Antitumor Effects and the Potential Mechanism of 10-HDA against SU-DHL-2 Cells
Abstract
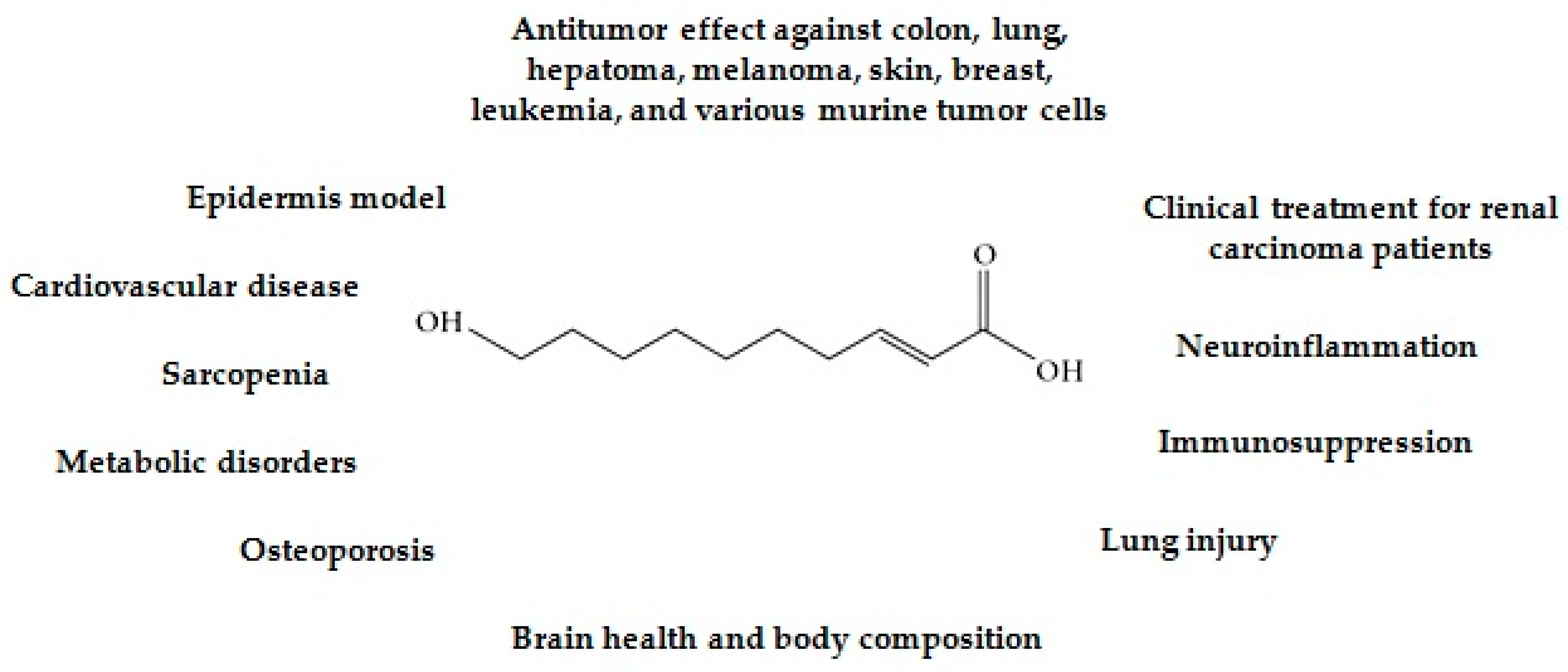
:1. Introduction
2. Results
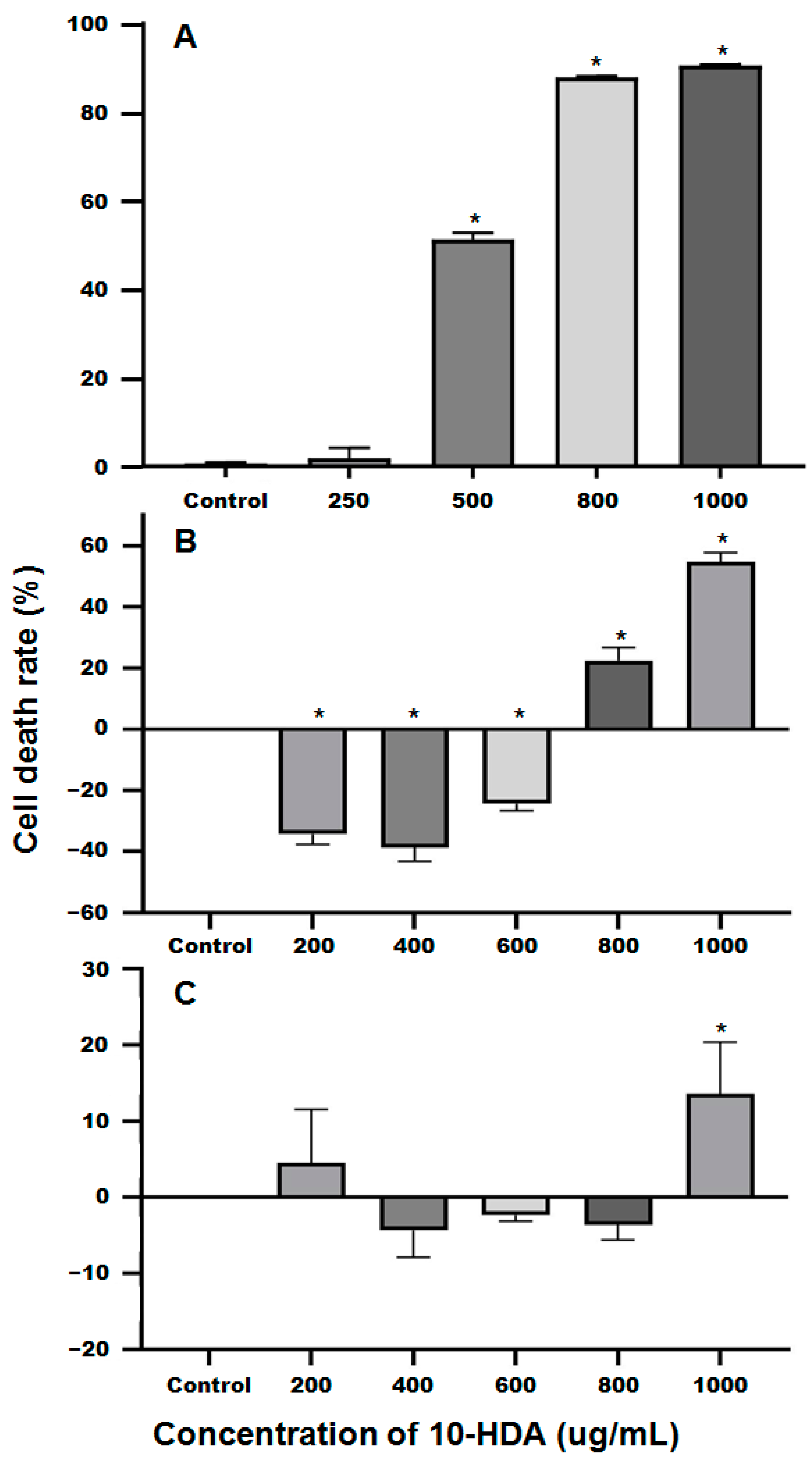
2.1. The Inhibitory Effects of 10-HDA on the Survival of SU-DHL-2 Cells
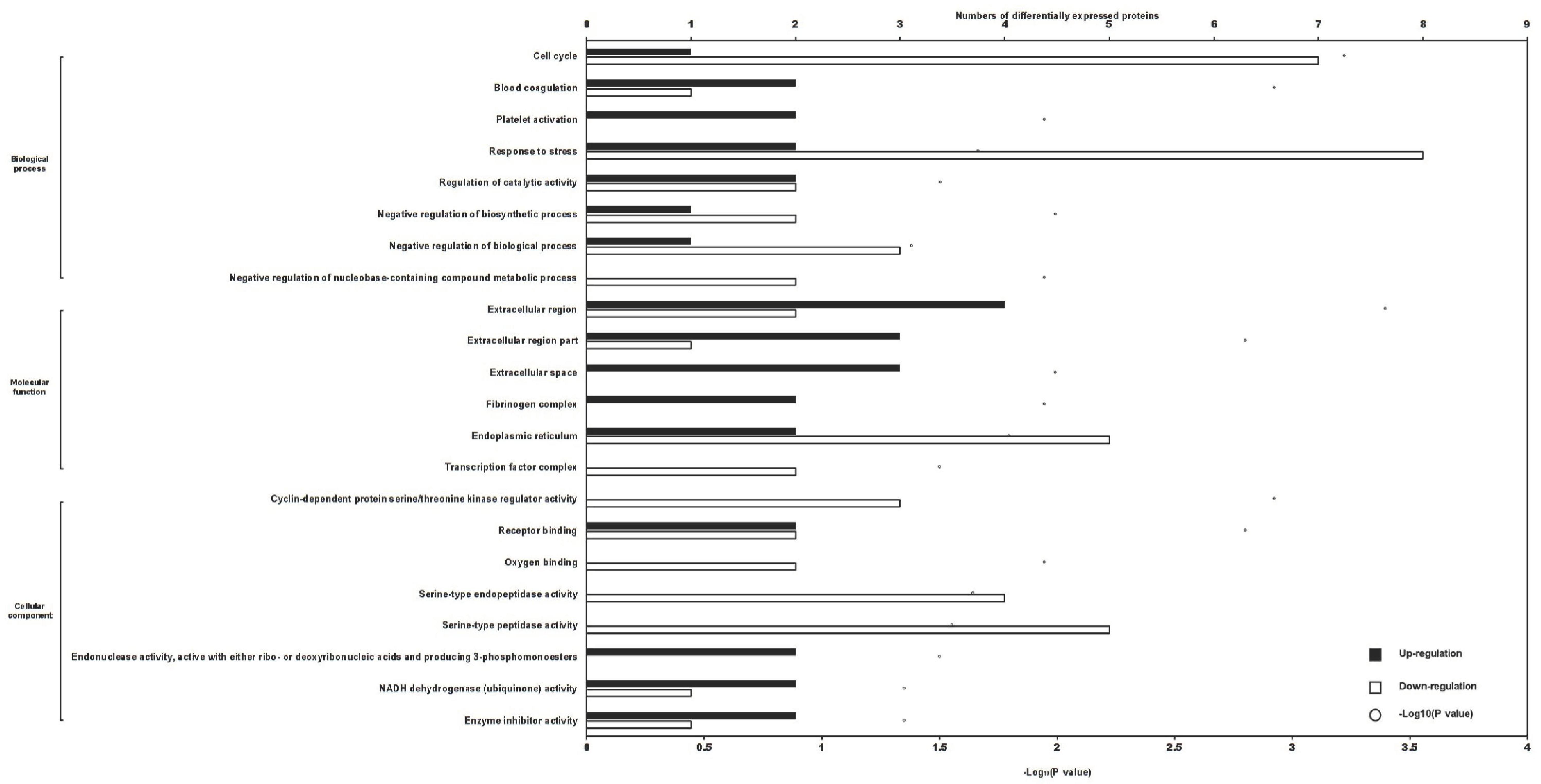
2.2. Differentially Expressed Proteins in Treated Cells
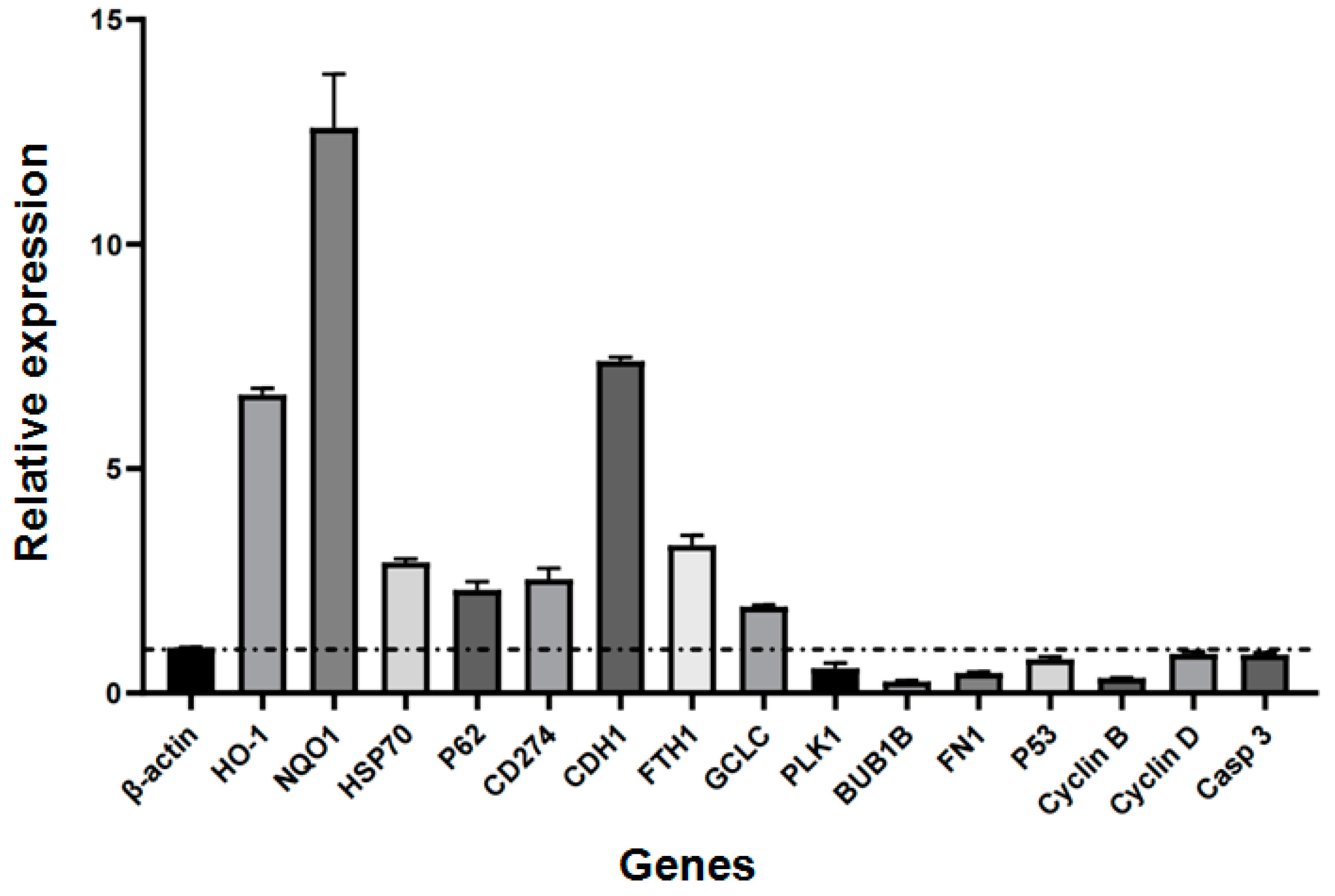
2.3. Relative Expression of Selected Genes
3. Discussion
4. Materials and Methods
4.1. Determination of the IC50 of 10-HDA against SU-DHL-2 Cells
4.2. Proteomic Determination of Differentially Expressed Proteins in Cells Following Different Treatments
4.3. Detection of the Relative Expression of Genes
4.4. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Cancer: Key Facts. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 3 February 2024).
- Sehn, L.H.; Salles, G. Diffuse large B-cell lymphoma. N. Engl. J. Med. 2021, 384, 842–858. [Google Scholar] [CrossRef]
- Teras, L.R.; DeSantis, C.E.; Cerhan, J.R.; Morton, L.M.; Jemal, A.; Flowers, C.R. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. Ca-Cancer J. Clin. 2016, 66, 443–459. [Google Scholar] [CrossRef]
- Dickinson, M.J.; Carlo-Stella, C.; Morschhauser, F.; Bachy, E.; Corradini, P.; Iacoboni, G.; Khan, C.; Wróbel, T.; Offner, F.; Trněný, M.; et al. Glofitamab for relapsed or refractory diffuse large B-Cell lymphoma. N. Engl. J. Med. 2022, 387, 2220–2231. [Google Scholar] [CrossRef]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, L.; Young, K. New agents and regimens for diffuse large B cell lymphoma. J. Hematol. Oncol. 2020, 13, 175. [Google Scholar] [CrossRef]
- Susanibar-Adaniya, S.; Barta, S.K. 2021 Update on Diffuse large B cell lymphoma: A review of currentdata and potential applications on risk stratification andmanagement. Am. J. Hematol. 2021, 96, 617–629. [Google Scholar] [CrossRef]
- He, Y.; Li, J.; Ding, N.; Wang, X.; Deng, L.; Xie, Y.; Ying, Z.; Liu, W.; Ping, L.; Zhang, C.; et al. Combination of Enzastaurin and Ibrutinib synergistically induces anti-tumor effects in diffuse large B cell lymphoma. J. Exp. Clin. Cancer Res. 2019, 38, 86. [Google Scholar] [CrossRef]
- Jeon, B.; Lee, Y.J.; Shin, J.; Choi, M.J.; Lee, C.E.; Son, M.K.; Park, J.H.; Kim, B.; Kim, H.R.; Jung, K.H.; et al. A combination of BR101801 and venetoclax enhances antitumor effect in DLBCL cells via c-Myc/Bcl-2/Mcl-1 triple targeting. Am. J. Cancer Res. 2023, 13, 452. [Google Scholar] [PubMed] [PubMed Central]
