In Silico Identification of Peanut Peptides Suitable for Allergy Immunotherapy in HLA-DRB1*03:01-Restricted Patients
Abstract
1. Introduction
2. Results
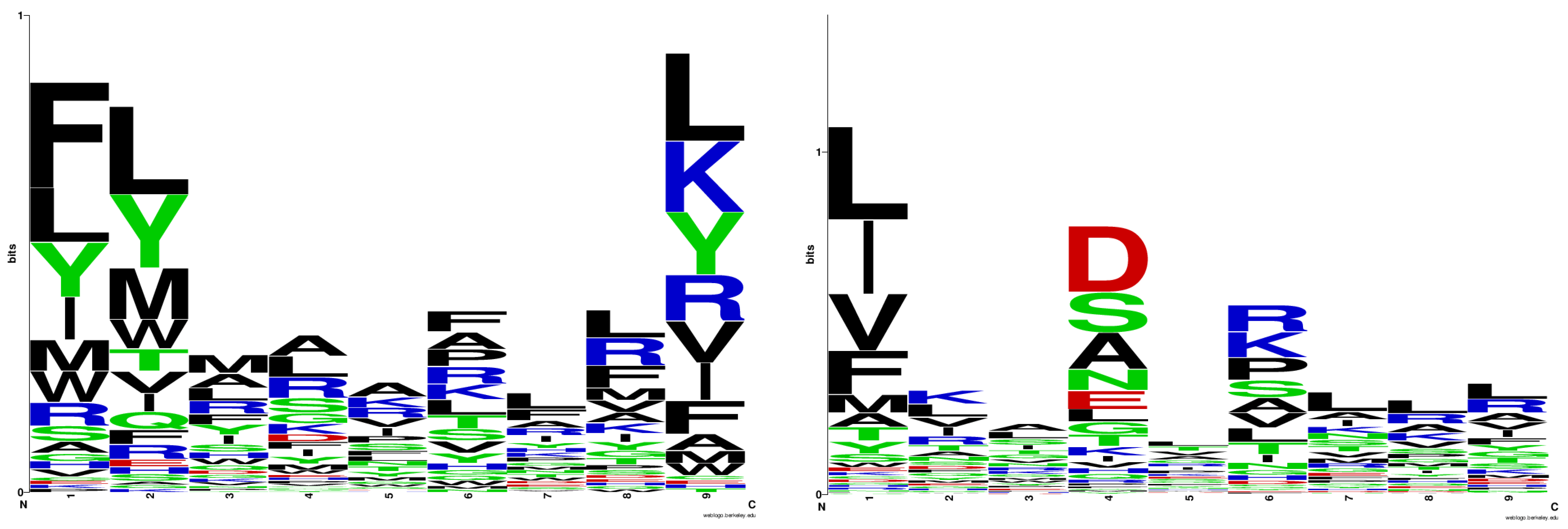
2.1. Logo Model Generation
2.2. Logo Model Validation
2.3. Prediction of Peptide Binders to HLA-DRB1*03:01 among Peanut Allergens
3. Discussion
- Mode of quantification: Schneider and Stephens use Shannon entropy to quantify the nucleic acid/amino acid frequency at a given position. In contrast, we apply mean normalization;
- Functionality: While the sequence logo method was developed as a graphical tool for visualizing patterns in aligned sequences, our method generates quantitative matrices used to calculate binding and non-binding scores. Based on these scores, peptides are classified as binders or non-binders to a given protein.
4. Materials and Methods
4.1. Datasets
4.2. Statistical Analysis
4.3. Docking Protocol
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Peters, R.L.; Krawiec, M.; Koplin, J.J.; Santos, A.F. Update on Food Allergy. Pediatr. Allergy Immunol. 2021, 32, 647–657. [Google Scholar]
- Muraro, A.; de Silva, D.; Halken, S.; Worm, M.; Khaleva, E.; Arasi, S.; Dunn-Galvin, A.; Nwaru, B.I.; De Jong, N.W.; Rodríguez Del Río, P.; et al. Managing Food Allergy: GA2LEN Guideline 2022. World Allergy Organ. J. 2022, 15, 100687. [Google Scholar]
- Mahr, T.A.; Lieberman, J.A.; Haselkorn, T.; Damle, V.; Ali, Y.; Chidambaram, A.; Griffin, N.M.; Sublett, J.W. Characteristics of Peanut Allergy Diagnosis in a US Health Care Claims Database (2011–2017). J. Allergy Clin. Immunol. Pract. 2021, 9, 1683–1694.e5. [Google Scholar]
- Supported by WHO/IUIS Allergen Nomenclature Sub-Committee. Available online: www.allergen.org (accessed on 20 March 2024).
