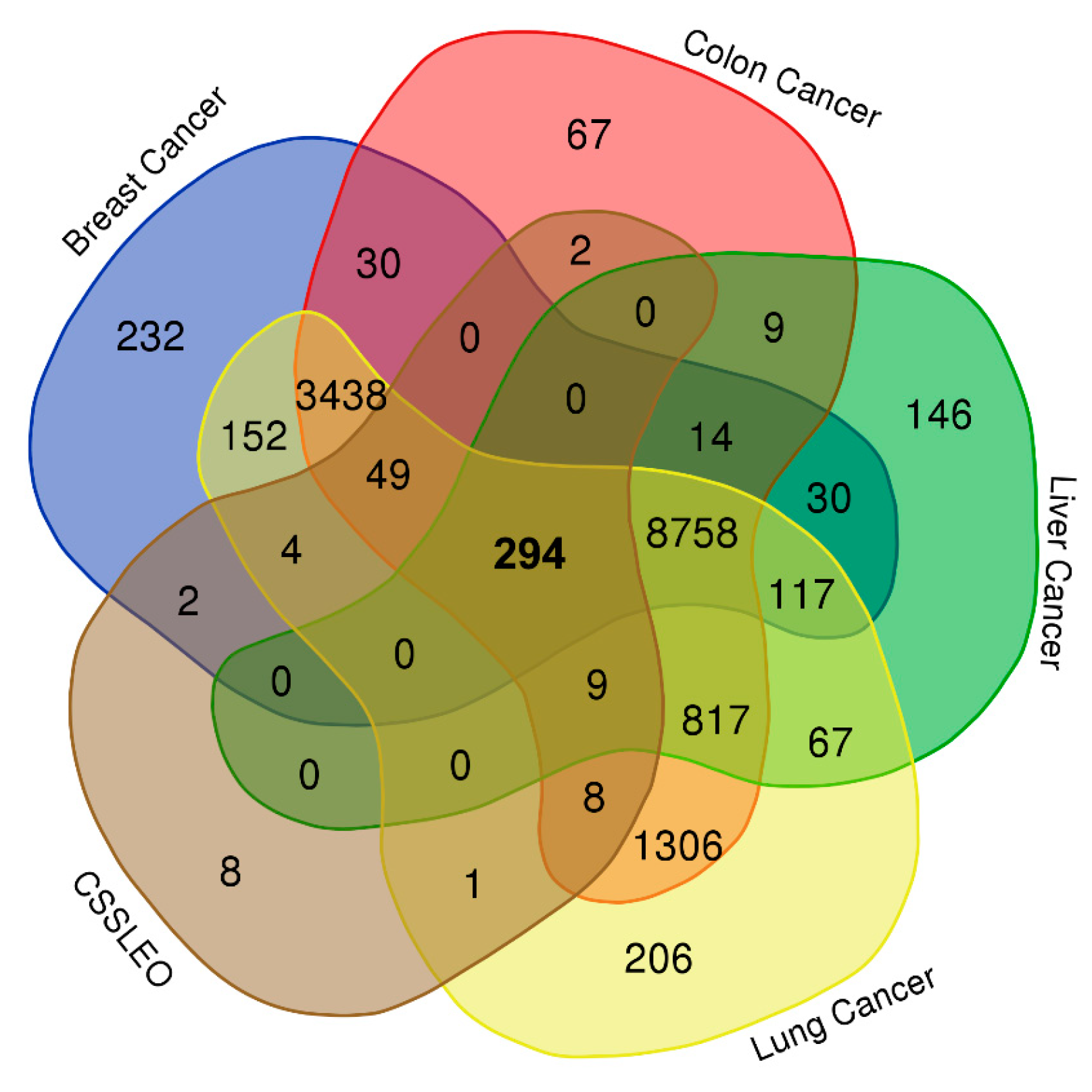
2.3. Network Pharmacology-Based Analysis
2.3.1. Drug-Likeness Screening of the Identified Compounds and Acquiring Compound and Disease-Related Targets
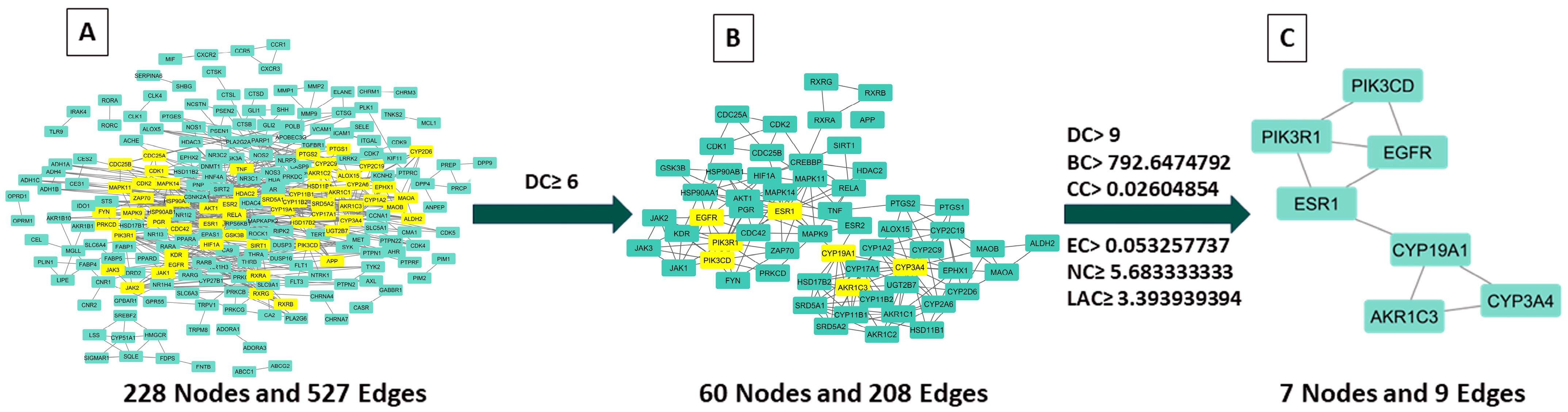
2.3.2. Examination of Hub Genes Using Protein–Protein Interactions and Topological Analysis
2.3.3. Key CSSLEO Compounds Associated with Cancer Targets
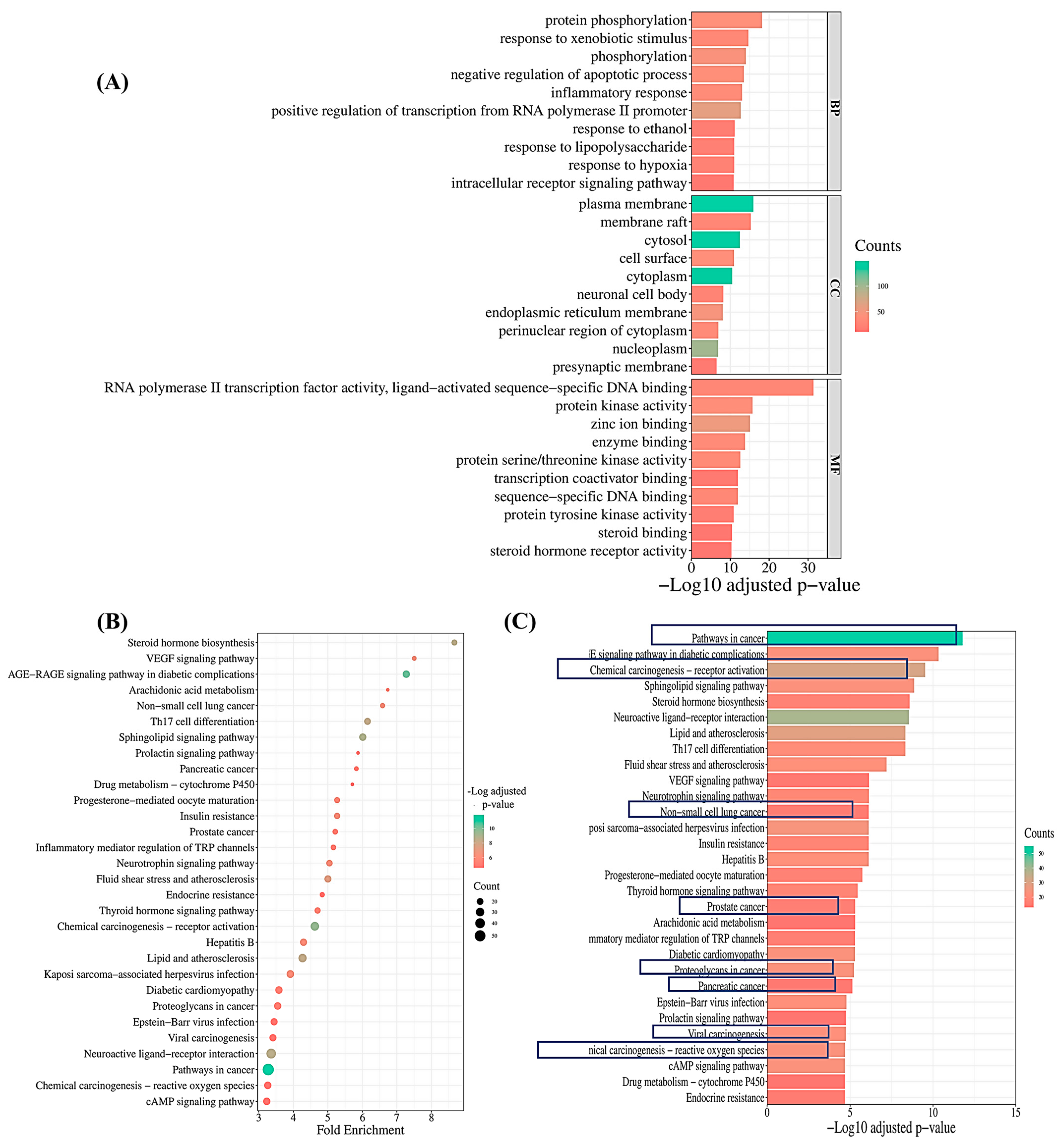
2.3.4. Pathway Enrichment Analysis
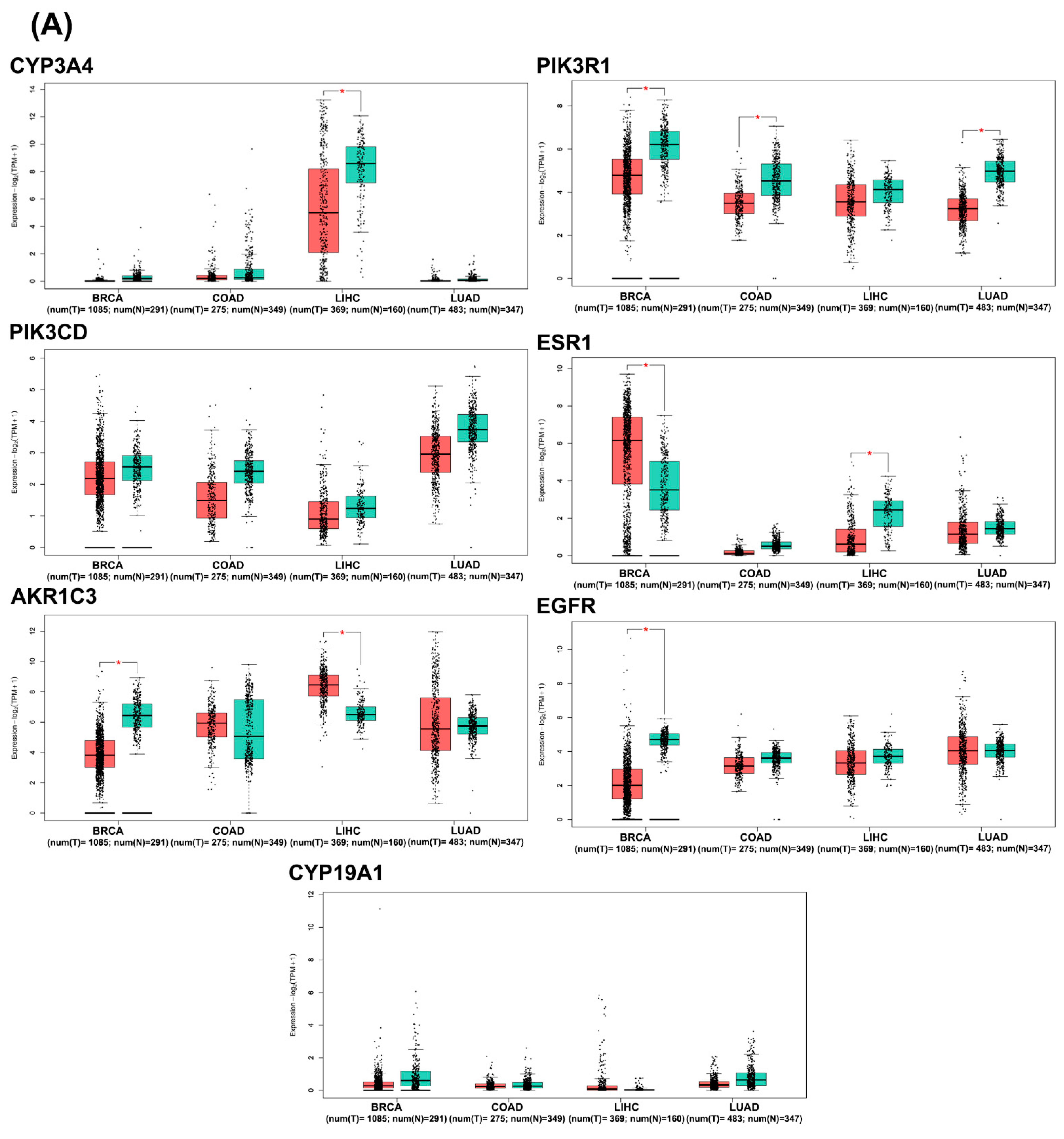
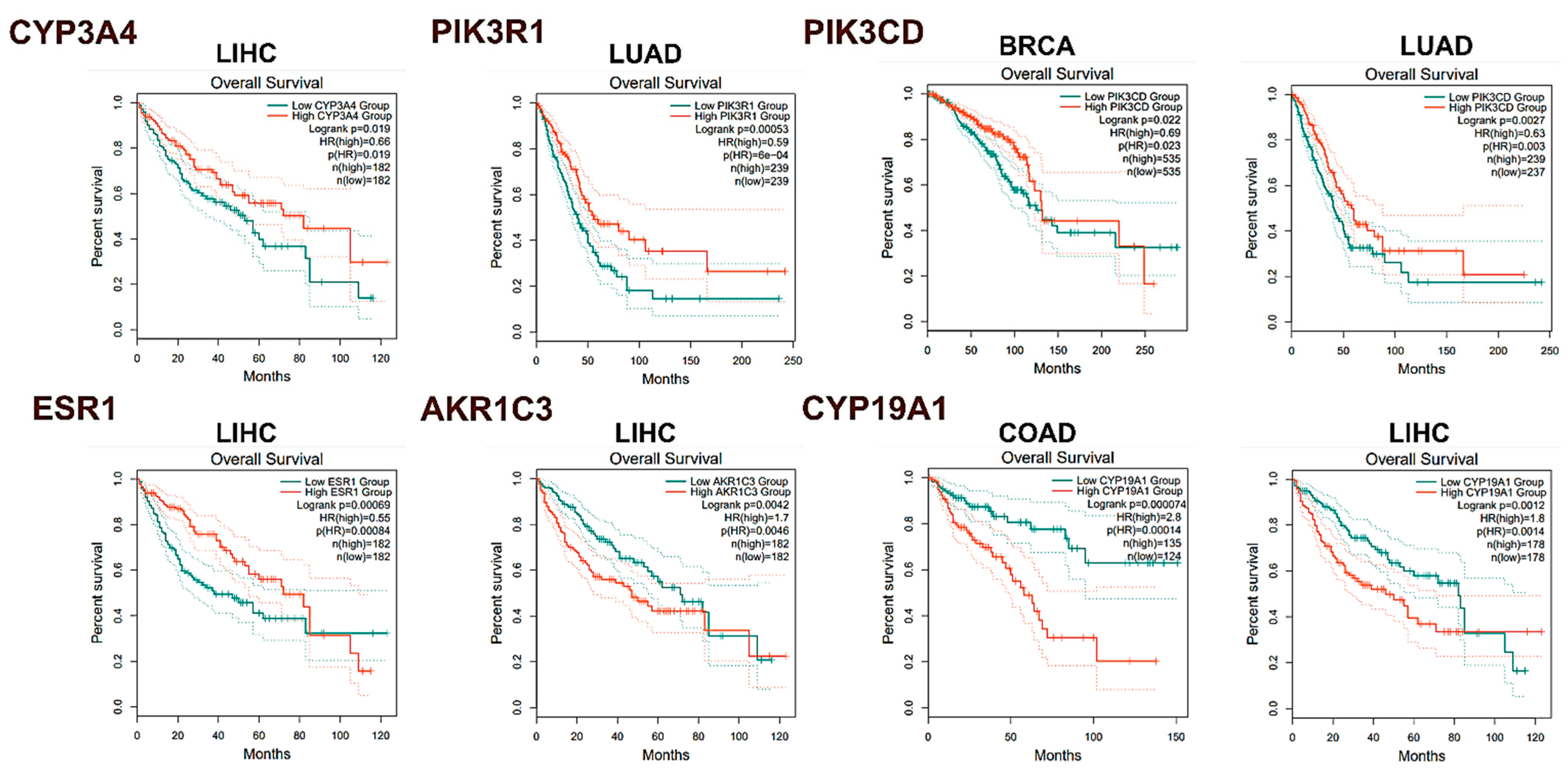
2.3.5. In Silico Analysis of Hub Gene Expression and Prognostic Significance
2.4. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.1.1. For Cytotoxic Assay
4.1.2. For Antioxidant and Antidiabetic Assay
4.2. Plant Material and Sample Preparation
4.3. GC–MS Analysis of CSSLEO
4.4. In Vitro Studies
4.4.1. Assessment of Cytotoxic Activity
Cell Line Propagation
Assessment of Cytotoxicity Using MTT-Based Cell Viability Assay
Data Analysis and Statistical Procedures
Quantifying CSSLEO’s Cytotoxic Selectivity
4.4.2. Assessment of Antioxidant Activity
4.4.3. Assessment of Antidiabetic Activity
α-Glucosidase Inhibition Activity
α-Amylase Inhibition Activity
Statistical Analysis
4.4.4. Assessment of Antimicrobial Activity
Antifungal and Antibacterial Activities (Well Diffusion Method)
Determination of Minimum Inhibitory Concentration (MIC)
4.5. Network Pharmacology
4.5.1. Evaluating the Pharmacokinetic Properties of CSSLEO Phytoconstituents
4.5.2. Identification of the Intersection Genes between Breast, Colon, Liver, and Lung Cancers and CSSLEO Bioactive Compounds
4.5.3. Constructing the Protein–Protein Interaction (PPI) Network
4.5.4. Establishing the Compound-Target Interaction Network
4.5.5. Enrichment Analysis of GO and KEGG Pathways
4.5.6. Computational Analysis of Gene Expression in Cancer Datasets
4.6. Molecular Docking Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ali, B.; Al-Wabel, N.A.; Shams, S.; Ahamad, A.; Khan, S.A.; Anwar, F. Essential oils used in aromatherapy: A systemic review. Asian Pac. J. Trop. Biomed. 2015, 5, 601–611. [Google Scholar] [CrossRef]
- Eslahi, H.; Fahimi, N.; Sardarian, A.R. Chemical composition of essential oils. Essent. Oils Food Process. Chem. Saf. Appl. 2017, 119–171. [Google Scholar] [CrossRef]
- Bhavaniramya, S.; Vishnupriya, S.; Al-Aboody, M.S.; Vijayakumar, R.; Baskaran, D. Role of essential oils in food safety: Antimicrobial and antioxidant applications. Grain Oil Sci. Technol. 2019, 2, 49–55. [Google Scholar] [CrossRef]
- Alves-Silva, J.M.; Zuzarte, M.; Girão, H.; Salgueiro, L. The role of essential oils and their main compounds in the management of cardiovascular disease risk factors. Molecules 2021, 26, 3506. [Google Scholar] [CrossRef] [PubMed]
- Aljaafari, M.N.; AlAli, A.O.; Baqais, L.; Alqubaisy, M.; AlAli, M.; Molouki, A.; Ong-Abdullah, J.; Abushelaibi, A.; Lai, K.-S.; Lim, S.-H.E. An overview of the potential therapeutic applications of essential oils. Molecules 2021, 26, 628. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, T.; Hong, J.-H.; Asnaashari, S.; Farajnia, S.; Delazar, A.; Hamishehkar, H.; Kim, K.H. Natural phytochemicals derived from gymnosperms in the prevention and treatment of cancers. Int. J. Mol. Sci. 2021, 22, 6636. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Sati, S. Antifungal potential of gymnosperms: A review. Microbes Divers. Biotechnol. 2012, 333–345. [Google Scholar]
- Ancuceanu, R.; Anghel, A.I.; Hovaneț, M.V.; Ciobanu, A.-M.; Lascu, B.E.; Dinu, M. Antioxidant Activity of Essential Oils from Pinaceae Species. Antioxidants 2024, 13, 286. [Google Scholar] [CrossRef] [PubMed]
- Swor, K.; Satyal, P.; Poudel, A.; Setzer, W.N. Gymnosperms of Idaho: Chemical compositions and enantiomeric distributions of essential oils of Abies lasiocarpa, Picea engelmannii, Pinus contorta, Pseudotsuga menziesii, and Thuja plicata. Molecules 2023, 28, 2477. [Google Scholar] [CrossRef]
- Schulz, C.; Knopf, P.; Stützel, T. Identification key to the Cypress family (Cupressaceae). Feddes Repert. Z. Für Bot. Taxon. Und Geobot. 2005, 116, 96–146. [Google Scholar] [CrossRef]