- He, W.; Xu, Z.; Song, D.; Zhang, H.; Li, B.; Gao, L.; Zhang, Y.; Feng, Q.; Yu, D.; Hu, L.; et al. Antitumor effects of ra-foxanide in Diffuse Large B Cell Lymphoma via the PTEN/PI3K/Akt and JNK/c-Jun Pathways. Life Sci. 2020, 243, 117249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, Q.; Yang, C.; Yang, H.; Rao, J.; Zhang, X. Curcumin exerts anti-tumor effects on diffuse large B cell lymphoma via regulating PPARγ expression. Biochem. Biophys. Res. Commun. 2020, 524, 70–76. [Google Scholar] [CrossRef]
- Han, J.; Guo, X.; Yu, X.; Liu, S.; Cui, X.; Zhang, B.; Liang, H. 25-Hydroxyvitamin D and total cancer incidence and mortality: A meta-analysis of prospective cohort studies. Nutrients 2019, 11, 2295. [Google Scholar] [CrossRef]
- Zhu, Y.; Lei, C.; Jiang, Q.; Yu, Q.; Qiu, L. DSF/Cu induces antitumor effect against diffuse large B-cell lymphoma through suppressing NF-κB/BCL6 pathways. Cancer Cell Int. 2022, 22, 236. [Google Scholar] [CrossRef]
- Kita, A.; Nakahara, T.; Yamanaka, K.; Nakano, K.; Nakata, M.; Mori, M.; Kaneko, N.; Koutoku, H.; Izumisawa, N.; Sasamata, M. Antitumor effects of YM155, a novel survivin suppressant, against human aggressive non-Hodgkin lymphoma. Leuk. Res. 2011, 35, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Ding, X. miR-145-5p exerts anti-tumor effects in diffuse large B-cell lymphoma by regulating S1PR1/STAT3/AKT pathway. Leuk. Lymphoma 2021, 62, 1884–1891. [Google Scholar] [CrossRef]
- Yang, S.; Sheng, L.; Xu, K.; Wang, Y.; Zhu, H.; Zhang, P.; Mu, Q.; Ouyang, G. Anticancer effect of quinacrine on diffuse large B-cell lymphoma via inhibition of MSI2-NUMB signaling pathway. Mol. Med. Rep. 2018, 17, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Othman, N.H.; Ahmed, S.; Sulaiman, S.A. Inhibitory effects of Malaysian Tualang honey and Australian/New Zealand Manuka honey in modulating experimental breast cancers induced by N-methyl-N-nitrosourea (mnu): A comparative study. Pathology 2016, 48, S148. [Google Scholar] [CrossRef]
- Kadir, E.A.; Sulaiman, S.A.; Yahya, N.K.; Othman, N.H. Inhibitory effects of Tualang honey on experimental breast cancer in rats: A preliminary study. Asian Pac. J. Cancer Prev. 2013, 14, 2249–2254. [Google Scholar] [CrossRef]
- Fauzi, A.N.; Norazmi, M.N.; Yaacob, N.S. Tualang honey induces apoptosis and disrupts the mitochondrial membrane potential of human breast and cervical cancer cell lines. Food Chem. Toxicol. 2011, 49, 871–878. [Google Scholar] [CrossRef]
- Milani, S.M.; Najafi, M.; Alizadeh, P.; Rezazadeh, H. Inhibitory effect of honey on 7,12-dimethylbenz(a)anthracene- initiated and croton oil-promoted skin carcinogenesis. Jundishapur J. Nat. Pharm. Prod. 2018, 13, e57992. [Google Scholar] [CrossRef]
- Hassan, M.I.; Mabrouk, G.M.; Shehata, H.H.; Aboelhussein, M.M. Antineoplastic effects of bee honey and Nigella sativa on hepatocellular carcinoma cells. Integr. Cancer Ther. 2012, 11, 354–363. [Google Scholar] [CrossRef]