- Vereda, A.; van Hage, M.; Ahlstedt, S.; Ibañez, M.D.; Cuesta-Herranz, J.; van Odijk, J.; Wickman, M.; Sampson, H.A. Peanut allergy: Clinical and immunologic differences among patients from 3 different geographic regions. J. Allergy Clin. Immunol. 2011, 127, 603–607. [Google Scholar] [PubMed]
- Hemmings, O.; Du Toit, G.; Radulovic, S.; Lack, G.; Santos, A.F. Ara h 2 is the dominant peanut allergen despite similarities with Ara h 6. J. Allergy Clin. Immunol. 2020, 146, 621–630. [Google Scholar] [PubMed]
- Rabjohn, P.; Helm, E.M.; Stanley, J.S.; West, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Investig. 1999, 103, 535–542. [Google Scholar] [PubMed]
- Howell, W.M.; Turner, S.J.; Hourihane, J.O.; Dean, T.P.; Warner, J.O. HLA Class II DRB1, DQB1 and DPB1 Genotypic Associations with Peanut Allergy: Evidence from a Family-Based and Case-Control Study. Clin. Exp. Allergy 1998, 28, 156–162. [Google Scholar]
- Pascal, M.; Konstantinou, G.N.; Masilamani, M.; Lieberman, J.; Sampson, H.A. In Silico Prediction of Ara h 2 T Cell Epitopes in Peanut-Allergic Children. Clin. Exp. Allergy 2013, 43, 116–127. [Google Scholar]
- Hemler, J.A.; Phillips, E.J.; Mallal, S.A.; Kendall, P.L. The Evolving Story of Human Leukocyte Antigen and the Immunogenetics of Peanut Allergy. Ann. Allergy Asthma Immunol. 2015, 115, 471–476. [Google Scholar]
- Martino, D.J.; Ashley, S.; Koplin, J.; Ellis, J.; Saffery, R.; Dharmage, S.C.; Gurrin, L.; Matheson, M.C.; Kalb, B.; Marenholz, I.; et al. Genome-wide Association Study of Peanut Allergy Reproduces Association with Amino Acid Polymorphisms in HLA-DRB1. Clin. Exp. Allergy 2017, 47, 217–223. [Google Scholar]
- Asai, Y.; Eslami, A.; van Ginkel, C.D.; Akhabir, L.; Wan, M.; Yin, D.; Ellis, G.; Ben-Shoshan, M.; Marenholz, I.; Martino, D.; et al. A Canadian Genome-Wide Association Study and Meta-Analysis Confirm HLA as a Risk Factor for Peanut Allergy Independent of Asthma. J. Allergy Clin. Immunol. 2018, 141, 1513–1516. [Google Scholar]
- Kostara, M.; Chondrou, V.; Sgourou, A.; Douros, K.; Tsabouri, S. HLA Polymorphisms and Food Allergy Predisposition. J. Pediatr. Genet. 2020, 9, 77–86. [Google Scholar]
- Kanchan, K.; Grinek, S.; Bahnson, H.T.; Ruczinski, I.; Shankar, G.; Larson, D.; Du Toit, G.; Barnes, K.C.; Sampson, H.A.; Suarez-Farinas, M.; et al. HLA Alleles and Sustained Peanut Consumption Promote IgG4 Responses in Subjects Protected from Peanut Allergy. J. Clin. Investig. 2022, 132, e152070. [Google Scholar]
- Jutel, M.; Jaeger, L.; Suck, R.; Meyer, H.; Fiebig, H.; Cromwell, O. Allergen-specific Immunotherapy with Recombinant Grass Pollen Allergens. J. Allergy Clin. Immunol. 2005, 116, 608–613. [Google Scholar] [PubMed]
- Yonekura, S.; Okamoto, Y.; Sakurai, D.; Horiguchi, S.; Hanazawa, T.; Nakano, A.; Kudou, F.; Nakamaru, Y.; Honda, K.; Hoshioka, A.; et al. Sublingual Immunotherapy with House Dust Extract for House Dust-Mite Allergic Rhinitis in Children. Allergol. Int. 2010, 59, 381–388. [Google Scholar]
