Lactoferrin Binds through Its N-Terminus to the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. In Vitro Binding Assay and Immunoblotting
4.3. Surface Plasmon Resonance (SPR)
4.4. In-Solution Binding and Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Makhluf, H.; Madany, H.; Kim, K. Long COVID: Long-Term Impact of SARS-CoV2. Diagnostics 2024, 14, 711. [Google Scholar] [CrossRef] [PubMed]
- Ohradanova-Repic, A.; Prazenicova, R.; Gebetsberger, L.; Moskalets, T.; Skrabana, R.; Cehlar, O.; Tajti, G.; Stockinger, H.; Leksa, V. Time to Kill and Time to Heal: The Multifaceted Role of Lactoferrin and Lactoferricin in Host Defense. Pharmaceutics 2023, 15, 1056. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-Converting Enzyme 2 Is a Functional Receptor for the SARS Coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Prior, P. Priming of SARS-CoV-2 S Protein by Several Membrane-Bound Serine Proteinases Could Explain Enhanced Viral Infectivity and Systemic COVID-19 Infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef] [PubMed]
- Dessie, G.; Malik, T. Role of Serine Proteases and Host Cell Receptors Involved in Proteolytic Activation, Entry of SARS-CoV-2 and Its Current Therapeutic Options. Infect. Drug Resist. 2021, 14, 1883–1892. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.; Uckeley, Z.M.; Doldan, P.; Stanifer, M.; Boulant, S.; Lozach, P.Y. TMPRSS2 Expression Dictates the Entry Route Used by SARS-CoV-2 to Infect Host Cells. EMBO J. 2021, 40, e107821. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.M.; Ashwaq, O.; Sarief, A.; Azad John Mohamed, A.K. A Comprehensive Review about SARS-CoV-2. Future Virol. 2020, 15, 625–648. [Google Scholar] [CrossRef] [PubMed]
- El-Fakharany, E.M. Nanoformulation of Lactoferrin Potentiates Its Activity and Enhances Novel Biotechnological Applications. Int. J. Biol. Macromol. 2020, 165 Pt A, 970–984. [Google Scholar] [CrossRef]
- Baker, E.N.; Baker, H.M. A Structural Framework for Understanding the Multifunctional Character of Lactoferrin. Biochimie 2009, 91, 3–10. [Google Scholar] [CrossRef]
- Lambert, L.A. Molecular Evolution of the Transferrin Family and Associated Receptors. Biochim. Biophys. Acta 2012, 1820, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a Context of Inflammation-Induced Pathology. Front. Immunol. 2017, 8, 1438. [Google Scholar] [CrossRef] [PubMed]