- Wen, C.T.P.; Hussein, S.Z.; Abdullah, S.; Karim, N.A.; Makpol, S.; Yusof, Y.A.M. Gelam and Nenas honeys inhibit proliferation of HT 29 colon cancer cells by inducing DNA damage and apoptosis while suppressing inflammation. Asian Pac. J. Cancer Prev. 2012, 13, 1605–1610. [Google Scholar] [CrossRef]
- Aliyu, M.; Odunola, O.A.; Farooq, A.D.; Mesaik, A.M.; Choudhary, M.I.; Fatima, B.; Qureshi, T.A.; Erukainure, O.L. Acacia honey modulates cell cycle progression, pro-inflammatory cytokines and calcium ions secretion in PC-3 cell line. J. Cancer Sci. Ther. 2012, 4, 401–407. [Google Scholar] [CrossRef]
- Swellam, T.; Miyanaga, N.; Onozawa, M.; Hattori, K.; Kawai, K.; Shimazui, T.; Akaza, H. Antineoplastic activity of honey in an experimental bladder cancer implantation model: In vivo and in vitro studies. Int. J. Urol. 2003, 10, 213–219. [Google Scholar] [CrossRef]
- Angst, E.; Park, J.L.; Moro, A.; Lu, Q.Y.; Lu, X.; Li, G.; King, J.; Chen, M.; Reber, H.A.; Go, V.L.; et al. The flavonoid quercetin inhibits pancreatic cancer growth in vitro and in vivo. Pancreas 2013, 42, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Aliyu, M.; Odunola, O.A.; Farooq, A.D.; Rasheed, H.; Mesaik, A.M.; Choudhary, M.I.; Channa, I.S.; Khan, S.A.; Erukainure, O.L. Molecular mechanism of antiproliferation potential of Acacia honey on NCI-H460 cell line. Nutr. Cancer 2013, 65, 296–304. [Google Scholar] [CrossRef]
- Pichichero, E.; Cicconi, R.; Mattei, M.; Muzi, M.G.; Canini, A. Acacia honey and chrysin reduce proliferation of melanoma cells through alterations in cell cycle progression. Int. J. Oncol. 2010, 37, 973–981. [Google Scholar] [CrossRef]
- Fernandez-Cabezudo, M.J.; El-Kharrag, R.; Torab, F.; Bashir, G.; George, J.A.; El-Taji, H.; al-Ramadi, B.K. Intravenous administration of Manuka honey inhibits tumor growth and improves host survival when used in combination with chemotherapy in a melanoma mouse model. PLoS ONE 2013, 8, e5599. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Dang, Q.; Xu, D.; Chen, Y.; Zhu, G.; Wu, K.; Zeng, J.; Long, Q.; Wang, X.; He, D.; et al. Kaempferol induces cell cycle arrest and apoptosis in renal cell carcinoma through EGFR/p38 signaling. Oncol. Rep. 2014, 31, 1350–1356. [Google Scholar] [CrossRef]
- Man, N.M.K.N.; Hassan, R.; Ang, C.Y.; Abdullah, A.D.; Radzi, M.A.R.M.; Sulaiman, S.A. Antileukemic effect of Tualang honey on acute and chronic leukemia cell lines. BioMed Res. Int. 2015, 2015, 307094. [Google Scholar] [CrossRef]