- Park, C.; Woo, H.; Park, M.-J. Development of Pinaceae and Cupressaceae Essential Oils from Forest Waste in South Korea. Plants 2023, 12, 3409. [Google Scholar] [CrossRef] [PubMed]
- Santos Filho, F.C.; da Silva Amaral, L.; Rodrigues-Filho, E. Composition of essential oils from Cupressus lusitanica and a Xylariaceous fungus found on its leaves. Biochem. Syst. Ecol. 2011, 39, 485–490. [Google Scholar] [CrossRef]
- Frezza, C.; De Vita, D.; Sciubba, F.; Toniolo, C.; Tomassini, L.; Nicoletti, M.; Franceschin, M.; Guiso, M.; Bianco, A.; Serafini, M.; et al. There Is Not Only Cupressus sempervirens L.: A Review on the Phytochemistry and Bioactivities of the Other Cupressus L. Species. Appl. Sci. 2022, 12, 7353. [Google Scholar] [CrossRef]
- Patgar, B.; Satish, S.; Shabaraya, A. Essential Oil of Cupressus sempervirens: A Brief Review. World J. Pharm. Pharm. Sci. 2021, 10, 864–872. [Google Scholar] [CrossRef]
- Hussain, K.M.; Saquib, M.; Ahamad, T.; Khatoon, S.; Khan, F.M. Mediterranean cypress “Cupressus sempervirens”: A Review on Phytochemical and Pharmacological Properties. Curr. Tradit. Med. 2019, 5, 278–297. [Google Scholar] [CrossRef]
- Orhan, I.E.; Tumen, I. Potential of Cupressus sempervirens (Mediterranean cypress) in health. In The Mediterranean Diet; Elsevier: Amsterdam, The Netherlands, 2015; pp. 639–647. [Google Scholar] [CrossRef]
- Al-Rajhi, A.M.H.; Bakri, M.M.; Qanash, H.; Alzahrani, H.Y.; Halawani, H.; Algaydi, M.A.; Abdelghany, T.M. Antimicrobial, Antidiabetic, Antioxidant, and Anticoagulant Activities of Cupressus sempervirens In Vitro and In Silico. Molecules 2023, 28, 7402. [Google Scholar] [CrossRef] [PubMed]
- Weick, C.W.; Aamir, N.; Reichart, J. The Ethnobotanical Evolution of the Mediterranean cypress (Cupressus sempervirens). Econ. Bot. 2023, 77, 203–221. [Google Scholar] [CrossRef]
- Skenfield, M.W. Historic Trees of the California Gold Rush Towns; Lulu Press: Morrisville, NC, USA, 2019. [Google Scholar]
- Rowell, R.J. Ornamental Conifers for Australian Gardens; UNSW Press: Sydney, Australia, 1996. [Google Scholar]
- Dirr, M.A. Dirr’s Encyclopedia of Trees and Shrubs; Timber Press: Portland, OR, USA, 2016. [Google Scholar]
- Boukhris, M.; Regane, G.; Yangui, T.; Sayadi, S.; Bouaziz, M. Chemical composition and biological potential of essential oil from Tunisian Cupressus sempervirens L. J. Arid Land Stud. 2012, 22, 329–332. [Google Scholar]
- Batiha, G.E.-S.; Teibo, J.O.; Shaheen, H.M.; Akinfe, O.A.; Awad, A.A.; Teibo, T.K.A.; Alexiou, A.; Papadakis, M. Bioactive compounds, pharmacological actions and pharmacokinetics of Cupressus sempervirens. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2023, 396, 389–403. [Google Scholar] [CrossRef]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry, 5th ed.; Texensis Publishing: Gruver, TX, USA, 2017. [Google Scholar]
- Hammoda, H.M.; Harraz, F.M.; Farag, M.A.; El-Aswad, A.F.; El-Hawiet, A.; Eid, A.M. Volatiles profiling and bioactivities of Cupressus spp. leaf and cone essential oils as analyzed via chemometrics tools. J. Essent. Oil Res. 2019, 31, 53–62. [Google Scholar] [CrossRef]
- Marongiu, B.; Porcedda, S.; Piras, A.; Sanna, G.; Murreddu, M.; Loddo, R. Extraction of Juniperus communis L. ssp. nana Willd. essential oil by supercritical carbon dioxide. Flavour Fragr. J. 2006, 21, 148–154. [Google Scholar] [CrossRef]
- Kamal, R.M.; Sabry, M.M.; El-Halawany, A.M.; Rabie, M.A.; El Sayed, N.S.; Hifnawy, M.S.; Younis, I.Y. GC-MS analysis and the effect of topical application of essential oils of Pinus canariensis C. Sm., Cupressus lusitanica Mill. and Cupressus arizonica Greene aerial parts in Imiquimod–Induced Psoriasis in Mice. J. Ethnopharmacol. 2024, 318, 116947. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, D.; Padhee, S.; Priyadarshini, A.; Champati, B.B.; Das, P.K.; Jena, S.; Sahoo, A.; Panda, P.C.; Nayak, S.; Ray, A. Elucidating the anti-cancer potential of Cinnamomum tamala essential oil against non-small cell lung cancer: A multifaceted approach involving GC-MS profiling, network pharmacology, and molecular dynamics simulations. Heliyon 2024, 10, e28026. [Google Scholar] [CrossRef]