- Clark, J.; White, N.D. Immunotherapy for Cat Allergies: A Potential Strategy to Scratch Back. Am. J. Lifestyle Med. 2017, 11, 310–313. [Google Scholar] [PubMed]
- Ruëff, F.; Bauer, A.; Becker, S.; Bircher, A.J.; Ebner, C.; Jung, K.; Klimek, L.; Koberne, F.; Koletzko, S.; Lepp, U.; et al. Diagnosis and Treatment of Hymenoptera Venom Allergy: S2k Guideline of the German Society of Allergology and Clinical Immunology (DGAKI) in Collaboration with the Arbeitsgemeinschaft für Berufs- und Umwelt-dermatologie e.V. (ABD), the Medical Association of German Allergologists (AeDA), the German Society of Dermatology (DDG), the German Society of Oto-Rhino-Laryngology, Head and Neck Surgery (DGHNOKC), the German Society of Pediatrics and Adolescent Medicine (DGKJ), the Society for Pediatric Allergy and Environmental Medicine (GPA), German Respiratory Society (DGP), and the Austrian Society for Allergy and Immunology (ÖGAI). Allergol. Select. 2023, 7, 154–190. [Google Scholar] [PubMed]
- Ponda, P.; Cerise, J.E.; Navetta-Modrov, B.; Zhang, C.; Goldfarb, J.; Irizar, H.; Goldberg, J.D.; Wang, M.; Zhang, S.; Sampson, H.A. The Age-Specific Microbiome of Children with Milk, Egg, and Peanut Allergy. Ann. Allergy Asthma Immunol. 2024, 133, 203–210.e6. [Google Scholar] [PubMed]
- Jones, S.M.; Kim, E.H.; Nadeau, K.C.; Nowak-Wegrzyn, A.; Wood, R.A.; Sampson, H.A.; Scurlock, A.M.; Chinthrajah, S.; Wang, J.; Pesek, R.D.; et al. Efficacy and Safety of Oral Immunotherapy in Children Aged 1–3 Years with Peanut Allergy (the Immune Tolerance Network IMPACT Trial): A Randomised Placebo-Controlled Study. Lancet 2022, 399, 359–371. [Google Scholar]
- Kim, E.H.; Keet, C.A.; Virkud, Y.V.; Bird, J.A.; Beyer, K.; Leickly, F.; Liu, A.H.; Petroni, D.; Sampson, H.A.; Wood, R.A.; et al. Open-Label Study of the Efficacy, Safety, and Durability of Peanut Sublingual Immunotherapy in Peanut-Allergic Children. J. Allergy Clin. Immunol. 2023, 151, 1558–1565.e6. [Google Scholar]
- Du Toit, G.; Brown, K.R.; Vereda, A.; Irani, A.M.; Tilles, S.; Ratnayake, A.; Jones, S.M.; Vickery, B.P. Oral Immunotherapy for Peanut Allergy in Children 1 to Less Than 4 Years of Age. NEJM Evid. 2023, 2, EVIDoa2300145. [Google Scholar] [PubMed]
- Greenhawt, M.; Shaker, M.; Abrams, E.M. Peanut Oral Immunotherapy in Very Young Children. Lancet 2022, 399, 336–337. [Google Scholar]
- Kim, E.H.; Bird, J.A.; Keet, C.A.; Virkud, Y.V.; Herlihy, L.; Ye, P.; Smeekens, J.M.; Guo, R.; Yue, X.; Penumarti, A.; et al. Desensitization and Remission after Peanut Sublingual Immunotherapy in 1- to 4-Year-Old Peanut-Allergic Children: A Randomized, Placebo-Controlled Trial. J. Allergy Clin. Immunol. 2024, 153, 173–181.e10. [Google Scholar]
- Tedner, S.G.; Asarnoj, A.; Thulin, H.; Westman, M.; Konradsen, J.R.; Nilsson, C. Food Allergy and Hypersensitivity Reactions in Children and Adults—A Review. J. Intern. Med. 2022, 291, 283–302. [Google Scholar] [PubMed]