- Tsiapara, A.V.; Jaakkola, M.; Chinou, I.; Graikou, K.; Tolonen, T.; Virtanen, V.; Moutsatsou, P. Bioactivity of Greek honey extracts on breast cancer (MCF-7), prostate cancer (PC-3) and endometrial cancer (Ishikawa) cells: Profile analysis of extracts. Food Chem. 2009, 116, 702–708. [Google Scholar] [CrossRef]
- Kyselova, Z. Toxicological aspects of the use of phenolic compounds in disease prevention. Interdiscip. Toxicol. 2011, 4, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.F.; Han, Y.C.; Shen, Y.C.; Golovinskaia, O.; Venkatakrishnan, K.; Wang, C.K. Chemopreventive and chemotherapeutic effect of propolis and its constituents: A mini-review. J. Cancer Prev. 2020, 25, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, M.G.M.; Hanafy, N.A.N. Targeted therapies for breast and lung cancers by using propolis loaded albumin protein nanoparticles. Int. J. Biol. Macromol. 2024, 260, 129338. [Google Scholar] [CrossRef]
- Kocyigit, A.; Guler, E.M.; Durmus, E.; Yenigun, V.B.; Kanimdan, E.; Ozman, Z.; Yasar, O.; Goren, A.C.; Hekimoglu, E.R.; Oruc, H.H.; et al. Propolis enhances 5-fluorouracil mediated antitumor efficacy and reduces side effects in colorectal cancer: An in vitro and in vivo study. Chem. Biodivers. 2023, 20, e202300591. [Google Scholar] [CrossRef]
- Shen, M.; Liu, C.; Chang, K.; Lai, C.; Chang, S.; Huang, C. Propolis has an anticancer effect on early stage colorectal cancer by affecting epithelial differentiation and gut immunity in the tumor microenvironment. Nutrients 2023, 15, 4494. [Google Scholar] [CrossRef] [PubMed]
- Salavatipour, M.S.; Kouhbananinejad, S.M.; Lashkari, M.; Bardsiri, M.S.; Moghadari, M.; Kashani, B.; Farsinejad, A.; Vahidi, R. Kermanian propolis induces apoptosis through upregulation of Bax/Bcl-2 ratio in acute myeloblastic leukemia cell line (NB4). J. Cancer Res. Ther. 2023, 19, 327–334. [Google Scholar] [CrossRef]
- Liu, X.; Tian, Y.; Yang, A.; Zhang, C.; Miao, X.; Yang, W. Antitumor effects of poplar propolis on DLBCL SU-DHL-2 cells. Foods 2023, 12, 283. [Google Scholar] [CrossRef]
- Aylanc, V.; Larbi, S.; Calhelha, R.; Barros, L.; Rezouga, F.; Rodríguez-Flores, M.S.; Seijo, M.C.; El Ghouizi, A.; Lyoussi, B.; Falcão, S.I.; et al. Evaluation of antioxidant and anticancer activity of mono- and polyfloral moroccan bee pollen by characterizing phenolic and volatile compounds. Molecules 2023, 28, 835. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pólit, C.; Gonzalez-Pastor, R.; Heredia-Moya, J.; Carrera-Pacheco, S.E.; Castillo-Solis, F.; Vallejo-Imbaquingo, R.; Barba-Ostria, C.; Guamán, L.P. Chemical properties and biological activity of bee pollen. Molecules 2023, 28, 7768. [Google Scholar] [CrossRef] [PubMed]
- Małek, A.; Strzemski, M.; Kurzepa, J.; Kurzepa, J. Can bee venom be used as anticancer agent in modern medicine? Cancers 2023, 15, 3714. [Google Scholar] [CrossRef]