- Lan, T.; Wang, J.; Zeng, R.; Gao, C.; Liu, X.; Luo, L.; Liang, Y.; Guo, Z.; Wang, W.; Hong, M. Therapeutic targets and molecular mechanisms of Huangqin decoction in liver cancer: A network pharmacology and molecular docking approach. J. Herb. Med. 2024, 43, 100822. [Google Scholar] [CrossRef]
- Elansary, H.O.; Salem, M.Z.; Ashmawy, N.A.; Yacout, M.M. Chemical composition, antibacterial and antioxidant activities of leaves essential oils from Syzygium cumini L., Cupressus sempervirens L. and Lantana camara L. from Egypt. J. Agric. Sci. 2012, 4, 144–152. [Google Scholar] [CrossRef]
- Fayed, S.A. Chemical composition, antioxidant, anticancer properties and toxicity evaluation of leaf essential oil of Cupressus sempervirens. Not. Bot. Horti Agrobot. Cluj-Napoca 2015, 43, 320–326. [Google Scholar] [CrossRef]
- Selim, S.A.; Adam, M.E.; Hassan, S.M.; Albalawi, A.R. Chemical composition, antimicrobial and antibiofilm activity of the essential oil and methanol extract of the Mediterranean cypress (Cupressus sempervirens L.). BMC Complement. Altern. Med. 2014, 14, 179. [Google Scholar] [CrossRef]
- Al-Snafi, A.E. Medical importance of Cupressus sempervirens—A review. IOSR J. Pharm. 2016, 6, 66–76. [Google Scholar]
- Alorabi, M.; Elghazawy, H.; Alorabi, M.; Elghazawy, H. Cancer control in Egypt: Investing in health. ASCO Post 2021. [Google Scholar]
- Mohamed Abdoul-Latif, F.; Ainane, A.; Houmed Aboubaker, I.; Mohamed, J.; Ainane, T. Exploring the potent anticancer activity of essential oils and their bioactive compounds: Mechanisms and prospects for future cancer therapy. Pharmaceuticals 2023, 16, 1086. [Google Scholar] [CrossRef]
- Fikry, E.; Orfali, R.; Elbaramawi, S.S.; Perveen, S.; El-Shafae, A.M.; El-Domiaty, M.M.; Tawfeek, N. Chamaecyparis lawsoniana leaf essential oil as a potential anticancer agent: Experimental and computational studies. Plants 2023, 12, 2475. [Google Scholar] [CrossRef] [PubMed]
- Bajes, H.R.; Oran, S.A.; Bustanji, Y.K. Chemical composition and antiproliferative and antioxidant activities of essential oil from Juniperus phoenicea L. cupressaceae. Res. J. Pharm. Technol. 2022, 15, 153–159. [Google Scholar] [CrossRef]
- Sahin Yaglioglu, A.; Eser, F.; Yaglioglu, M.S.; Demirtas, I. The antiproliferative and antioxidant activities of the essential oils of Juniperus species from Turkey. Flavour Fragr. J. 2020, 35, 511–523. [Google Scholar] [CrossRef]
- Wajs-Bonikowska, A.; Sienkiewicz, M.; Stobiecka, A.; Maciąg, A.; Szoka, Ł.; Karna, E. Chemical composition and biological activity of Abies alba and A. koreana seed and cone essential oils and characterization of their seed hydrolates. Chem. Biodivers. 2015, 12, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Sertel, S.; Eichhorn, T.; Plinkert, P.K.; Efferth, T. Chemical composition and antiproliferative activity of essential oil from the leaves of a medicinal herb, Levisticum officinale, against UMSCC1 head and neck squamous carcinoma cells. Anticancer Res. 2011, 31, 185–191. [Google Scholar] [PubMed]
- Fekrazad, R.; Afzali, M.; Pasban-Aliabadi, H.; Esmaeili-Mahani, S.; Aminizadeh, M.; Mostafavi, A. Cytotoxic effect of Thymus caramanicus Jalas on human oral epidermoid carcinoma KB cells. Braz. Dent. J. 2017, 28, 72–77. [Google Scholar] [CrossRef]
- Ojah, E.O.; Moronkola, D.O.; Akintunde, A.-A.M. α-amylase and α-glucosidase antidiabetic potential of ten essential oils from Calophyllum inophyllum Linn. Iberoam. J. Med. 2020, 2, 253–260. [Google Scholar] [CrossRef]
- Adefegha, S.A.; Olasehinde, T.A.; Oboh, G. Essential oil composition, antioxidant, antidiabetic and antihypertensive properties of two Afromomum species. J. Oleo Sci. 2017, 66, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Mechchate, H.; Es-Safi, I.; Louba, A.; Alqahtani, A.S.; Nasr, F.A.; Noman, O.M.; Farooq, M.; Alharbi, M.S.; Alqahtani, A.; Bari, A. In vitro alpha-amylase and alpha-glucosidase inhibitory activity and in vivo antidiabetic activity of Withania frutescens L. Foliar extract. Molecules 2021, 26, 293. [Google Scholar] [CrossRef]
- Rana, S.; Dixit, S.; Mittal, A. Anticancer effects of chemotherapy and nature products. J. Med. Discov. 2017, 2, 1–8. [Google Scholar] [CrossRef]
