Investigating Potential Anti-Bacterial Natural Products Based on Ayurvedic Formulae Using Supervised Network Analysis and Machine Learning Approaches
Abstract
:1. Introduction
2. Results
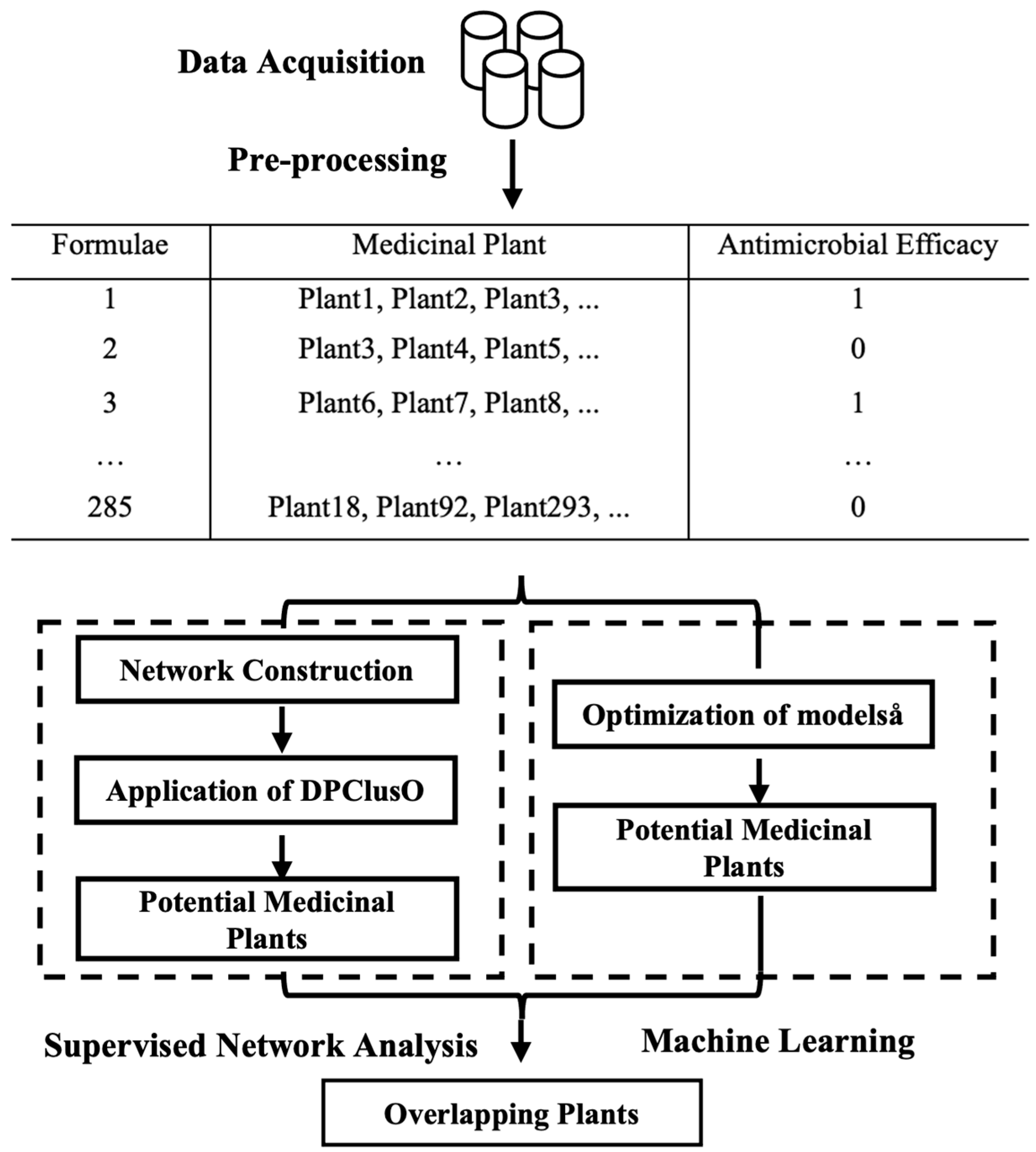
2.1. Data Preprocessing
2.2. Supervised Network
2.3. Machine Learning for Antimicrobial Phenotype Decision
2.4. Overlapping Results
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Preprocessing
4.2. Supervised Network Clustering
4.3. Learning Models
4.4. Validation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. Mayo Clin. Proc. 2011, 86, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Gopal Rao, G. Risk factors for the spread of antibiotic-resistant bacteria. Drugs 1998, 55, 323–330. [Google Scholar]
- Muteeb, G.; Rehman, T.; Shahwan, M.; Aatif, M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.K.; Hussein, S.; Qurbani, K.; Ibrahim, R.H.; Fareeq, A.; Mahmood, K.A.; Mohamed, M.G. Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health 2024, 2, 100081. [Google Scholar] [CrossRef]
- Lewis, K. The science of antibiotic discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Gangwal, A.; Ansari, A.; Ahmad, I.; Azad, A.K.; Sulaiman, W.M.A.W. Current strategies to address data scarcity in artificial intelligence-based drug discovery: A comprehensive review. Comput. Biol. Med. 2024, 179, 108734. [Google Scholar] [CrossRef]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Vaou, N.; Stavropoulou, E.; Voidarou, C.; Tsigalou, C.; Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms 2021, 9, 2041. [Google Scholar] [CrossRef]
- Johnston, C.W.; Badran, A.H. Natural and engineered precision antibiotics in the context of resistance. Curr. Opin. Chem. Biol. 2022, 69, 102160. [Google Scholar] [CrossRef]
- Ahad, B.; Shahri, W.; Rasool, H.; Reshi, Z.A.; Rasool, S.; Hussain, T. Medicinal plants and herbal drugs: An overview. In Medicinal and Aromatic Plants: Healthcare and Industrial Applications; Springer: Cham, Switzerland, 2021; pp. 1–40. [Google Scholar]
- Uwineza, P.A.; Waśkiewicz, A. Recent advances in supercritical fluid extraction of natural bioactive compounds from natural plant materials. Molecules 2020, 25, 3847. [Google Scholar] [CrossRef]