- Paredes-Barquero, M.; Niso-Santano, M.; Fuentes, J.M.; Martínez-Chacón, G. In vitro and in vivo models to study the biological and pharmacological properties of queen bee acid (QBA, 10-hydroxy-2-decenoic acid): A systematic review. J. Funct. Foods 2022, 94, 105143. [Google Scholar] [CrossRef]
- Yang, Y.; Chou, W.; Widowati, D.; Lin, I.; Peng, C. 10-Hydroxy-2-decenoic acid of royal jelly exhibits bactericide and anti-inflammatory activity in human colon cancer cells. BMC Complement. Altern. Med. 2018, 18, 202. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, S.; Luo, Y.; Xu, W.; Zhang, Y.; Zhang, T.; Xue, H.; Zuo, W.; Li, Y.; Lu, B.; et al. 10-HDA induces ROS-Mediated apoptosis in A549 human lung cancer cells by regulating the MAPK, STAT3, NF-κB, and TGF-β1 signaling pathways. Biomed Res. Int. 2020, 2020, 3042636. [Google Scholar] [CrossRef]
- Saad Al Shehri, Z.; Alanazi, A.D.; Alnomasy, S.F. Anti-cancer effects of queen bee acid (10-Hydroxy-2-decenoic acid) and its cellular mechanisms against human hepatoma cells. Molecules 2023, 28, 1972. [Google Scholar] [CrossRef]
- Peng, C.; Sun, H.; Lin, I.; Kuo, P.; Li, J. The functional property of royal jelly 10-Hydroxy-2-decenoic acid as a melanogenesis inhibitor. BMC Complement. Altern. Med. 2017, 17, 392. [Google Scholar] [CrossRef] [PubMed]
- Atef, B.; Ishak, R.A.H.; Badawy, S.S.; Osman, R. 10-Hydroxy decanoic acid-based vesicles as a novel topical delivery system: Would it be a better platform than conventional oleic acid ufasomes for skin cancer treatment? Pharmaceutics 2023, 15, 1461. [Google Scholar] [CrossRef]
- Pengpanich, S.; Srisupabh, D.; Tanechpongtamb, W.U. Potential role of royal jelly and 10-Hydroxy-2-decenoic acid as metastasis inhibitors in triple-negative breast cancer cells. J. Med. Assoc. Thail. 2019, 102, 17. [Google Scholar]
- Miyata, Y.; Sakai, H. Anti-cancer and protective effects of royal jelly for therapy-induced toxicities in malignancies. Int. J. Mol. Sci. 2018, 19, 3270. [Google Scholar] [CrossRef]
- Albalawi, A.E.; Althobaiti, N.A.; Alrdahe, S.S.; Alhasani, R.H.; Alaryani, F.S.; BinMowyna, M.N. Anti-tumor effects of queen bee acid (10-Hydroxy-2-decenoic acid) alone and in combination with cyclophosphamide and its cellular mechanisms against ehrlich solid tumor in mice. Molecules 2021, 26, 7021. [Google Scholar] [CrossRef] [PubMed]
- Pengpanich, S.; Tanechpongtamb, W. The Inhibitory Effect of 10-Hydroxy-2decenoic acid (10-HDA) on Breast Cancer Cell Proliferation and Metastasis. Doctoral Dissertation, Srinakharinwirot University, Bangkok, Thailand, 2018. Available online: http://ir-ithesis.swu.ac.th/dspace/handle/123456789/129 (accessed on 4 May 2024).