- Asmat, U.; Abad, K.; Ismail, K. Diabetes mellitus and oxidative stress—A concise review. Saudi Pharm. J. 2016, 24, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Galovičová, L.; Čmiková, N.; Schwarzová, M.; Vukic, M.D.; Vukovic, N.L.; Kowalczewski, P.Ł.; Bakay, L.; Kluz, M.I.; Puchalski, C.; Obradovic, A.D.; et al. Biological Activity of Cupressus sempervirens Essential Oil. Plants 2023, 12, 1097. [Google Scholar] [CrossRef] [PubMed]
- Akermi, S.; Smaoui, S.; Elhadef, K.; Fourati, M.; Louhichi, N.; Chaari, M.; Chakchouk Mtibaa, A.; Baanannou, A.; Masmoudi, S.; Mellouli, L. Cupressus sempervirens Essential Oil: Exploring the Antibacterial Multitarget Mechanisms, Chemcomputational Toxicity Prediction, and Safety Assessment in Zebrafish Embryos. Molecules 2022, 27, 2630. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Jurado, F.; Franco-Vega, A.; Ramírez-Corona, N.; Palou, E.; López-Malo, A. Essential Oils: Antimicrobial Activities, Extraction Methods, and Their Modeling. Food Eng. Rev. 2015, 7, 275–297. [Google Scholar] [CrossRef]
- Abers, M.; Schroeder, S.; Goelz, L.; Sulser, A.; St. Rose, T.; Puchalski, K.; Langland, J. Antimicrobial activity of the volatile substances from essential oils. BMC Complement. Med. Ther. 2021, 21, 124. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, X.; Wang, Y.; Chen, Y.; Lu, H.; Meng, X.; Ye, X.; Chen, W. Activation/inactivation of anticancer drugs by CYP3A4: Influencing factors for personalized cancer therapy. Drug Metab. Dispos. 2023, 51, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Cobleigh, M.A.; Layng, K.V.; Mauer, E.; Mahon, B.; Hockenberry, A.J.; Abukhdeir, A.M. Comparative genomic analysis of PIK3R1-mutated and wild-type breast cancers. Breast Cancer Res. Treat. 2024, 204, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, D.; Li, Z.; Li, X.; Jin, M.; Jia, N.; Cui, X.; Hu, G.; Tang, T.; Yu, Q. Pan-cancer analysis on the role of PIK3R1 and PIK3R2 in human tumors. Sci. Rep. 2022, 12, 5924. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, H.; Cheng, H.; Wen, J.; Li, D. PIK3CD correlates with prognosis, epithelial–mesenchymal transition and tumor immune infiltration in breast carcinoma. Discov. Oncol. 2023, 14, 187. [Google Scholar] [CrossRef]
- Zundelevich, A.; Dadiani, M.; Kahana-Edwin, S.; Itay, A.; Sella, T.; Gadot, M.; Cesarkas, K.; Farage-Barhom, S.; Saar, E.G.; Eyal, E.; et al. ESR1 mutations are frequent in newly diagnosed metastatic and loco-regional recurrence of endocrine-treated breast cancer and carry worse prognosis. Breast Cancer Res. 2020, 22, 16. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Yu, J.; Wang, X.; Cheng, L.; Ma, Z.; Chen, X.; Wang, L.; Goh, B.C. AKR1C3 in carcinomas: From multifaceted roles to therapeutic strategies. Front. Pharmacol. 2024, 15, 1378292. [Google Scholar] [CrossRef]
- Uribe, M.L.; Marrocco, I.; Yarden, Y. EGFR in cancer: Signaling mechanisms, drugs, and acquired resistance. Cancers 2021, 13, 2748. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Mo, M.; Chen, X.; Chao, D.; Zhang, Y.; Chen, X.; Wang, Y.; Zhang, N.; He, N.; Yuan, X.; et al. Targeting inhibition of prognosis-related lipid metabolism genes including CYP19A1 enhances immunotherapeutic response in colon cancer. J. Exp. Clin. Cancer Res. 2023, 42, 85. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Riegel, K.; Vijayarangakannan, P.; Kechagioglou, P.; Bogucka, K.; Rajalingam, K. Recent advances in targeting protein kinases and pseudokinases in cancer biology. Front. Cell Dev. Biol. 2022, 10, 942500. [Google Scholar] [CrossRef]
- Singh, P.; Lim, B. Targeting Apoptosis in Cancer. Curr. Oncol. Rep. 2022, 24, 273–284. [Google Scholar] [CrossRef]
- Tuli, H.S.; Kaur, J.; Vashishth, K.; Sak, K.; Sharma, U.; Choudhary, R.; Behl, T.; Singh, T.; Sharma, S.; Saini, A.K.; et al. Molecular mechanisms behind ROS regulation in cancer: A balancing act between augmented tumorigenesis and cell apoptosis. Arch. Toxicol. 2023, 97, 103–120. [Google Scholar] [CrossRef] [PubMed]
- Razaghi, A.; Heimann, K.; Schaeffer, P.M.; Gibson, S.B. Negative regulators of cell death pathways in cancer: Perspective on biomarkers and targeted therapies. Apoptosis 2018, 23, 93–112. [Google Scholar] [CrossRef]