- Legrand, D.; Mazurier, J. A Critical Review of the Roles of Host Lactoferrin in Immunity. Biometals 2010, 23, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Vogel, H.J. Lactoferrin, a Bird’s Eye View. Biochem. Cell Biol. 2012, 90, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, P.; Kaczynska, K.; Kleczkowska, P.; Bukowska-Osko, I.; Kramkowski, K.; Sulejczak, D. The Lactoferrin Phenomenon—A Miracle Molecule. Molecules 2022, 27, 2941. [Google Scholar] [CrossRef] [PubMed]
- Zarzosa-Moreno, D.; Avalos-Gomez, C.; Ramirez-Texcalco, L.S.; Torres-Lopez, E.; Ramirez-Mondragon, R.; Hernandez-Ramirez, J.O.; Serrano-Luna, J.; de la Garza, M. Lactoferrin and Its Derived Peptides: An Alternative for Combating Virulence Mechanisms Developed by Pathogens. Molecules 2020, 25, 5763. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Fernandez, E.; Egia-Mendikute, L.; Vila-Vecilla, L.; Bosch, A.; Barreira-Manrique, A.; Lee, S.Y.; Garcia-Del Rio, A.; Antonana-Vildosola, A.; Jimenez-Lasheras, B.; Moreno-Cugnon, L.; et al. Hypoxia Reduces Cell Attachment of SARS-CoV-2 Spike Protein by Modulating the Expression of ACE2, Neuropilin-1, Syndecan-1 and Cellular Heparan Sulfate. Emerg. Microbes Infect. 2021, 10, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Meng, X.; Zhang, F.; Xiang, Y.; Wang, J. The In Vitro Antiviral Activity of Lactoferrin against Common Human Coronaviruses and SARS-CoV-2 is Mediated by Targeting the Heparan Sulfate Co-Receptor. Emerg. Microbes Infect. 2021, 10, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.; Chen, S.; Fogliano, V.; Madadlou, A. Hydrolysis Improves the Inhibition Efficacy of Bovine Lactoferrin against Infection by SARS-CoV-2 Pseudovirus. Int. Dairy J. 2023, 137, 105488. [Google Scholar] [CrossRef]
- Ohradanova-Repic, A.; Skrabana, R.; Gebetsberger, L.; Tajti, G.; Barath, P.; Ondrovicova, G.; Prazenicova, R.; Jantova, N.; Hrasnova, P.; Stockinger, H.; et al. Blockade of TMPRSS2-Mediated Priming of SARS-CoV-2 by Lactoferricin. Front. Immunol. 2022, 13, 958581. [Google Scholar] [CrossRef]
- Lai, X.; Yu, Y.; Xian, W.; Ye, F.; Ju, X.; Luo, Y.; Dong, H.; Zhou, Y.H.; Tan, W.; Zhuang, H.; et al. Identified Human Breast Milk Compositions Effectively Inhibit SARS-CoV-2 and Variants Infection and Replication. iScience 2022, 25, 104136. [Google Scholar] [CrossRef] [PubMed]
- He, S.T.; Qin, H.; Guan, L.; Liu, K.; Hong, B.; Zhang, X.; Lou, F.; Li, M.; Lin, W.; Chen, Y.; et al. Bovine Lactoferrin Inhibits SARS-CoV-2 and SARS-CoV-1 by Targeting the RdRp Complex and Alleviates Viral Infection in the Hamster Model. J. Med. Virol. 2023, 95, e28281. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Yang, N.; Deng, J.; Liu, K.; Yang, P.; Zhang, G.; Jiang, C. Inhibition of SARS Pseudovirus Cell Entry by Lactoferrin Binding to Heparan Sulfate Proteoglycans. PLoS ONE 2011, 6, e23710. [Google Scholar] [CrossRef] [PubMed]