- Izah, S.C.; Ogidi, O.I.; Ogwu, M.C.; Salimon, S.S.; Yusuf, Z.M.; Akram, M.; Raimi, M.O.; Iyingiala, A.-A. Historical perspectives and overview of the value of herbal medicine. In Herbal Medicine Phytochemistry: Applications and Trends; Springer International Publishing: Cham, Switzerland, 2024; pp. 3–35. [Google Scholar]
- Theodoridis, S.; Drakou, E.G.; Hickler, T.; Thines, M.; Nogues-Bravo, D. Evaluating natural medicinal resources and their exposure to global change. Lancet Planet. Health 2023, 7, e155–e163. [Google Scholar] [CrossRef]
- Ansari, S. Overview of traditional systems of medicine in different continents. In Preparation of Phytopharmaceuticals for the Management of Disorders; Academic Press: Cambridge, MA, USA, 2021; pp. 431–473. [Google Scholar]
- Talukdar, A.D.; Patra, J.K.; Das, G.; Nath, D. (Eds.) Traditional Resources and Tools for Modern Drug Discovery: Ethnomedicine and Pharmacology; Springer Nature: Berlin/Heidelberg, Germany, 2024. [Google Scholar]
- Pratibha, N.; Mukesh, E.; VinodKumar, M.V. Ayurvedic practice, education and research, beyond dilemmas and confessions. J. Ayurveda Integr. Med. 2023, 14, 100814. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.; Bamne, F.; Ali, A.; Momin, M.; Khan, T. Herbal medicine: Exploring its scope across belief systems of the Indian medicine. In Herbal Medicine Phytochemistry: Applications and Trends; Springer International Publishing: Cham, Switzerland, 2024; pp. 1279–1304. [Google Scholar]
- Jansen, C.; Baker, J.; Kodaira, E.; Ang, L.; Bacani, A.; Aldan, J.; Shimoda, L.; Salameh, M.; Small-Howard, A.; Stokes, A.; et al. Medicine in motion: Opportunities, challenges and data analytics-based solutions for traditional medicine integration into western medical practice. J. Ethnopharmacol. 2021, 267, 113477. [Google Scholar] [CrossRef]
- Wijaya, S.H.; Afendi, F.M.; Batubara, I.; Huang, M.; Ono, N.; Kanaya, S.; Amin, A.U. Identification of Targeted Proteins by Jamu Formulas for Different Efficacies Using Machine Learning Approach. Life 2021, 11, 866. [Google Scholar] [CrossRef]
- Gao, P.; Nasution, A.K.; Yang, S.; Chen, Z.; Ono, N.; Kanaya, S.; Amin, A.U. On finding natural antibiotics based on TCM formulae. Methods 2023, 214, 35–45. [Google Scholar] [CrossRef]
- Amin, A.U.; Wada, M.; Kanaya, S. Partitioning a PPI Network into Overlapping Modules Constrained by High-Density and Periphery Tracking. ISRN Biomath. 2012, 2012, 726429. [Google Scholar] [CrossRef]