- Vucevic, D.; Melliou, E.; Vasilijic, S.; Gasic, S.; Ivanovski, P.; Chinou, I.; Colic, M. Fatty acids isolated from royal jelly modulate dendritic cell-mediated immune response in vitro. Int. Immunopharmacol. 2007, 7, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Perminaite, K.; Marksa, M.; Stančiauskaitė, M.; Juknius, T.; Grigonis, A.; Ramanauskiene, K. Formulation of Ocular In Situ Gels with Lithuanian Royal Jelly and Their Biopharmaceutical Evaluation In Vitro. Molecules 2021, 26, 3552. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Kagan, J.; Sidransky, D.; Srivastava, S. Proteomic analysis of cancer-cell mitochondria. Nat. Rev. Cancer 2003, 3, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Geva-Zatorsky, N.; Eden, E.; Frenkel-Morgenstern, M.; Issaeva, I.; Sigal, A.; Milo, R.; Cohen-Saidon, C.; Liron, Y.; Kam, Z.; et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 2008, 322, 1511–1516. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Zhu, Y.; Kang, K.; Luo, B.; He, J.; Wu, Y. Protein corona of magnetic PEI/siRNA complex under the influence of a magnetic field improves transfection efficiency via complement and coagulation cascades. J. Mater. Chem. B 2019, 7, 4207–4216. [Google Scholar] [CrossRef]
- Sharma, S.; Ray, S.; Moiyadi, A.; Sridhar, E.; Srivastava, S. Quantitative proteomic analysis of meningiomas for the identification of surrogate protein markers. Sci. Rep. 2014, 4, 7140. [Google Scholar] [CrossRef]
- Zhang, W.; Peng, P.; Ou, X.; Shen, K.; Wu, X. Ovarian cancer circulating extracelluar vesicles promote coagulation and have a potential in diagnosis: An iTRAQ based proteomic analysis. BMC Cancer 2019, 19, 1095. [Google Scholar] [CrossRef]
- Zhang, C.; Peng, L.; Zhang, Y.; Liu, Z.; Li, W.; Chen, S.; Li, G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 2017, 34, 101. [Google Scholar] [CrossRef]
- Gao, X.; Wang, X.; Zhang, S. Bioinformatics identification of crucial genes and pathways associated with hepatocellular carcinoma. Biosci. Rep. 2018, 38, BSR20181441. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Cao, Y.; Yang, Z. C8B in complement and coagulation cascades signaling pathway is a predictor for survival in HBV-related hepatocellular carcinoma patients. Cancer Manag. Res. 2021, 13, 3503–3515. [Google Scholar] [CrossRef]
- Gao, X.; Chen, Y.; Chen, M.; Wang, S.; Wen, X.; Zhang, S. Identification of key candidate genes and biological pathways in bladder cancer. PeerJ 2018, 6, e6036. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.R.; Islam, T.; Al-Mamun, M.A.; Zaman, T.; Karim, M.R.; Moni, M.A. The influence of depression on ovarian cancer: Discovering molecular pathways that identify novel biomarkers and therapeutic targets. Inform. Med. Unlocked 2019, 16, 100207. [Google Scholar] [CrossRef]
- Gong, Z.; He, Y.; Mi, X.; Li, C.; Sun, X.; Wang, G.; Li, L.; Han, Y.; Xu, C.; Xu, C.; et al. Complement and coagulation cascades pathway-related signature as a predictor of immunotherapy in metastatic urothelial cancer. Aging 2023, 15, 9479. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, E.; Yoon, D.; Kim, J.; Rhim, T.Y.; Kim, S.S. Recombinant human prothrombin kringles have potent anti-angiogenic activities and inhibit Lewis lung carcinoma tumor growth and metastases. Angiogenesis 2002, 5, 191–201. [Google Scholar] [CrossRef]
- Sun, W.Y.; Witte, D.P.; Degen, J.L.; Colbert, M.C.; Burkart, M.C.; Holmbäck, K.; Xiao, Q.; Bugge, T.H.; Degen, S.J. Prothrombin deficiency results in embryonic and neonatal lethality in mice. Proc. Natl. Acad. Sci. USA 1998, 95, 7597–7602. [Google Scholar] [CrossRef]
- Xue, J.; Wu, Q.; Westfield, L.A.; Tuley, E.A.; Lu, D.; Zhang, Q.; Shim, K.; Zheng, X.; Sadler, J.E. Incomplete embryonic lethality and fatal neonatal hemorrhage caused by prothrombin deficiency in mice. Proc. Natl. Acad. Sci. USA 1998, 95, 7603–7607. [Google Scholar] [CrossRef] [PubMed]
- Krishnaswamy, S. The transition of prothrombin to thrombin. J. Thromb. Haemost. 2013, 11, 265–276. [Google Scholar] [CrossRef]
- Alexander, E.T.; Minton, A.R.; Hayes, C.S.; Goss, A.; Van Ryn, J.; Gilmour, S.K. Thrombin inhibition and cyclophosphamide synergistically block tumor progression and metastasis. Cancer Biol. Ther. 2015, 16, 1802–1811. [Google Scholar] [CrossRef]
- Alexander, E.T.; Minton, A.R.; Peters, M.C.; van Ryn, J.; Gilmour, S.K. Thrombin inhibition and cisplatin block tumor progression in ovarian cancer by alleviating the immunosuppressive microenvironment. Oncotarget 2016, 7, 85291–85305. [Google Scholar] [CrossRef]
- Bharadwaj, A.G.; Holloway, R.W.; Miller, V.A.; Waisman, D.M. Plasmin and plasminogen system in the tumor microenvironment: Implications for cancer diagnosis, prognosis, and therapy. Cancers 2021, 13, 1838. [Google Scholar] [CrossRef]
- Kwaan, H.C.; McMahon, B. The role of plasminogen-plasmin system in cancer. Cancer Treat. Res. 2009, 148, 43–66. [Google Scholar] [CrossRef]