- Iles, J.; Zmuidinaite, R.; Sadee, C.; Gardiner, A.; Lacey, J.; Harding, S.; Ule, J.; Roblett, D.; Heeney, J.; Baxendale, H.; et al. SARS-CoV-2 Spike Protein Binding of Glycated Serum Albumin-Its Potential Role in the Pathogenesis of the COVID-19 Clinical Syndromes and Bias Towards Individuals with Pre-Diabetes/Type 2 Diabetes and Metabolic Diseases. Int. J. Mol. Sci. 2022, 23, 4126. [Google Scholar] [CrossRef]
- Zekri-Nechar, K.; Zamorano-Leon, J.J.; Segura-Fragoso, A.; Alcaide, J.R.; Reche, C.; Andres-Castillo, A.; Martinez-Martinez, C.H.; Giner, M.; Jimenez-Garcia, R.; Lopez-de-Andres, A.; et al. Albumin Binds COVID-19 Spike 1 Subunit and Predicts in-Hospital Survival of Infected Patients-Possible Alteration by Glucose. J. Clin. Med. 2022, 11, 587. [Google Scholar] [CrossRef] [PubMed]
- Zwirzitz, A.; Reiter, M.; Skrabana, R.; Ohradanova-Repic, A.; Majdic, O.; Gutekova, M.; Cehlar, O.; Petrovcikova, E.; Kutejova, E.; Stanek, G.; et al. Lactoferrin Is a Natural Inhibitor of Plasminogen Activation. J. Biol. Chem. 2018, 293, 8600–8613. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.H.; Jenssen, H.; Sandvik, K.; Gutteberg, T.J. Anti-HSV Activity of Lactoferrin and Lactoferricin Is Dependent on the Presence of Heparan Sulphate at the Cell Surface. J. Med. Virol. 2004, 74, 262–271. [Google Scholar] [CrossRef]
- Shestakov, A.; Jenssen, H.; Nordstrom, I.; Eriksson, K. Lactoferricin but Not Lactoferrin Inhibit Herpes Simplex Virus Type 2 Infection in Mice. Antivir. Res. 2012, 93, 340–345. [Google Scholar] [CrossRef]
- Groot, F.; Geijtenbeek, T.B.; Sanders, R.W.; Baldwin, C.E.; Sanchez-Hernandez, M.; Floris, R.; van Kooyk, Y.; de Jong, E.C.; Berkhout, B. Lactoferrin Prevents Dendritic Cell-Mediated Human Immunodeficiency Virus Type 1 Transmission by Blocking the DC-SIGN—gp120 Interaction. J. Virol. 2005, 79, 3009–3015. [Google Scholar] [CrossRef]
- Takayama, Y.; Aoki, R.; Uchida, R.; Tajima, A.; Aoki-Yoshida, A. Role of CXC Chemokine Receptor Type 4 as a Lactoferrin Receptor. Biochem. Cell Biol. 2017, 95, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Becquart, P.; Hocini, H.; Kazatchkine, M.D.; Bouhlal, H.; Belec, L. Modulation of HIV Binding to Epithelial Cells and HIV Transfer from Immature Dendritic Cells to CD4 T Lymphocytes by Human Lactoferrin and Its Major Exposed LF-33 Peptide. Open Virol. J. 2011, 5, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Stax, M.J.; Mouser, E.E.; van Montfort, T.; Sanders, R.W.; de Vries, H.J.; Dekker, H.L.; Herrera, C.; Speijer, D.; Pollakis, G.; Paxton, W.A. Colorectal Mucus Binds DC-SIGN and Inhibits HIV-1 Trans-Infection of CD4+ T-Lymphocytes. PLoS ONE 2015, 10, e0122020. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Liu, Z.Y.; Groopman, J.E. The Alpha-Chemokine Receptor CXCR4 is Expressed on the Megakaryocytic Lineage from Progenitor to Platelets and Modulates Migration and Adhesion. Blood 1998, 92, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Fan, Y.C.; Lin, J.W.; Chen, Y.Y.; Hsu, W.L.; Chiou, S.S. Bovine Lactoferrin Inhibits Dengue Virus Infectivity by Interacting with Heparan Sulfate, Low-Density Lipoprotein Receptor, and DC-SIGN. Int. J. Mol. Sci. 2017, 18, 1957. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Chavez, S.A.; Arevalo-Gallegos, S.; Rascon-Cruz, Q. Lactoferrin: Structure, Function and Applications. Int. J. Antimicrob. Agents 2009, 33, 301.e1–301.e8. [Google Scholar] [CrossRef] [PubMed]
- van Berkel, P.H.; Geerts, M.E.; van Veen, H.A.; Mericskay, M.; de Boer, H.A.; Nuijens, J.H. N-Terminal Stretch Arg2, Arg3, Arg4 and Arg5 of Human Lactoferrin Is Essential for Binding to Heparin, Bacterial Lipopolysaccharide, Human Lysozyme and DNA. Biochem. J. 1997, 328 Pt 1, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Rosa, L.; Cutone, A.; Conte, M.P.; Campione, E.; Bianchi, L.; Valenti, P. An Overview on In Vitro and In Vivo Antiviral Activity of Lactoferrin: Its Efficacy against SARS-CoV-2 Infection. Biometals 2022, 36, 417–436. [Google Scholar] [CrossRef] [PubMed]
- El Yazidi-Belkoura, I.; Legrand, D.; Nuijens, J.; Slomianny, M.C.; van Berkel, P.; Spik, G. The Binding of Lactoferrin to Glycosaminoglycans on Enterocyte-Like HT29-18-C1 Cells is Mediated through Basic Residues Located in the N-Terminus. Biochim. Biophys. Acta-Gen. Subj. 2001, 1568, 197–204. [Google Scholar] [CrossRef]
- Gifford, J.L.; Hunter, H.N.; Vogel, H.J. Lactoferricin: A Lactoferrin-Derived Peptide with Antimicrobial, Antiviral, Antitumor and Immunological Properties. Cell. Mol. Life Sci. 2005, 62, 2588–2598. [Google Scholar] [CrossRef]
- Hunter, H.N.; Demcoe, A.R.; Jenssen, H.; Gutteberg, T.J.; Vogel, H.J. Human Lactoferricin is Partially Folded in Aqueous Solution and Is Better Stabilized in a Membrane Mimetic Solvent. Antimicrob. Agents Chemother. 2005, 49, 3387–3395. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, F.; Panella, G.; Leboffe, L.; Antonini, G. Lactoferrin from Milk: Nutraceutical and Pharmacological Properties. Pharmaceuticals 2016, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Vorland, L.H.; Ulvatne, H.; Andersen, J.; Haukland, H.; Rekdal, O.; Svendsen, J.S.; Gutteberg, T.J. Lactoferricin of Bovine Origin Is More Active than Lactoferricins of Human, Murine and Caprine Origin. Scand. J. Infect. Dis. 1998, 30, 513–517. [Google Scholar] [PubMed]