- Anonymous. Bangladesh National Formulary of Ayurvedic Medicine 1992; National Unani and Ayurvedic Formulary Committee Bangladesh Board of Unani and Ayurvedic Systems of Medicine: Dhaka, Bangladesh, 2011; Volume 38, ISBN 978-984-33-3254-7. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Moo, C.L.; Yang, S.K.; Osman, M.A.; Yuswan, M.H.; Loh, J.Y.; Lim, W.M.; Swee-Hua-Erin, L.I.M.; Lai, K.S. Antibacterial Activity and Mode of Action of β-caryophyllene on Bacillus cereus. Pol. J. Microbiol. 2020, 69, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, W.; Sun, Z. Antimicrobial activity and mechanism of limonene against Staphylococcus aureus. J. Food Saf. 2021, 41, e12918. [Google Scholar] [CrossRef]
- Elbestawy, M.K.; El-Sherbiny, G.M.; Moghannem, S.A.; Farghal, E.E. Antibacterial, Antibiofilm, and Anti-Inflammatory Activities of Ginger Extract against Helicobacter pylori. Microbiol. Res. 2023, 14, 1124–1138. [Google Scholar] [CrossRef]
- Bai, X.; Li, X.; Liu, X.; Xing, Z.; Su, R.; Wang, Y.; Xia, X.; Shi, C. Antibacterial Effect of Eugenol on Shigella flexneri and Its Mechanism. Foods 2022, 11, 2565. [Google Scholar] [CrossRef]
- Majdi, C.; Duvauchelle, V.; Meffre, P.; Benfodda, Z. An overview on the antibacterial properties of juglone, naphthazarin, plumbagin and lawsone derivatives and their metal complexes. Biomed. Pharmacother. 2023, 162, 114690. [Google Scholar] [CrossRef]
- Vyas, A.; Sharma, B. Therapeutic Potential of Trichosanthes dioica Plant (Pointed Gourd)—A Review. Authorea Prepr. 2022. [Google Scholar] [CrossRef]
- Saki, M.; Seyed-Mohammadi, S.; Montazeri, E.A.; Siahpoosh, A.; Moosavian, M.; Latifi, S.M. In vitro antibacterial properties of Cinnamomum zeylanicum essential oil against clinical extensively drug-resistant bacteria. Eur. J. Integr. Med. 2020, 37, 101146. [Google Scholar] [CrossRef]
- Borges, M.F.d.A.; Lacerda, R.d.S.; Correia, J.P.d.A.; de Melo, T.R.; Ferreira, S.B. Potential antibacterial action of α-pinene. Med. Sci. Forum 2022, 12, 11. [Google Scholar] [CrossRef]
- Farhanghi, A.; Aliakbarlu, J.; Tajik, H.; Mortazavi, N.; Manafi, L.; Jalilzadeh-Amin, G. Antibacterial interactions of pulegone and 1, 8-cineole with monolaurin ornisin against Staphylococcus aureus. Food Sci. Nutr. 2022, 10, 2659–2666. [Google Scholar] [CrossRef]