- Fleischer, D.M.; Shreffler, W.G.; Campbell, D.E.; Green, T.D.; Anvari, S.; Assa’ad, A.; Bégin, P.; Beyer, K.; Bird, J.A.; Brown-Whitehorn, T.; et al. Long-Term, Open-Label Extension Study of the Efficacy and Safety of Epicutaneous Immunotherapy for Peanut Allergy in Children: PEOPLE 3-Year Results. J. Allergy Clin. Immunol. 2020, 146, 863–874. [Google Scholar] [PubMed]
- Schneider, T.D.; Stephens, R.M. Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [PubMed]
- Doytchinova, I.; Atanasova, M.; Fernandez, A.; Moreno, F.J.; Koning, F.; Dimitrov, I. Modeling Peptide-Protein Interactions by a Logo-Based Method: Application in Peptide-HLA Binding Predictions. Molecules 2024, 29, 284. [Google Scholar] [CrossRef]
- Nielsen, M.; Andreatta, M. NNAlign: A Platform to Construct and Evaluate Artificial Neural Network Models of Receptor-Ligand Interactions. Nucleic Acids Res. 2017, 45, W344–W349. [Google Scholar]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar]
- Chicco, D.; Jurman, G. The Matthews Correlation Coefficient (MCC) Should Replace the ROC AUC as the Standard Metric for Assessing Binary Classification. BioData Min. 2023, 16, 4. [Google Scholar]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar]
- Celis, E.; Tsai, V.; Crimi, C.; DeMars, R.; Wentworth, P.A.; Chesnut, R.W.; Grey, H.M.; Sette, A.; Serra, H.M. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc. Natl. Acad. Sci. USA 1994, 91, 2105–2109. [Google Scholar] [PubMed]
- Barker, D.J.; Maccari, G.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Robinson, J.; Marsh, S.G.E. The IPD-IMGT/HLA Database. Nucleic Acids Res. 2023, 51, D1053–D1060. [Google Scholar] [PubMed]
- MacDonald, K.S.; Embree, J.E.; Nagelkerke, N.J.; Castillo, J.; Ramhadin, S.; Njenga, S.; Oyug, J.; Ndinya-Achola, J.; Barber, B.H.; Bwayo, J.J.; et al. The HLA A2/6802 supertype is associated with reduced risk of perinatal human immunodeficiency virus type 1 transmission. J. Infect. Dis. 2001, 183, 503–506. [Google Scholar]
- Rallón, N.; Restrepo, C.; Vicario, J.L.; Del Romero, J.; Rodríguez, C.; García-Samaniego, J.; García, M.; Cabello, A.; Górgolas, M.; Benito, J.M. Human leucocyte antigen (HLA)-DQB103:02 and HLA-A02:01 have opposite patterns in their effects on susceptibility to HIV infection. HIV Med. 2017, 18, 587–594. [Google Scholar] [PubMed]
- MacDonald, K.S.; Fowke, K.R.; Kimani, J.; Dunand, V.A.; Nagelkerke, N.J.; Ball, T.B.; Oyugi, J.; Njagi, E.; Gaur, L.K.; Brunham, R.C.; et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 2000, 181, 1581–1589. [Google Scholar]
- Noble, J.A.; Valdes, A.M.; Thomson, G.; Erlich, H.A. The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes 2000, 49, 121–125. [Google Scholar]
- Zhang, L.; Zhang, Y.J.; Chen, J.; Yang, M.; Wei, W.; Wang, Y.; Lu, J. The Association of HLA-B27 and Klebsiella pneumoniae in Ankylosing Spondylitis: A Systematic Review. Microb. Pathog. 2018, 117, 49–54. [Google Scholar]
- Lobos, C.A.; Downing, J.; D’Orsogna, L.J.; Chatzileontiadou, D.S.M.; Gras, S. Protective HLA-B57: T Cell and Natural Killer Cell Recognition in HIV Infection. Biochem. Soc. Trans. 2022, 50, 1329–1339. [Google Scholar]