- Arredondo-Beltran, I.G.; Ramirez-Sanchez, D.A.; Zazueta-Garcia, J.R.; Canizalez-Roman, A.; Angulo-Zamudio, U.A.; Velazquez-Roman, J.A.; Bolscher, J.G.M.; Nazmi, K.; Leon-Sicairos, N. Antitumor Activity of Bovine Lactoferrin and Its Derived Peptides against HepG2 Liver Cancer Cells and Jurkat Leukemia Cells. Biometals 2023, 36, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Han, F.F.; Gao, Y.H.; Luan, C.; Xie, Y.G.; Liu, Y.F.; Wang, Y.Z. Comparing Bacterial Membrane Interactions and Antimicrobial Activity of Porcine Lactoferricin-Derived Peptides. J. Dairy Sci. 2013, 96, 3471–3487. [Google Scholar] [CrossRef] [PubMed]
- Eliassen, L.T.; Berge, G.; Sveinbjornsson, B.; Svendsen, J.S.; Vorland, L.H.; Rekdal, O. Evidence for a Direct Antitumor Mechanism of Action of Bovine Lactoferricin. Anticancer Res. 2002, 22, 2703–2710. [Google Scholar]
- El-Baky, N.A.; Elkhawaga, M.A.; Abdelkhalek, E.S.; Sharaf, M.M.; Redwan, E.M.; Kholef, H.R. De Novo Expression and Antibacterial Potential of Four Lactoferricin Peptides in Cell-Free Protein Synthesis System. Biotechnol. Rep. 2021, 29, e00583. [Google Scholar] [CrossRef]
- Andersen, J.H.; Osbakk, S.A.; Vorland, L.H.; Traavik, T.; Gutteberg, T.J. Lactoferrin and Cyclic Lactoferricin Inhibit the Entry of Human Cytomegalovirus into Human Fibroblasts. Antivir. Res. 2001, 51, 141–149. [Google Scholar] [CrossRef]
- Hwang, S.A.; Kruzel, M.L.; Actor, J.K. Immunomodulatory Effects of Recombinant Lactoferrin during MRSA Infection. Int. Immunopharmacol. 2014, 20, 157–163. [Google Scholar] [CrossRef]
- Alves, N.S.; Azevedo, A.S.; Dias, B.M.; Horbach, I.S.; Setatino, B.P.; Denani, C.B.; Schwarcz, W.D.; Lima, S.M.B.; Missailidis, S.; Ano Bom, A.P.D.; et al. Inhibition of SARS-CoV-2 Infection in Vero Cells by Bovine Lactoferrin under Different Iron-Saturation States. Pharmaceuticals 2023, 16, 1352. [Google Scholar] [CrossRef]
- Cutone, A.; Rosa, L.; Bonaccorsi di Patti, M.C.; Iacovelli, F.; Conte, M.P.; Ianiro, G.; Romeo, A.; Campione, E.; Bianchi, L.; Valenti, P.; et al. Lactoferrin Binding to SARS-CoV-2 Spike Glycoprotein Blocks Pseudoviral Entry and Relieves Iron Protein Dysregulation in Several In Vitro Models. Pharmaceutics 2022, 14, 2111. [Google Scholar] [CrossRef] [PubMed]