- Li, B.; Qin, Y.; Yu, X.; Xu, X.; Yu, W. Lipid raft involvement in signal transduction in cancer cell survival, cell death and metastasis. Cell Prolif. 2022, 55, e13167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yue, P.; Lu, T.; Wang, Y.; Wei, Y.; Wei, X. Role of lysosomes in physiological activities, diseases, and therapy. J. Hematol. Oncol. 2021, 14, 79. [Google Scholar] [CrossRef]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Linzer, N.; Trumbull, A.; Nar, R.; Gibbons, M.D.; Yu, D.T.; Strouboulis, J.; Bungert, J. Regulation of RNA polymerase II transcription initiation and elongation by transcription factor TFII-I. Front. Mol. Biosci. 2021, 8, 681550. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, J.D.; Subramanian, T.; Liu, K.; King, M.R. Rafting down the metastatic cascade: The role of lipid rafts in cancer metastasis, cell death, and clinical outcomes. Cancer Res. 2021, 81, 5–17. [Google Scholar] [CrossRef]
- Skotland, T.; Kavaliauskiene, S.; Sandvig, K. The role of lipid species in membranes and cancer-related changes. Cancer Metastasis Rev. 2020, 39, 343–360. [Google Scholar] [CrossRef] [PubMed]
- Aouiche, C.; Chen, B.; Shang, X. Predicting stage-specific cancer related genes and their dynamic modules by integrating multiple datasets. BMC Bioinform. 2019, 20, 194. [Google Scholar] [CrossRef] [PubMed]
- Sarathi, A.; Palaniappan, A. Novel significant stage-specific differentially expressed genes in hepatocellular carcinoma. BMC Cancer 2019, 19, 663. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Azam, Z.; Guo, J.; To, S.S.T. Oncogenic potential of PIK3CD in glioblastoma is exerted through cytoskeletal proteins PAK3 and PLEK2. Lab. Investig. 2022, 102, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Barros-Oliveira, M.d.C.; Costa-Silva, D.R.; Dos Santos, A.R.; Pereira, R.O.; Soares-Junior, J.M.; Silva, B.B.d. Influence of CYP19A1 gene expression levels in women with breast cancer: A systematic review of the literature. Clinics 2021, 76, e2846. [Google Scholar] [CrossRef]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef]
- Li, Z.; Wu, Y.; Yates, M.E.; Tasdemir, N.; Bahreini, A.; Chen, J.; Levine, K.M.; Priedigkeit, N.M.; Nasrazadani, A.; Ali, S. Hotspot ESR1 mutations are multimodal and contextual modulators of breast cancer metastasis. Cancer Res. 2022, 82, 1321–1339. [Google Scholar] [CrossRef]
- Reinert, T.; Gonçalves, R.; Bines, J. Implications of ESR1 Mutations in Hormone Receptor-Positive Breast Cancer. Curr. Treat. Options Oncol. 2018, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, H.; Hashemnia, V.; Nazemalhosseini-Mojarad, E.; Ghasemi, M.R.; Mirfakhraie, R. Correlated downregulation of VDR and CYP3A4 in colorectal cancer. Mol. Biol. Rep. 2023, 50, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Huang, J.Q.; Luo, B.; Dong, S.H.; Wang, R.C.; Jiang, Z.k.; Xie, Y.K.; Yi, W.; Wen, G.M.; Zhong, J.F. PIK 3 CD induces cell growth and invasion by activating AKT/GSK-3β/β-catenin signaling in colorectal cancer. Cancer Sci. 2019, 110, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Lung, D.K.; Reese, R.M.; Alarid, E.T. Intrinsic and extrinsic factors governing the transcriptional regulation of ESR1. Horm. Cancer 2020, 11, 129–147. [Google Scholar] [CrossRef]
- Ali, R.; Wendt, M.K. The paradoxical functions of EGFR during breast cancer progression. Signal Transduct. Target. Ther. 2017, 2, 1–7. [Google Scholar] [CrossRef]
- Bisson, J.; McAlpine, J.B.; Friesen, J.B.; Chen, S.-N.; Graham, J.; Pauli, G.F. Can invalid bioactives undermine natural product-based drug discovery? J. Med. Chem. 2016, 59, 1671–1690. [Google Scholar] [CrossRef]
- Baell, J.B. Feeling nature’s PAINS: Natural products, natural product drugs, and pan assay interference compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [Google Scholar] [CrossRef]
- Baell, J.; Walters, M.A. Chemistry: Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef]
- Gomha, S.M.; Riyadh, S.M.; Mahmmoud, E.A.; Elaasser, M.M. Synthesis and anticancer activity of arylazothiazoles and 1,3,4-thiadiazoles using chitosan-grafted-poly(4-vinylpyridine) as a novel copolymer basic catalyst. Chem. Heterocycl. Compd. 2015, 51, 1030–1038. [Google Scholar] [CrossRef]