- Ge, C.; Weisse, S.; Xu, B.; Thiel, M.; Chalouni, C.; Travis, A.M.; Kowal, C.; Elwood, J.; Brawn-Cinani, B.; Turner, D.; et al. Key Interactions in the Trimolecular Complex Consisting of the Rheumatoid Arthritis-Associated DRB1*04:01 Molecule, the Major Glycosylated Collagen II Peptide and the T-Cell Receptor. Ann. Rheum. Dis. 2022, 81, 480–489. [Google Scholar]
- Capittini, C.; De Silvestri, A.; Terzaghi, M.; Pasi, A.; Zucconi, M.; Luisi, C.; De Amici, M.; Beri, R.; Tinelli, C.; Milani, G.P.; et al. Correlation between HLA-DQB1*06:02 and Narcolepsy with and without Cataplexy: Approving a Safe and Sensitive Genetic Test in Four Major Ethnic Groups. A Systematic Meta-Analysis. Sleep Med. 2018, 52, 150–157. [Google Scholar] [PubMed]
- D’Amato, M.; Scotto d’Abusco, A.; Maggi, E.; Parmiani, S.; Ferrara, A.; Pene, J.; Richter, A.; Kontou-Fili, K.; Dente, F.L.; Capron, F.; et al. Association of Responsiveness to the Major Pollen Allergen of Parietaria officinalis with HLA-DRB1* Alleles: A Multicenter Study. Hum. Immunol. 1996, 46, 100–106. [Google Scholar]
- Aboulaghras, S.; Piancatelli, D.; Taghzouti, K.; Benkirane, H.; Cantalupo, G.; Bellanti, F.; Lancellotti, S.; Strohmenger, L.; Caserta, V.; Sorrentino, M.C.; et al. Meta-Analysis and Systematic Review of HLA DQ2/DQ8 in Adults with Celiac Disease. Int. J. Mol. Sci. 2023, 24, 1188. [Google Scholar] [CrossRef]
- Howell, W.M.; Holgate, S.T. HLA genetics and allergic disease. Thorax 1995, 50, 815–818. [Google Scholar] [PubMed]
- Kontakioti, E.; Domvri, K.; Papakosta, D.; Daniilidis, M. HLA and asthma phenotypes/endotypes: A review. Hum. Immunol. 2014, 75, 930–939. [Google Scholar]
- Dimitrov, I.; Doytchinova, I. Associations between main food allergens and HLA-DR/DQ polymorphism. Int. Arch. Allergy Immunol. 2016, 169, 33–39. [Google Scholar]
- Gheerbrant, H.; Guillien, A.; Vernet, R.; Lupinek, C.; Pison, C.; Pin, I.; Demenais, F.; Nadif, R.; Bousquet, J.; Pickl, W.F.; et al. Associations between specific IgE sensitization to 26 respiratory allergen molecules and HLA class II alleles in the EGEA cohort. Allergy 2021, 76, 2575–2586. [Google Scholar]
- Germundson, D.L.; Nookala, S.; Smith, N.A.; Warda, Y.; Nagamoto-Combs, K. HLA-II Alleles Influence Physical and Behavioral Responses to a Whey Allergen in a Transgenic Mouse Model of Cow’s Milk Allergy. Front. Allergy 2022, 3, 870513. [Google Scholar]
- Southwood, S.; Sidney, J.; Kondo, A.; del Guercio, M.F.; Appella, E.; Hoffman, S.; Kubo, R.T.; Chesnut, R.W.; Grey, H.M.; Sette, A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1998, 160, 3363–3373. [Google Scholar]
- Doytchinova, I.A.; Flower, D.R. In silico identification of supertypes for Class II Major Histocompatibility Complexes. J. Immunol. 2005, 174, 7085–7095. [Google Scholar]
- Stern, L.J.; Calvo-Calle, J.M. HLA-DR: Molecular insights and vaccine design. Curr. Pharm. Des. 2009, 15, 3249–3261. [Google Scholar]