- Vylliotis, A.; Yapijakis, C.; Nkenke, E.; Nisyrios, T.; Avgoustidis, D.; Adamopoulou, M.; Ragos, V.; Vassiliou, S.; Koronellos, N.; Vairakraris, E. Effect of thrombosis-related gene polymorphisms upon oral cancer: A regression analysis. Anticancer. Res. 2013, 33, 4033–4039. [Google Scholar]
- Wu, Q.; Wang, X.; Wu, W.; Chen, Y.; Wang, J.; Zhang, X.; Qian, Y.; Du, S.; Sun, J.; Zeng, Z. Molecular mechanisms investigation for liver metastasis of colorectal cancer by combined bioinformatic gene expression profile analysis. Cancer Treat. Res. Commun. 2023, 35, 100694. [Google Scholar] [CrossRef]
- An, R.; Yu, H.; Wang, Y.; Lu, J.; Gao, Y.; Xie, X.; Zhang, J. Integrative analysis of plasma metabolomics and proteomics reveals the metabolic landscape of breast cancer. Cancer Metab. 2022, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Luan, Y.; Wang, Z.; Zhao, J.; Xu, C. Suppression of TAFI by siRNA inhibits invasion and migration of breast cancer cells. Mol. Med. Rep. 2017, 16, 3469–3474. [Google Scholar] [CrossRef]
- Monden, T.; Morimoto, H.; Shimano, T.; Yagyu, T.; Murotani, M.; Nagaoka, H.; Kawasaki, Y.; Kobayashi, T.; Mori, T. Use of fibrinogen to enhance the antitumor effect of OK-432. A new approach to immunotherapy for colorectal carcinoma. Cancer 1992, 69, 636–642. [Google Scholar] [CrossRef]
- Zhang, S.; Rao, G.; Heimberger, A.; Li, S. Fibrinogen-like protein 2: Its biological function across cell types and the potential to serve as an immunotherapy target for brain tumors. Cytokine Growth Factor Rev. 2023, 69, 73–79. [Google Scholar] [CrossRef]
- Patadia, H.; Priyadarshini, A.; Gangawane, A. Integrated proteomic, transcriptomic, and genomic analysis identifies fibrinogen beta and fibrinogen gamma as key modulators of breast cancer progression and metastasis. Biomed. Biotechnol. Res. J. 2022, 6, 266–277. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, Z.; Xia, Y.; Xie, X.; Zhou, K.; Xu, L.; Shi, Y.; Wang, Q.; Bi, J. Ligustrazine reverts anthracycline chemotherapy resistance of human breast cancer by inhibiting JAK2/STAT3 signaling and decreasing fibrinogen gamma chain (FGG) expression. Am. J. Cancer Res. 2020, 10, 939–952. [Google Scholar] [PubMed]
- Peng, H.; Wang, J.; Xiao, L.; Yan, M.; Chen, S.; Wang, L.; Yang, K. Elevated serum fgg levels prognosticate and promote the disease progression in prostate cancer. Front. Genet. 2021, 12, 651647. [Google Scholar] [CrossRef] [PubMed]
- Tinholt, M.; Stavik, B.; Tekpli, X.; Garred, Ø.; Borgen, E.; Kristensen, V.; Sahlberg, K.K.; Sandset, P.M.; Iversen, N. Coagulation factor V is a marker of tumor-infiltrating immune cells in breast cancer. Oncoimmunology 2020, 9, 1824644. [Google Scholar] [CrossRef]
- Andresen, M.S.; Sletten, M.; Sandset, P.M.; Iversen, N.; Stavik, B.; Tinholt, M. Coagulation factor V (F5) is an estrogen-responsive gene in breast cancer cells. Thromb. Haemost. 2022, 122, 1288–1295. [Google Scholar] [CrossRef] [PubMed]
- Lind, S.M. The Role of Coagulation Factor V in Breast Cancer: Effect on Treatment Response. Master’s Thesis, Norwegian University of Life Sciences, Ås, Norway, 2021. Available online: https://hdl.handle.net/11250/2826527 (accessed on 6 February 2024).
- Tong, Y.; Tan, Z.; Wang, P.; Gao, X. A machine learning method for predicting biomarkers associated with prostate cancer. Front. Biosci.-Landmark 2023, 28, 333. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, X.; Qin, Y.; Mo, X.; Luo, S. Identification of F5 as a prognostic biomarker in patients with gastric cancer. BioMed Res. Int. 2020, 2020, 9280841. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Xu, B.; Sui, Y.; Chen, Z.; Luan, Y.; Jiang, Y.; Wei, L.; Long, W.; Zhao, S.; Han, L.; et al. Pan-cancer analysis and validation reveals that D-Dimer-Related genes are prognostic and downregulate CD8+ T cells via TGF-Beta signaling in gastric cancer. Front. Mol. Biosci. 2022, 9, 790706. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Lu, H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004, 279, 44475–44482. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, W.; Liao, J.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell. Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Wilson-Edell, K.A.; Kehasse, A.; Scott, G.K.; Yau, C.; Rothschild, D.E.; Schilling, B.; Gabriel, B.S.; Yevtushenko, M.A.; Hanson, I.M.; Held, J.M.; et al. RPL24: A potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget 2014, 5, 5165–5176. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Akbani, R.; Ng, P.K.S.; Werner, H.M.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on the Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Li, H.; Jin, H.; Jin, J.; Yu, M.; Ma, C.; Tong, Y.; Zhou, L.; Lei, H.; Xu, H.; et al. Identification of 11(13)-dehydroivaxillin as a potent therapeutic agent against non-Hodgkin’s lymphoma. Cell Death Dis. 2017, 8, e3050. [Google Scholar] [CrossRef]