- Elkomy, N.M.; El-Shaibany, A.; Elnagar, G.M.; Abdelkhalek, A.S.; Al-Mahbashi, H.; Elaasser, M.M.; Raweh, S.M.; Aldiyarbi, M.A.; Raslan, A.E. Evaluation of acute oral toxicity, anti-diabetic and antioxidant effects of Aloe vera flowers extract. J. Ethnopharmacol. 2023, 309, 116310. [Google Scholar] [CrossRef]
- Ling, L.T.; Yap, S.-A.; Radhakrishnan, A.K.; Subramaniam, T.; Cheng, H.M.; Palanisamy, U.D. Standardised Mangifera indica extract is an ideal antioxidant. Food Chem. 2009, 113, 1154–1159. [Google Scholar] [CrossRef]
- Elaasser, M.; Morsi, M.; Galal, S.; Abd El-Rahman, M.; Katry, M. Antioxidant, anti-inflammatory and cytotoxic activities of the unsaponifiable fraction of extra virgin olive oil. Grasas Y Aceites 2020, 71, e386. [Google Scholar] [CrossRef]
- Jaradat, N.; Dacca, H.; Hawash, M.; Abualhasan, M.N. Ephedra alata fruit extracts: Phytochemical screening, anti-proliferative activity and inhibition of DPPH, α-amylase, α-glucosidase, and lipase enzymes. BMC Chem. 2021, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Al-Maharik, N.; Jaradat, N.; Al-Hajj, N.; Jaber, S. Myrtus communis L.: Essential oil chemical composition, total phenols and flavonoids contents, antimicrobial, antioxidant, anticancer, and α-amylase inhibitory activity. Chem. Biol. Technol. Agric. 2023, 10, 41. [Google Scholar] [CrossRef]
- Hindler, J.; Howard, B.; Keiser, J. Antimicrobial agents and antimicrobial susceptibility testing. In Howard BJ. Clinical and Pathogenic Microbiology, 2nd ed.; Mosby: St Louis, MO, USA, 1994. [Google Scholar]
- Choudhary, M.I.; Thomsen, W.J. Bioassay Techniques for Drug Development; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Yoshida, T.; Jono, K.; Okonogi, K. Modified agar dilution susceptibility testing method for determining in vitro activities of antifungal agents, including azole compounds. Antimicrob. Agents Chemother. 1997, 41, 1349–1351. [Google Scholar] [CrossRef]
- Wayne, P. Reference method for broth dilution antifungal susceptibility testing of yeasts, approved standard. CLSI Doc. M27-A2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2002. [Google Scholar]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Yao, Z.J.; Dong, J.; Che, Y.J.; Zhu, M.F.; Wen, M.; Wang, N.N.; Wang, S.; Lu, A.P.; Cao, D.S. TargetNet: A web service for predicting potential drug-target interaction profiling via multi-target SAR models. J. Comput. Aided Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef]
- Rebhan, M.; Chalifa-Caspi, V.; Prilusky, J.; Lancet, D. GeneCards: Integrating information about genes, proteins and diseases. Trends Genet. TIG 1997, 13, 163. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Zaru, R.; Orchard, S.; Consortium, U. Uniprot tools: bLAST, align, peptide search, and ID mapping. Curr. Protoc. 2023, 3, e697. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.X. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015, 127, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, H.; Yang, X.; Li, Z.; Zhan, W.; Zhu, H.; Zhang, T. Network pharmacology analysis reveals potential targets and mechanisms of proton pump inhibitors in breast cancer with diabetes. Sci. Rep. 2023, 13, 7623. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Duan, J.; Zhao, M.; Huang, S.; Mu, F.; Su, J.; Liu, K.; Pan, Y.; Lu, X.; Li, J. A network pharmacology approach to reveal the protective mechanism of Salvia miltiorrhiza-Dalbergia odorifera coupled-herbs on coronary heart disease. Sci. Rep. 2019, 9, 19343. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, 1–11. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Digre, A.; Lindskog, C. The Human Protein Atlas—Spatial localization of the human proteome in health and disease. Protein Sci. 2021, 30, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. In Protein Crystallography: Methods and Protocols; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Springer: New York, NY, USA, 2017; pp. 627–641. [Google Scholar]
- Yano, J.K.; Wester, M.R.; Schoch, G.A.; Griffin, K.J.; Stout, C.D.; Johnson, E.F. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-Å resolution. J. Biol. Chem. 2004, 279, 38091–38094. [Google Scholar] [CrossRef] [PubMed]