- Liu, X.; Bian, L.; Duan, X.; Zhuang, X.; Sui, Y.; Yang, L. Alantolactone: A sesquiterpene lactone with diverse pharmacological effects. Chem. Biol. Drug Des. 2021, 98, 1131–1145. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Huang, C.; Huang, H.; Zhao, Y.; Khan, M.R.U.; Zhao, H.; Huang, L. Antibacterial mechanism of curcumin: A review. Chem. Biodivers. 2020, 17, 2000171. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Dipti, T.T.; Islam, M.N.; Abdullah, A.T.M.; Jahan, S.; Alam, M.; Karim, M.R. Chemical composition, antioxidant and antibacterial activity of Piper chaba stem extracts with preservative effects on storage of raw beef patties. Saudi J. Biol. Sci. 2023, 30, 103663. [Google Scholar] [CrossRef]
- Bouyahya, A.; Mechchate, H.; Benali, T.; Ghchime, R.; Charfi, S.; Balahbib, A.; Burkov, P.; Shariati, M.A.; Lorenzo, J.M.; El Omari, N. Health benefits and pharmacological properties of carvone. Biomolecules 2021, 11, 1803. [Google Scholar] [CrossRef]
- Nam, Y.J.; Hwang, Y.S. Antibacterial and antioxidant effect of ethanol extracts of Terminalia chebula on Streptococcus mutans. Clin. Exp. Dent. Res. 2021, 7, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Wani, Z.A.; Pant, S. Aconitum heterophyllum Wall. ex Royle: An endemic, highly medicinal and critically endangered plant species of northwestern himalaya in peril. Curr. Tradit. Med. 2021, 7, e240921196789. [Google Scholar] [CrossRef]
- Bane, S.P.; Thosar, N.R.; Rathi, N.V.; A Deshpande, M.; Deulkar, P.V. Comparative Evaluation of Antibacterial Efficacy of Emblica officinalis Lollipop Against Streptococcus mutans Counts in Institutionalized Visually Impaired Children. Cureus 2022, 14, e28207. [Google Scholar] [CrossRef]
- Yadav, A.N.; Singh, P.; Upadhyay, S.; Tyagi, U.P.; Singh, A.K.; Singh, P.; Srivastava, A. Amla (Emblica officinalis)-Derived Bionanosilver (Ag NPs) for Excellent Antibacterial Activity. Plasmonics 2024. [Google Scholar] [CrossRef]
- Rafiq, M.; Tunio, A.A.; Qureshi, A.S.; Rehman, T.; Bhutto, M.A.; Lasharı, Z. Determination of Phytochemicals, Antimicrobial, Antioxidant and Allelopathic Effects of Fagonia cretica L., collected from Jamshoro, Pakistan. Yuz. Yıl Univ. J. Agric. Sci. 2022, 32, 785–794. [Google Scholar] [CrossRef]
- Dongare, G.; Kale, A.; Band, P.; Chavan, A.; Aher, A.N.; Gawali, S. A review on phytochemical and pharmacological action of Clerodendrum serratum Linn. (Bharangi). World J. Pharm. Res. 2020, 9, 229–236. [Google Scholar]