- Jiang, L.; Sun, Y.; Wang, J.; He, Q.; Chen, X.; Lan, X.; Chen, J.; Dou, Q.P.; Shi, X.; Liu, J. Proteasomal cysteine deubiquitinase inhibitor b-AP15 suppresses migration and induces apoptosis in diffuse large B cell lymphoma. J. Exp. Clin. Cancer Res. 2019, 38, 453. [Google Scholar] [CrossRef]
- Chauhan, D.; Catley, L.; Li, G.; Podar, K.; Hideshima, T.; Velankar, M.; Mitsiades, C.; Mitsiades, N.; Yasui, H.; Letai, A. A novel orally active proteasome inhibitor induces apoptosis in multiple myeloma cells with mechanisms distinct from Bortezomib. Cancer Cell 2005, 8, 407–419. [Google Scholar] [CrossRef] [PubMed]


| Pathway | p-Value | Adjusted p-Value | Differentially Expressed Proteins | |
|---|---|---|---|---|
| Upregulated | Downregulated | |||
| Complement and coagulation cascades | 4.9 × 10−5 | 0.0124 | A0A0A0MRJ7, P02675, C9JC84 | P00734, A0A0F7G8J1, B2R7F8, Q96IY4 |
| Basal cell carcinoma | 0.0013 | 0.1615 | A0A3B3ITW1, Q92466, Q6TKP8 | A0A0U1RQC9 |
| Mineral absorption | 0.0028 | 0.2324 | B2R7U4, P02795, P04733, A0A140VJP7 | A0A0U1RQC9 |
| Neuroactive ligand–receptor interaction | 0.0037 | 0.2360 | - | P00734, A0A0F7G8J1, B2R7F8 |
| Pathways in cancer | 0.0131 | 0.5780 | K7EP08, P15559, A0A3B3ITW1, B2R7U4, P02751, A0A024R728, Q92466, P63218, Q6TKP8, A0A2 × 0SFF5 | B3KT21, A0A0U1RQC9, Q9HAV0, U6FVB0, X5D945, P11802, A8K725, P33552, A0A024R8H5, Q5T178, A0A024QYW7, B3KNJ3 |
| Small-cell lung cancer | 0.0147 | 0.5780 | P02751, Q92466 | A0A0U1RQC9, P11802, P33552, A0A024R8H5, Q5T178 |
| Staphylococcus aureus infection | 0.0161 | 0.5780 | C9JC84 | A0A0F7G8J1, B2R7F8 |
| Pyrimidine metabolism | 0.0210 | 0.6627 | P36954 | P23921, D6W4Z6, Q9BZX2, A0A024R8N6, O14802, A0A5F9ZHU7, P56282, Q3B726, Q9NR33, Q7Z3R8, A8K9A5 |
| Melanoma | 0.0285 | 0.7929 | A0A024R728, Q92466 | A0A0U1RQC9, X5D945, P11802 |
| Thyroid cancer | 0.0346 | 0.7929 | Q92466 | A0A0U1RQC9, U6FVB0, X5D945 |
| Bladder cancer | 0.0346 | 0.7929 | - | A0A0U1RQC9, X5D945, P11802, B3KNJ3 |
| Cell Type | Cell Line | IC50 Value (μg/mL) | Method | References |
|---|---|---|---|---|
| Lung cancer cell | A549 | 4.22 | CCK-8 | [43] |
| Lung cancer cell | NCI-H460 | 8.20 | CCK-8 | [43] |
| Lung cancer cell | NCI-H23 | 8.34 | CCK-8 | [43] |
| Human hepatoma cell | HepG2 | 59.6 | MTT | [44] |
| Human colorectal adenocarcinoma cell | CaCo-2 | 37.5 | MTT | [50] |
| Breast cancer cell | MDA-MB231 | 651.88 | - | [53] |
| Breast cancer cell | MDA-MB436 | 949.88 | - | [53] |
| Breast cancer cell | HCC1937 | 979.68 | - | [53] |
| Breast cancer cell | MCF-7 | 972.225 | - | [53] |
| Human epithelial breast cell | MCF-10a | 931.25 | - | [53] |
| Human normal liver cell line | THLE-3 | 106.4 | MTT | [44] |
| African green monkey kidney cell | Vero | 3445.63 | - | [53] |
| Fibroblast | NIH3T3 (Wt) | 1862.5 | - | [53] |
| Fibroblast | BRAF V600E mutation | 1303.75 | - | [53] |
| SIRC (Statens Seruminstitut Rabbit Cornea) cell | SIRC | 2.38 | MTT | [52] |
| Primer | Sequences (5′→3′) |
|---|---|
| β-actin-F | GATCATTGCTCCTCCTGAGC |
| β-actin-R | ACTCCTGCTTGCTGATCCAC |
| HO-1-F | TCTTGGCTGGCTTCCTTACC |
| HO-1-R | GGATGTGCTTTTCGTTGGGG |
| NQO1-F | TGAAAGGCTGGTTTGAGCGA |
| NQO1-R | TCCAGGCGTTTCTTCCATCC |
| GCL-F | AGGTCAAACCCAACCCAGT |
| GCL-R | TGTTAAGGTACTGAAGCGAGG |
| BUB1B-F | GGATGGGTCCTTCTGGAAACT |
| BUB1B-R | GTGGCCTCATCATTGGCATTC |
| FTH1-F | CAGAACTACCACCAGGACTCA |
| FTH1-R | TCAAAGCCACATCATCGCGG |
| HSP70-F | GTGTAACCCCATCATCAGCG |
| HSP70-R | GCTCCAAAACAAAAACAGCAATCT |
| p62-F | TACCAGGACAGCGAGAGGAAG |
| p62-R | ATCCTTTCTCAAGCCCCATGT |
| Cyclin B-F | GATACTGCCTCTCCAAGCC |
| Cyclin B-R | GCACACAATTATTCTCAAGTTGTC |
| Cyclin D-F | GCCGGGGACCGAAACT |
| Cyclin D-R | GCAGTGGCGAAGTGTTTACAAAG |
| CD274-F | TTTGCTGAACGCCCCATACA |
| CD274-R | TCCAGATGACTTCGGCCTTG |
| CDH1-F | GCTGGACCGAGAGAGTTTCC |
| CDH1-R | CAAAATCCAAGCCCGTGGTG |
| p53-F | ACACGCTTCCCTGGATTGG |
| p53-R | TCATCCATTGCTTGGGACGG |
| Casp 3-F | CTCTGGTTTTCGGTGGGTGT |
| Casp 3-R | CTTCCATGTATGATCTTTGGTTCC |
| FN1-F | CAAGCATGTCTCTCTGCCAAG |
| FN-R | CAGAACAGGCAATGTGCAGC |
| PLK1-F | CCTGCACCGAAACCGAGTTA |
| PLK1-R | ACCTCGAAACTGTGCCCTTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Liu, X.; Wang, J.; Zhang, C.; Yang, W. Antitumor Effects and the Potential Mechanism of 10-HDA against SU-DHL-2 Cells. Pharmaceuticals 2024, 17, 1088. https://doi.org/10.3390/ph17081088
Tian Y, Liu X, Wang J, Zhang C, Yang W. Antitumor Effects and the Potential Mechanism of 10-HDA against SU-DHL-2 Cells. Pharmaceuticals. 2024; 17(8):1088. https://doi.org/10.3390/ph17081088
Chicago/Turabian StyleTian, Yuanyuan, Xiaoqing Liu, Jie Wang, Chuang Zhang, and Wenchao Yang. 2024. "Antitumor Effects and the Potential Mechanism of 10-HDA against SU-DHL-2 Cells" Pharmaceuticals 17, no. 8: 1088. https://doi.org/10.3390/ph17081088