- Furet, P.; Guagnano, V.; Fairhurst, R.A.; Imbach-Weese, P.; Bruce, I.; Knapp, M.; Fritsch, C.; Blasco, F.; Blanz, J.; Aichholz, R. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorganic Med. Chem. Lett. 2013, 23, 3741–3748. [Google Scholar] [CrossRef] [PubMed]
- Fradera, X.; Methot, J.L.; Achab, A.; Christopher, M.; Altman, M.D.; Zhou, H.; McGowan, M.A.; Kattar, S.D.; Wilson, K.; Garcia, Y. Design of selective PI3Kδ inhibitors using an iterative scaffold-hopping workflow. Bioorganic Med. Chem. Lett. 2019, 29, 2575–2580. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Wu, J.Y.; Birzin, E.T.; Frisch, K.; Chan, W.; Pai, L.-Y.; Yang, Y.T.; Mosley, R.T.; Fitzgerald, P.M.; Sharma, N. Estrogen receptor ligands. II. Discovery of benzoxathiins as potent, selective estrogen receptor α modulators. J. Med. Chem. 2004, 47, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, D.M.; Flanagan, J.U.; Jamieson, S.M.; Silva, S.; Rigoreau, L.J.; Trivier, E.; Raynham, T.; Turnbull, A.P.; Denny, W.A. Synthesis and structure–activity relationships for 1-(4-(piperidin-1-ylsulfonyl) phenyl) pyrrolidin-2-ones as novel non-carboxylate inhibitors of the aldo-keto reductase enzyme AKR1C3. Eur. J. Med. Chem. 2013, 62, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Ouyang, X.; Wan, X.; Gajiwala, K.S.; Kath, J.C.; Jones, L.H.; Burlingame, A.L.; Taunton, J. Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes. J. Am. Chem. Soc. 2017, 139, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Lo, J.; Morton, D.; Valette, D.; Xi, J.; Griswold, J.; Hubbell, S.; Egbuta, C.; Jiang, W.; An, J.; et al. Novel Aromatase Inhibitors by Structure-Guided Design. J. Med. Chem. 2012, 55, 8464–8476. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. Discovery Studio Visualizer v21. 1.0. 20298; Dassault Systèmes: San Diego, CA, USA, 2021. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Fikry, E.; Orfali, R.; El-Sayed, S.S.; Perveen, S.; Ghafar, S.; El-Shafae, A.M.; El-Domiaty, M.M.; Tawfeek, N. Potential Hepatoprotective Effects of Chamaecyparis lawsoniana against Methotrexate-Induced Liver Injury: Integrated Phytochemical Profiling, Target Network Analysis, and Experimental Validation. Antioxidants 2023, 12, 2118. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
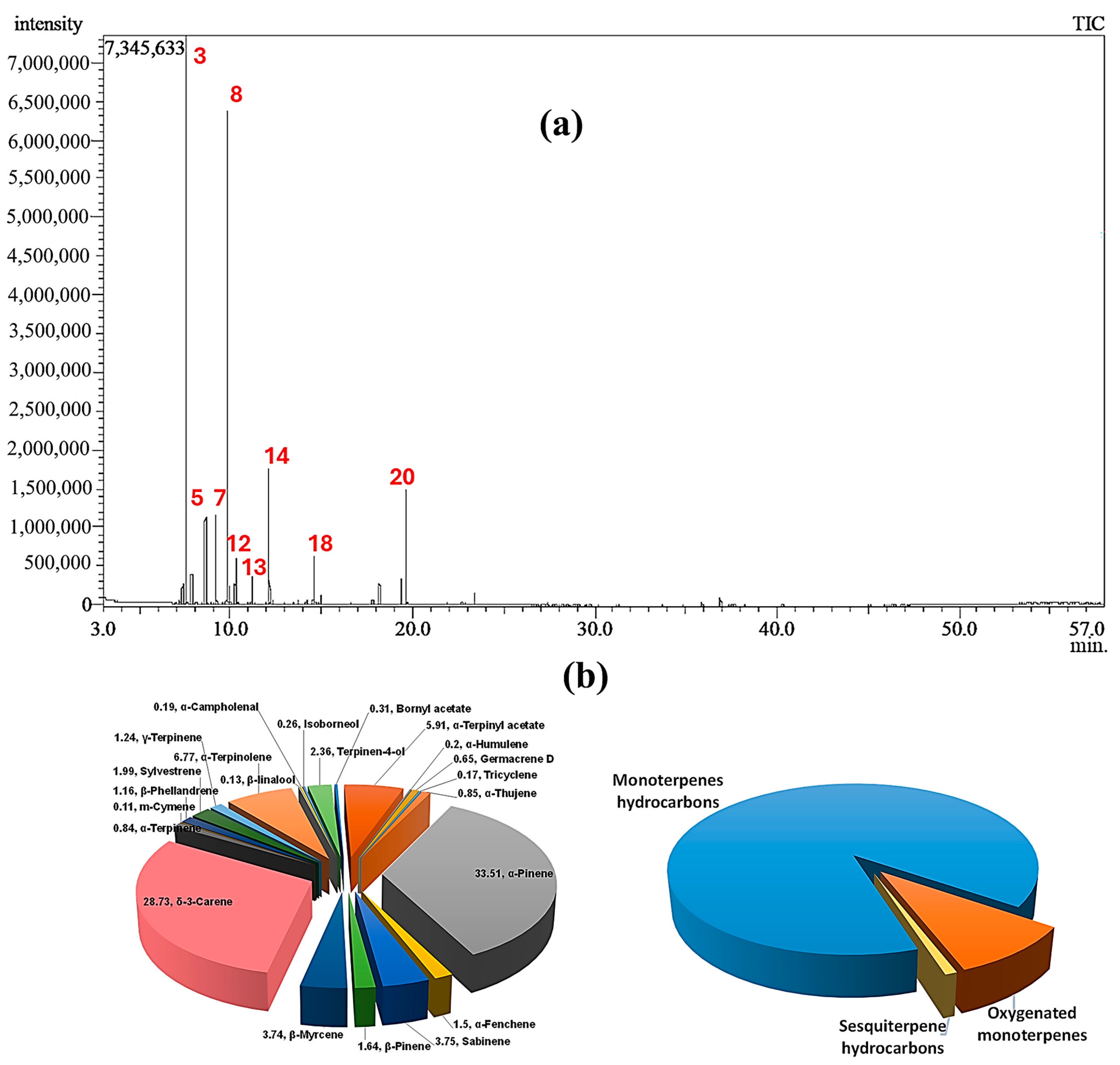




| Peak | Rt | Compound Name | Molecular Formula | Chemical Class | RIExp. a | RILit b | Area % | Identification c |
|---|---|---|---|---|---|---|---|---|
| 1 | 7.205 | Tricyclene | C10H16 | Tricyclic monoterpene hydrocarbon | 911 | 902 | 0.17 | MS, RI |
| 2 | 7.370 | α-Thujene | C10H16 | Bicyclic monoterpene hydrocarbon | 917 | 908 | 0.85 | MS, RI |
| 3 | 7.575 | α-Pinene | C10H16 | Bicyclic monoterpene hydrocarbon | 923 | 916 | 33.51 | MS, RI |
| 4 | 7.855 | α-Fenchene | C10H16 | Bicyclic monoterpene hydrocarbon | 933 | 929 | 1.50 | MS, RI |
| 5 | 8.615 | Sabinene | C10H16 | Bicyclic monoterpene hydrocarbon | 958 | 958 | 3.75 | MS, RI |
| 6 | 8.705 | β-Pinene | C10H16 | Bicyclic monoterpene hydrocarbon | 961 | 961 | 1.64 | MS, RI |
| 7 | 9.210 | β-Myrcene | C10H16 | Acyclic monoterpene hydrocarbon | 978 | 978 | 3.74 | MS, RI |
| 8 | 9.825 | δ-3-Carene | C10H16 | Bicyclic monoterpene hydrocarbon | 998 | 999 | 28.73 | MS, RI |
| 9 | 9.945 | α-Terpinene | C10H16 | Monocyclic monoterpene hydrocarbon | 1002 | 1006 | 0.84 | MS, RI |
| 10 | 10.040 | m-Cymene | C10H14 | Aromatic monoterpene hydrocarbon | 1005 | 1012 | 0.11 | MS, RI |
| 11 | 10.255 | β-Phellandrene | C10H16 | Monocyclic monoterpene hydrocarbon | 1012 | 1013 | 1.16 | MS, RI |
| 12 | 10.320 | Sylvestrene | C10H16 | Monocyclic monoterpene hydrocarbon | 1014 | 1018 | 1.99 | MS, RI |
| 13 | 11.200 | γ-Terpinene | C10H16 | Monocyclic monoterpene hydrocarbon | 1042 | 1048 | 1.24 | MS, RI |
| 14 | 12.115 | α-Terpinolene | C10H16 | Monocyclic monoterpene hydrocarbon | 1071 | 1078 | 6.77 | MS, RI |
| 15 | 12.945 | β-linalool | C10H18O | Acyclic monoterpene alcohol | 1098 | 1090 | 0.13 | MS, RI |
| 16 | 13.725 | α-Campholenal | C10H16O | Monocyclic monoterpene aldehyde | 1123 | 1125 | 0.19 | MS, RI |
| 17 | 14.525 | Isoborneol | C10H18O | Bicyclic monoterpene alcohol | 1149 | 1147 | 0.26 | MS, RI |
| 18 | 14.610 | Terpinen-4-ol | C10H18O | Monocyclic monoterpene alcohol | 1152 | 1167 | 2.36 | MS |