- Din, M.S.U.; Gohar, U.F.; Mukhtar, H.; Khan, I.; Morris, J.; Pornpukdeewattana, S.; Massa, S. Antibiotic Reversal Activity of Piper longum Fruit Extracts against Staphylococcus aureus Multi-Drug Resistant Phenotype. Microbiol. Biotechnol. Lett. 2023, 51, 432–440. [Google Scholar] [CrossRef]
- Wijaya, S.H.; Husnawati, H.; Afendi, F.M.; Batubara, I.; Darusman, L.K.; Amin, A.U.; Sato, T.; Ono, N.; Sugiura, T.; Kanaya, S. Supervised Clustering Based on DPClusO: Prediction of Plant-Disease Relations Using Jamu Formulas of KNApSAcK Database. BioMed Res. Int. 2014, 2014, 831751. [Google Scholar] [CrossRef] [PubMed]
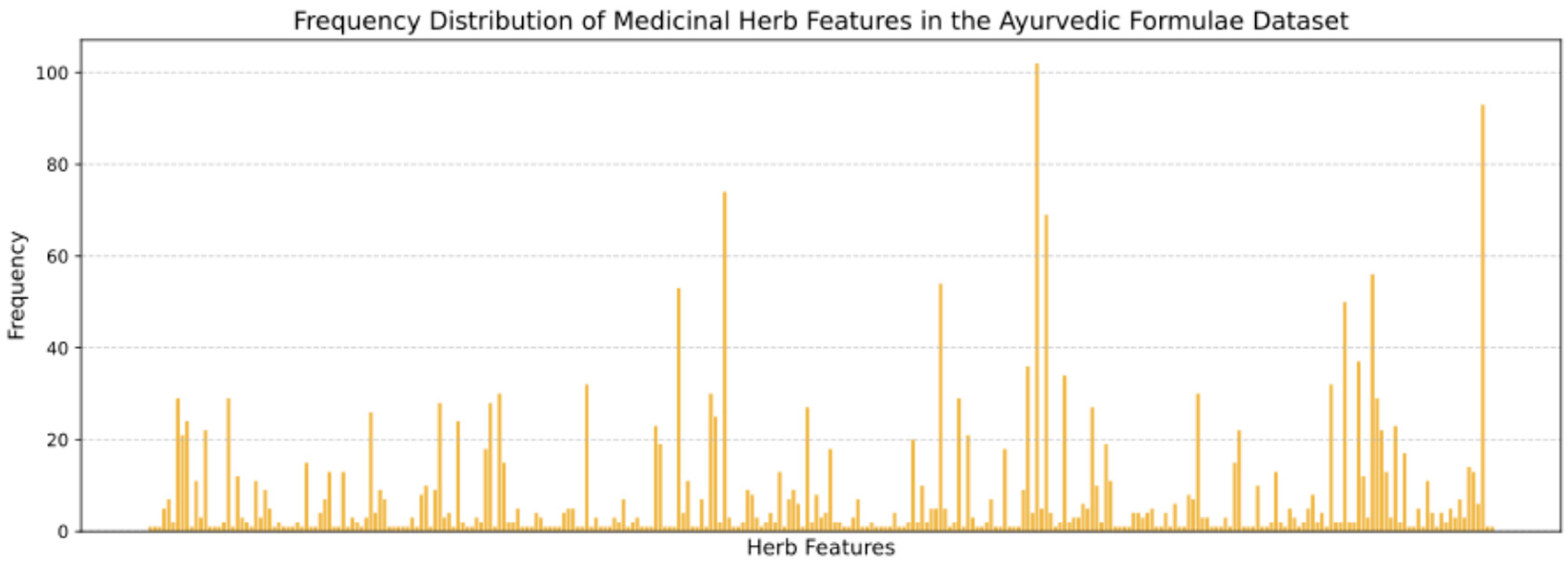
| No. | Medicinal Plant | Frequency |
|---|---|---|
| 1 | Piper longum | 102 |
| 2 | Zingiber officinale | 93 |
| 3 | Emblica officinalis | 74 |
| 4 | Piper nigrum | 69 |
| 5 | Terminalia chebula | 56 |
| 6 | Myristica fragrans | 54 |
| 7 | Cyperus rotundus | 53 |
| 8 | Tenninalia chebula | 50 |
| 9 | Terminalia bellcrica | 37 |
| 10 | Piper chaba | 36 |
| 11 | Plumbago zeylanica | 34 |
| 12 | Syzygium aromaticum | 32 |
| 13 | Coriandrum sativum | 32 |
| 14 | Cinnamomum zeylanicum | 30 |
| 15 | Elettaria cardamomum | 30 |
| 16 | Saussurea hypoleuca | 30 |
| 17 | Aconitum ferox | 29 |
| 18 | Aloe barbadensis | 29 |
| 19 | Tinospora cordifolia | 29 |
| 20 | Nigella sativa | 29 |
| 21 | Carum curvi | 28 |
| 22 | Cinnamomum tamala | 28 |
| 23 | Glycyrrhiza glabra | 27 |
| 24 | Pterocarpus santalinus | 27 |
| 25 | Berberis aristata | 26 |
| 26 | Embelia ribes | 25 |
| 27 | Cedrus deodara | 24 |
| 28 | Acorus calamus | 24 |
| 29 | Tribulus terresrris | 23 |
| 30 | Curcuma longa | 23 |
| 31 | Aegle marmelos | 22 |
| 32 | Trachyspermum ammi | 22 |
| 33 | Sida cordifolia | 22 |
| 34 | Aconitum heterophyllum | 21 |
| 35 | Operculina turpethum | 21 |
| 36 | Mesua ferrea | 20 |
| 37 | Rhus succedanea | 19 |
| 38 | Curcuma zedoaria | 19 |
| 39 | Hollerrhena antidysenterica | 18 |
| 40 | Picrorhiza kurroa | 18 |
| Machine Learning Models | Filtering | SMOTE | Filtering + SMOTE |
|---|---|---|---|
| Decision Tree Classifier | 0.673 ± 0.025 | 0.648 ± 0.015 | 0.635 ± 0.010 |
| Naïve Bayes Classifier | 0.626 ± 0.013 | 0.675 ± 0.028 | 0.648 ± 0.017 |
| Gradient Boosting Classifier | 0.748 ± 0.014 | 0.748 ± 0.011 | 0.775 ± 0.026 |
| K-Neighbors Classifier | 0.755 ± 0.009 | 0.709 ± 0.017 | 0.746 ± 0.012 |
| Logistic Regression | 0.790 ± 0.016 | 0.755 ± 0.008 | 0.797 ± 0.015 |