- Greaves, S.A.; Ravindran, A.; Santos, R.G.; Chen, L.; Falta, M.T.; Wang, Y.; Mitchell, A.M.; Atif, S.M.; Mack, D.G.; Tinega, A.N.; et al. CD4+ T cells in the lungs of acute sarcoidosis patients recognize an Aspergillus nidulans epitope. J. Exp. Med. 2021, 218, e20210785. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- DeLong, J.H.; Simpson, K.H.; Wambre, E.; James, E.A.; Robinson, D.; Kwok, W.W. Ara h 1-reactive T cells in individuals with peanut allergy. J. Allergy Clin. Immunol. 2011, 127, 1211–1218.e3. [Google Scholar] [PubMed]
- Birrueta, G.; Tripple, V.; Pham, J.; Manohar, M.; James, E.A.; Kwok, W.W.; Nadeau, K.C.; Sette, A.; Peters, B.; Schulten, V. Peanut-specific T cell responses in patients with different clinical reactivity. PLoS ONE 2018, 13, e0204620. [Google Scholar]
- Sette, A.; Vitiello, A.; Reherman, B.; Fowler, P.; Nayersina, R.; Kast, W.M.; Melief, C.J.; Oseroff, C.; Yuan, L.; Ruppert, J.; et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994, 153, 5586–5592. [Google Scholar]
- Baumgaertner, P.; Schmidt, J.; Costa-Nunes, C.; Bordry, N.; Guillaume, P.; Luescher, I.; Speiser, D.; Rufer, N.; Hebeisen, M. CD8 T Cell Function and Cross-Reactivity Explored by Stepwise Increased Peptide-HLA versus TCR Affinity. Front. Immunol. 2022, 13, 973986. [Google Scholar]
- Arunachalam, A.B. Vaccines Induce Homeostatic Immunity, Generating Several Secondary Benefits. Vaccines 2024, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Peptides for Vaccine Development. ACS Appl. Bio Mater. 2022, 5, 905–944. [Google Scholar]
- Gupta, K.; Kumar, S.; Das, M.; Dwivedi, P.D. Peptide Based Immunotherapy: A Pivotal Tool for Allergy Treatment. Int. Immunopharmacol. 2014, 19, 391–398. [Google Scholar] [PubMed]

| aa | p1 | p2 | p3 | p4 | p5 | p6 | p7 | p8 | p9 |
|---|---|---|---|---|---|---|---|---|---|
| Ala | −0.083 | −0.157 | 0.213 | 0.324 | 0.324 | 0.176 | 0.102 | 0.028 | −0.046 |
| Arg | 0.028 | −0.046 | 0.176 | 0.324 | 0.176 | 0.176 | 0.102 | 0.398 | 0.213 |
| Asn | −0.194 | −0.194 | −0.120 | −0.157 | −0.009 | −0.120 | −0.046 | −0.194 | −0.194 |
| Asp | −0.194 | −0.194 | −0.083 | −0.009 | −0.157 | −0.194 | −0.083 | −0.120 | −0.194 |
| Cys | −0.157 | −0.194 | −0.194 | −0.157 | −0.194 | −0.194 | −0.194 | −0.194 | −0.157 |
| Gln | −0.194 | −0.009 | −0.046 | −0.157 | −0.120 | −0.083 | −0.157 | −0.120 | −0.194 |
| Glu | −0.157 | −0.120 | −0.194 | −0.120 | −0.157 | −0.194 | −0.083 | −0.120 | −0.157 |
| Gly | −0.120 | −0.157 | −0.083 | 0.065 | −0.083 | −0.083 | −0.046 | −0.046 | −0.194 |
| His | −0.120 | −0.120 | −0.009 | −0.120 | −0.157 | −0.046 | −0.009 | −0.120 | −0.157 |
| Ile | 0.213 | −0.009 | 0.065 | −0.009 | 0.102 | −0.194 | 0.065 | −0.009 | 0.139 |
| Leu | 0.324 | 0.694 | 0.176 | 0.324 | −0.009 | 0.139 | 0.435 | 0.398 | 0.583 |
| Lys | −0.157 | −0.157 | −0.083 | 0.065 | 0.176 | 0.139 | −0.009 | 0.028 | 0.435 |
| Met | 0.102 | 0.324 | 0.324 | −0.046 | −0.046 | −0.120 | −0.046 | 0.065 | −0.083 |
| Phe | 0.806 | −0.046 | 0.102 | −0.009 | −0.009 | 0.250 | 0.250 | 0.287 | 0.102 |
| Pro | −0.157 | −0.194 | −0.120 | −0.120 | 0.065 | 0.176 | −0.009 | −0.083 | −0.194 |