- Campione, E.; Lanna, C.; Cosio, T.; Rosa, L.; Conte, M.P.; Iacovelli, F.; Romeo, A.; Falconi, M.; Del Vecchio, C.; Franchin, E.; et al. Lactoferrin against SARS-CoV-2: In Vitro and In Silico Evidences. Front. Pharmacol. 2021, 12, 666600. [Google Scholar] [CrossRef] [PubMed]
- Kovacech, B.; Fialova, L.; Filipcik, P.; Skrabana, R.; Zilkova, M.; Paulenka-Ivanovova, N.; Kovac, A.; Palova, D.; Rolkova, G.P.; Tomkova, K.; et al. Monoclonal Antibodies Targeting Two Immunodominant Epitopes on the Spike Protein Neutralize Emerging SARS-CoV-2 Variants of Concern. EBioMedicine 2022, 76, 103818. [Google Scholar] [CrossRef] [PubMed]
- Myszka, D.G. Improving Biosensor Analysis. J. Mol. Recognit. 1999, 12, 279–284. [Google Scholar] [CrossRef]
- Leksa, V.; Pfisterer, K.; Ondrovicova, G.; Binder, B.; Lakatosova, S.; Donner, C.; Schiller, H.B.; Zwirzitz, A.; Mrvova, K.; Pevala, V.; et al. Dissecting Mannose 6-Phosphate-Insulin-Like Growth Factor 2 Receptor Complexes That Control Activation and Uptake of Plasminogen in Cells. J. Biol. Chem. 2012, 287, 22450–22462. [Google Scholar] [CrossRef]


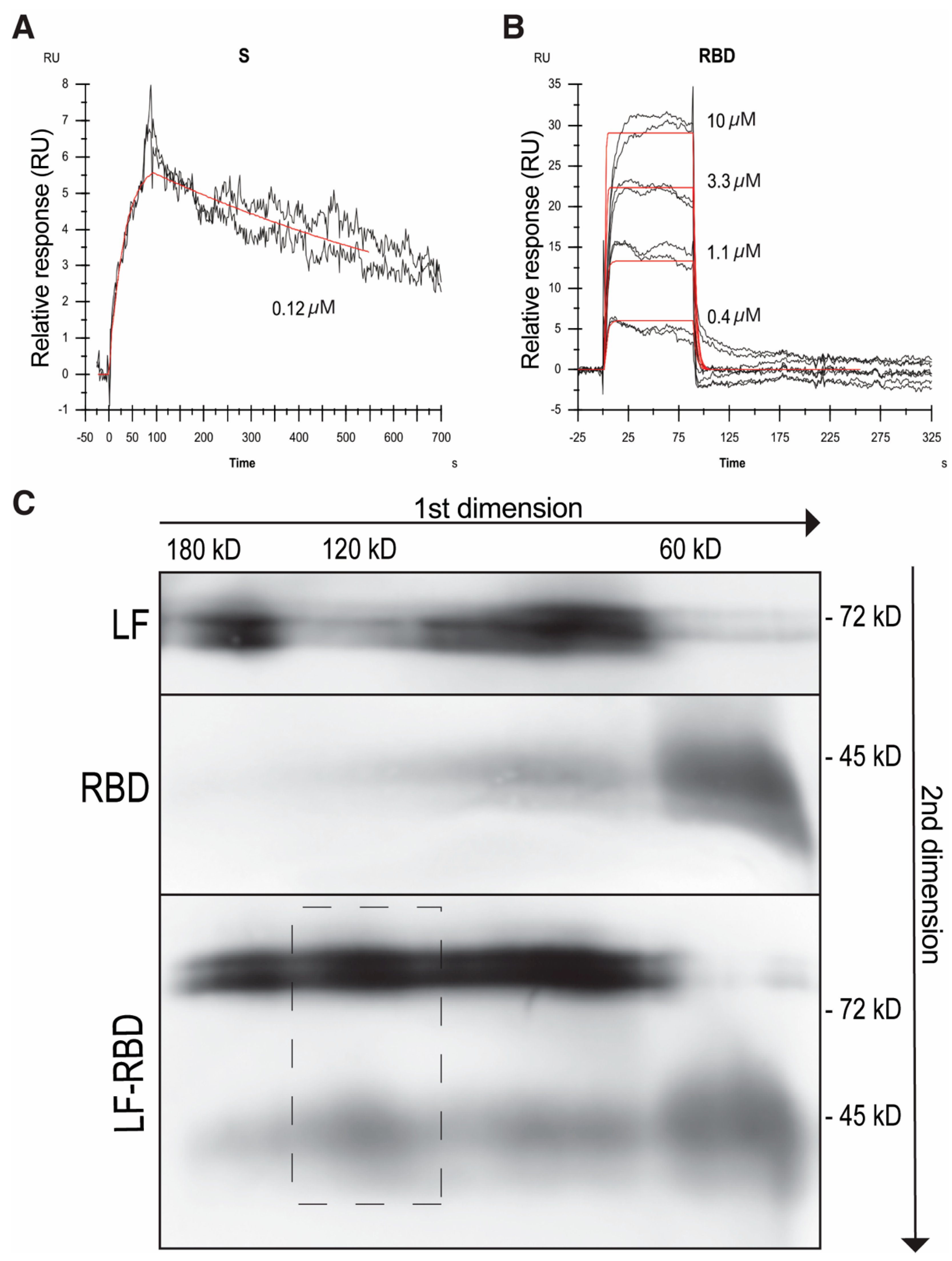
| ka [M−1s−1] | kd [s−1] | KD [M] | KD * [M] | |
|---|---|---|---|---|
| LF–RBD | (2.45 ± 0.45) × 105 | (3.08 ± 0.85) × 10−1 | (1.26 ± 0.42) × 10−6 | (1.31 ± 0.49) × 10−6 |
| LF–S-protein | (1.60 ± 1.23) × 105 | (6.93 ± 5.76) × 10−4 | (4.32 ± 4.88) × 10−9 | N.D. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babulic, P.; Cehlar, O.; Ondrovičová, G.; Moskalets, T.; Skrabana, R.; Leksa, V. Lactoferrin Binds through Its N-Terminus to the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein. Pharmaceuticals 2024, 17, 1021. https://doi.org/10.3390/ph17081021
Babulic P, Cehlar O, Ondrovičová G, Moskalets T, Skrabana R, Leksa V. Lactoferrin Binds through Its N-Terminus to the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein. Pharmaceuticals. 2024; 17(8):1021. https://doi.org/10.3390/ph17081021
Chicago/Turabian StyleBabulic, Patrik, Ondrej Cehlar, Gabriela Ondrovičová, Tetiana Moskalets, Rostislav Skrabana, and Vladimir Leksa. 2024. "Lactoferrin Binds through Its N-Terminus to the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein" Pharmaceuticals 17, no. 8: 1021. https://doi.org/10.3390/ph17081021
APA StyleBabulic, P., Cehlar, O., Ondrovičová, G., Moskalets, T., Skrabana, R., & Leksa, V. (2024). Lactoferrin Binds through Its N-Terminus to the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein. Pharmaceuticals, 17(8), 1021. https://doi.org/10.3390/ph17081021