| 19 | 17.810 | Bornyl acetate | C12H20O2 | Bicyclic monoterpene ester | 1259 | 1276 | 0.31 | MS |
| 20 | 19.640 | α-Terpinyl acetate | C12H20O2 | Monocyclic monoterpene ester | 1323 | 1339 | 5.91 | MS |
| 21 | 22.710 | α-Humulene | C15H24 | Sesquiterpene hydrocarbon | 1436 | 1445 | 0.20 | MS |
| 22 | 23.390 | Germacrene D | C15H24 | Sesquiterpene hydrocarbon | 1462 | 1473 | 0.65 | MS |
| Total identified | 96.01 | |||||||
| Monoterpene hydrocarbons | 86.00 | |||||||
| Oxygenated monoterpenes | 9.16 | |||||||
| Sesquiterpene hydrocarbons | 0.85 | |||||||
| Vero | MCF-7 | HCT-116 | HepG-2 | A-549 | |||||
|---|---|---|---|---|---|---|---|---|---|
| CC50 | IC50 | SI | IC50 | SI | IC50 | SI | IC50 | SI | |
| CSSLEO | 26.95 ± 1.06 | 7.20 ± 0.32 | 3.74 | 6.29 ± 0.29 | 4.35 | 4.44 ±0.21 | 6.07 | 6.71 ± 0.28 | 4.02 |
| Cisplatin | 40.68 ± 2.75 | 5.57 ± 0.68 | 7.30 | 2.75 ± 0.24 | 14.28 | 3.87 ± 0.55 | 10.52 | 7.68 ± 0.36 | 5.29 |
| IC50 ± SD (μg/mL) | |||
|---|---|---|---|
| DPPH | ABTS | FRAP | |
| CSSLEO | 212.08 ± 4.71 | 138.20 ± 3.69 | 307.35 ± 6.04 |
| Ascorbic acid | 10.19 ± 0.65 | 10.78 ± 0.65 | 20.88 ± 0.94 |
| Tested Sample | Inhibition Zone (IZ mm) Diameter (Mean ± SD)/Minimum Inhibitory Concentration (MIC mg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungi | Gram-Positive Bacteria | Gram-Negative Bacteria | ||||||||||
| Aspergillus fumigatus (RCMB 002008) | Candida albicans (RCMB 005003 (1) ATCC 10231) | Staphylococcus aureus (ATCC 25923) | Bacillus subtilis RCMB 015 (1) NRRL B-543 | Escherichia coli ATCC 25922 | Proteus vulgaris RCMB 004 (1) ATCC 13315 | |||||||
| IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | IZ | MIC | |
| CSSLEO | NA | - | NA | - | NA | - | 8 ± 0.56 | 25 ± 0.97 | 13 ± 0.62 | 6.25 ± 0.83 | NA | - |
| Ketoconazole (100 mg/mL) | 17 ± 0.36 | - | 20 ± 0.27 | - | - | - | - | - | - | - | - | - |
| Gentamycin (4 mg/mL) | - | - | - | - | 24 ± 0.58 | - | 26 ± 0.68 | - | 30 ± 0.73 | - | 25 ± 0.81 | - |
| Target Name | Description | DC | BC | CC | EC | NC | LAC |
|---|---|---|---|---|---|---|---|
| CYP3A4 | Cytochrome P450 3A4 | 22 | 5123.8018 | 0.026203394 | 0.328590542 | 14.08333333 | 4.363636364 |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 | 19 | 4439.497006 | 0.026478479 | 0.062474731 | 11.61941474 | 3.789473684 |
| PIK3CD | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | 18 | 2712.974904 | 0.026340218 | 0.054360334 | 11.06023046 | 3.888888889 |
| ESR1 | Estrogen receptor | 18 | 7909.706627 | 0.026640066 | 0.098252818 | 9.888965283 | 4.222222222 |
| AKR1C3 | Aldo-keto reductase family 1 member C3 | 16 | 2055.232626 | 0.026053024 | 0.310182363 | 12.1260101 | 5.875 |
| EGFR | Epidermal growth factor receptor | 15 | 3023.4256 | 0.026383078 | 0.058514997 | 6.031962482 | 3.466666667 |
| CYP19A1 | Cytochrome P450 19A1 (Human aromatase) | 12 | 6596.306973 | 0.026364692 | 0.249898016 | 7.346969697 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fikry, E.; Orfali, R.; Tawfeek, N.; Perveen, S.; Ghafar, S.; El-Domiaty, M.M.; El-Shafae, A.M. Unveiling the Bioactive Efficacy of Cupressus sempervirens ‘Stricta’ Essential Oil: Composition, In Vitro Activities, and In Silico Analyses. Pharmaceuticals 2024, 17, 1019. https://doi.org/10.3390/ph17081019
Fikry E, Orfali R, Tawfeek N, Perveen S, Ghafar S, El-Domiaty MM, El-Shafae AM. Unveiling the Bioactive Efficacy of Cupressus sempervirens ‘Stricta’ Essential Oil: Composition, In Vitro Activities, and In Silico Analyses. Pharmaceuticals. 2024; 17(8):1019. https://doi.org/10.3390/ph17081019
Chicago/Turabian StyleFikry, Eman, Raha Orfali, Nora Tawfeek, Shagufta Perveen, Safina Ghafar, Maher M. El-Domiaty, and Azza M. El-Shafae. 2024. "Unveiling the Bioactive Efficacy of Cupressus sempervirens ‘Stricta’ Essential Oil: Composition, In Vitro Activities, and In Silico Analyses" Pharmaceuticals 17, no. 8: 1019. https://doi.org/10.3390/ph17081019
APA StyleFikry, E., Orfali, R., Tawfeek, N., Perveen, S., Ghafar, S., El-Domiaty, M. M., & El-Shafae, A. M. (2024). Unveiling the Bioactive Efficacy of Cupressus sempervirens ‘Stricta’ Essential Oil: Composition, In Vitro Activities, and In Silico Analyses. Pharmaceuticals, 17(8), 1019. https://doi.org/10.3390/ph17081019