| Ser | −0.046 | −0.120 | 0.028 | 0.176 | 0.065 | 0.065 | −0.083 | −0.083 | −0.194 |
| Thr | −0.194 | 0.028 | −0.157 | −0.157 | −0.009 | 0.102 | −0.046 | −0.046 | −0.157 |
| Trp | 0.102 | 0.102 | −0.009 | −0.083 | −0.083 | −0.083 | −0.046 | −0.120 | −0.083 |
| Tyr | 0.324 | 0.546 | 0.102 | −0.009 | −0.009 | 0.028 | 0.028 | −0.009 | 0.361 |
| Val | −0.120 | 0.028 | −0.083 | −0.120 | 0.139 | 0.065 | −0.120 | 0.065 | 0.176 |
| aa | p1 | p2 | p3 | p4 | p5 | p6 | p7 | p8 | p9 |
|---|---|---|---|---|---|---|---|---|---|
| Ala | −0.078 | 0.109 | 0.234 | 0.422 | 0.297 | −0.141 | 0.047 | 0.359 | 0.422 |
| Arg | −0.016 | −0.078 | 0.047 | −0.141 | −0.141 | −0.078 | −0.016 | 0.109 | −0.203 |
| Asn | −0.078 | 0.109 | 0.047 | −0.016 | −0.078 | −0.141 | −0.141 | −0.203 | −0.141 |
| Asp | 0.172 | −0.016 | 0.109 | −0.078 | −0.203 | −0.078 | −0.203 | −0.266 | −0.266 |
| Cys | −0.203 | 0.047 | −0.328 | −0.016 | −0.203 | 0.047 | −0.141 | −0.078 | −0.016 |
| Gln | −0.141 | −0.141 | 0.109 | −0.266 | −0.078 | −0.203 | −0.203 | −0.141 | −0.141 |
| Glu | 0.297 | 0.047 | 0.109 | −0.078 | −0.078 | −0.203 | 0.047 | −0.141 | −0.016 |
| Gly | 0.047 | 0.359 | 0.359 | 0.172 | 0.109 | −0.016 | 0.297 | 0.047 | 0.609 |
| His | −0.141 | −0.203 | −0.078 | −0.141 | −0.203 | 0.109 | −0.266 | −0.016 | −0.141 |
| Ile | 0.047 | −0.078 | −0.016 | 0.047 | 0.172 | 0.047 | 0.172 | 0.109 | 0.047 |
| Leu | −0.078 | 0.234 | −0.016 | 0.359 | −0.016 | 0.359 | 0.484 | 0.422 | 0.422 |
| Lys | 0.297 | 0.297 | −0.078 | −0.078 | 0.109 | −0.078 | −0.141 | 0.047 | −0.078 |
| Met | −0.328 | −0.078 | −0.078 | −0.016 | −0.078 | −0.078 | −0.203 | 0.047 | −0.141 |
| Phe | −0.078 | −0.141 | −0.203 | −0.016 | 0.234 | −0.141 | −0.078 | −0.203 | 0.047 |
| Pro | −0.141 | −0.078 | −0.203 | −0.141 | −0.078 | −0.266 | 0.172 | −0.016 | −0.266 |
| Ser | 0.484 | 0.234 | 0.234 | 0.359 | 0.359 | 0.672 | 0.172 | 0.109 | 0.234 |
| Thr | −0.141 | −0.078 | 0.234 | −0.141 | 0.047 | −0.141 | −0.078 | 0.047 | 0.172 |
| Trp | −0.328 | −0.328 | −0.266 | −0.266 | −0.203 | −0.141 | −0.266 | −0.203 | −0.266 |
| Tyr | −0.078 | −0.203 | −0.328 | −0.078 | −0.016 | 0.172 | 0.047 | −0.141 | −0.203 |
| Val | 0.484 | −0.016 | 0.109 | 0.109 | 0.047 | 0.297 | 0.297 | 0.109 | −0.078 |
| Training set of binders | 105 nonamers |
| 10 training sets of non-binders | 105 nonamers |
| Test set of binders | 7814 peptides of different length |
| 10 test set of non-binders | 1018 nonamers |
| True positives (TP) | 7658 |
| False positives (FP) | 365 |
| True negatives (TN) | 653 |
| False negatives (FN) | 155 |
| Sensitivity (Recall) | 0.980 |
| Specificity | 0.641 |
| Accuracy | 0.941 |
| Precision | 0.955 |
| Matthews’s correlation coefficient (MCC) | 0.689 |
| F1 score | 0.967 |
| Allergen Name | Allergen and Variants | GenBank Protein | Uniprot |
|---|---|---|---|
| Ara h 1 | Ara h 1.0101 | AAB00861 | P43238 |
| Ara h 2 | Ara h 2.0101 | AAK96887 | - |
| Ara h 2.0201 | AAN77576 | Q6PSU2-1 | |
| Ara h 3 | Ara h 3.0101 | AAC63045 | O82580 |
| Ara h 3.0201 | AAD47382 | Q9SQH7 | |
| Ara h 5 | Ara h 5.0101 | AAD55587 | Q9SQI9 |
| Ara h 6 | Ara h 6.0101 | AAD56337 | Q647G9 |
| Ara h 7 | Ara h 7.0101 | AAD56719 | Q9SQH1 |
| Ara h 7.0201 | ABW17159 | B4XID4 | |
| Ara h 7.0301 | - | Q647G8 | |
| Ara h 8 | Ara h 8.0101 | AAQ91847 | Q6VT83 |
| Ara h 8.0201 | ABP97433 | B0YIU5 | |
| Ara h 9 | Ara h 9.0101 | ABX56711 | B6CEX8 |
| Ara h 9.0201 | ABX75045 | B6CG41 | |
| Ara h 10 | Ara h 10.0101 | AAU21499 | Q647G5 |
| Ara h 10.0102 | AAU21500 | Q647G4 | |
| Ara h 11 | Ara h 11.0101 | AAZ20276 | Q45W87 |
| Ara h 11.0102 | AAZ20277 | Q45W86 | |
| Ara h 12 | Ara h 12.0101 | - | B3EWP3 |
| Ara h 13 | Ara h 13.0101 | - | B3EWP4 |
| Ara h 13.0102 | - | C0HJZ1 | |
| Ara h 14 | Ara h 14.0101 | AAK13449 | Q9AXI1 |
| Ara h 14.0102 | AAK13450 | Q9AXI0 | |
| Ara h 14.0103 | AAT11925 | Q6J1J8 | |
| Ara h 15 | Ara h 15.0101 | AAU21501 | Q647G3 |
| Ara h 16 | Ara h 16.0101 | ASU04353 | A0A509ZX51 |
| Ara h 17 | Ara h 17.0101 | ASU04352 | A0A510A9S3 |
| Ara h 18 | Ara h 18.0101 | XP_025675300 | A0A444XS96 |
| Allergen | Predicted Best Binder | BS |
|---|---|---|
| Ara h 1.0101 | MLLLGILVL | 2.102 |
| Ara h 2.0101 | LTILVALAL ILVALALFL LVALALFLL FLLAAHASA | 2.102 2.620 2.583 2.251 |
| Ara h 3.0101 | ALSRLVLRR VLRRNALRR | 2.065 2.287 |
| Ara h 3.0201 | LLILRWLGL | 2.472 |
| Ara h 6.0101 | LVALLALVL | 2.139 |
| Ara h 10.0101 | LLLFAGLAL GLALAGTLL | 2.472 2.287 |
| Ara h 11.0101 | LLILAGLVL FLASGGFGV | 2.731 2.103 |
| Ara h 14.0101 | LLLLSGLSL LLLSGLSLL | 2.435 2.324 |
| Ara h 15.0101 | FLILSGLIL GLIIATPLL | 2.880 2.028 |
| Allergen | Known T-Cell Epitope | Binding to HLA-DRB1*03:01 IC50 nM | Predicted Best Binder | BS |
|---|---|---|---|---|
| Ara h 1.0101 | NNFGKLFEVKPDKKNPQLQD [56] | na | KLFEVKPDK | 1.103 |
| Ara h 2.0101 | ARQQWELQGDRRCQS [57] | 1470 | QWELQGDRR | 0.363 |
| Ara h 3.0101 | EFLEQAFQVDDRQIV [57] | 147 | FLEQAFQVD | 1.437 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doytchinova, I.; Atanasova, M.; Sotirov, S.; Dimitrov, I. In Silico Identification of Peanut Peptides Suitable for Allergy Immunotherapy in HLA-DRB1*03:01-Restricted Patients. Pharmaceuticals 2024, 17, 1097. https://doi.org/10.3390/ph17081097
Doytchinova I, Atanasova M, Sotirov S, Dimitrov I. In Silico Identification of Peanut Peptides Suitable for Allergy Immunotherapy in HLA-DRB1*03:01-Restricted Patients. Pharmaceuticals. 2024; 17(8):1097. https://doi.org/10.3390/ph17081097
Chicago/Turabian StyleDoytchinova, Irini, Mariyana Atanasova, Stanislav Sotirov, and Ivan Dimitrov. 2024. "In Silico Identification of Peanut Peptides Suitable for Allergy Immunotherapy in HLA-DRB1*03:01-Restricted Patients" Pharmaceuticals 17, no. 8: 1097. https://doi.org/10.3390/ph17081097
APA StyleDoytchinova, I., Atanasova, M., Sotirov, S., & Dimitrov, I. (2024). In Silico Identification of Peanut Peptides Suitable for Allergy Immunotherapy in HLA-DRB1*03:01-Restricted Patients. Pharmaceuticals, 17(8), 1097. https://doi.org/10.3390/ph17081097