| Multi-layer Perceptron | 0.671 ± 0.020 | 0.752 ± 0.014 | 0.748 ± 0.025 |
| Random Forest | 0.727 ± 0.022 | 0.774 ± 0.014 | 0.824 ± 0.018 |
| No. | Medicinal Plant | Ayurvedic Formulae |
|---|---|---|
| 1 | Zingiber officinale | 6 |
| 2 | Cyperus rotundus | 5 |
| 3 | Piper longum | 5 |
| 4 | Nigella sativa | 4 |
| 5 | Rhus succedanea | 4 |
| 6 | Tinospora cordifolia | 4 |
| 7 | Terminalia chebula | 4 |
| 8 | Bharangi—Clerodendrum | 3 |
| 9 | Carum curvi | 3 |
| 10 | Cedrus deodara | 3 |
| 11 | Coriandrum sativum | 3 |
| 12 | Emblica officinalis | 3 |
| 13 | Myrica nagi/Myrica sapida | 3 |
| 14 | Piper nigrum | 3 |
| 15 | Saussurea hypoleuca | 3 |
| 16 | Fagonia cretica | 3 |
| 17 | Picrorhiza kurroa | 3 |
| 18 | Cinnamomum tamala | 2 |
| 19 | Cinnamomum zeylanicum | 2 |
| 20 | Croton polyandrum | 2 |
| 21 | Elettaria cardamomum | 2 |
| 22 | Glycyrrhiza glabra | 2 |
| 23 | Myristica fragrans | 2 |
| 24 | Piper chaba | 2 |
| 25 | Plumbago zeylanica | 2 |
| 26 | Syzygium aromaticum | 2 |
| 27 | Trachyspermum ammi carum copticum | 2 |
| 28 | Trianthema portulacastrum | 2 |
| 29 | Tribulus terrestris | 2 |
| 30 | Uraria lagopoides | 2 |
| 31 | Aegle marmelos | 2 |
| 32 | Solanum indicum | 2 |
| 33 | Aconitum heterophyllum | 2 |
| 34 | Cissampelos pareira | 2 |
| 35 | Azadirachta indica | 2 |
| 36 | Curcuma longa | 2 |
| 37 | Trichosanthes dioica | 2 |
| 38 | Inula racemosa | 2 |
| 39 | Nigella sativa | 2 |
| No. | Medicinal Plant | Weight |
|---|---|---|
| 1 | Cyperus rotundus | 0.03109277 |
| 2 | Piper longum | 0.0261781 |
| 3 | Aconitum ferox | 0.02305289 |
| 4 | Sida cordifolia | 0.01956573 |
| 5 | Piper nigrum | 0.01927294 |
| 6 | Zingiber officinale | 0.01859687 |
| 7 | Myristica fragrans | 0.01832107 |
| 8 | Plumbago zeylanica | 0.01659803 |
| 9 | Acacia leucophloea | 0.01593949 |
| 10 | Terminalia chebula | 0.01574221 |
| 11 | Trichosanthes dioica | 0.01567844 |
| 12 | Cinnamomum zeylanicum | 0.01544429 |
| 13 | Bambusa bambos | 0.01483098 |
| 14 | Elettaria cardamomum | 0.0143906 |
| 15 | Inula racemosa | 0.0140497 |
| 16 | Curcuma longa | 0.01371762 |
| 17 | Piper chaba | 0.0136643 |
| 18 | Punica granatum | 0.01346506 |
| 19 | Carum curvi | 0.01325254 |
| 20 | Femia foetida | 0.01210255 |
| 21 | Tenninalia chebula | 0.01149938 |
| 22 | Aconitum heterophyllum | 0.0111556 |
| 23 | Emblica officinalis | 0.01114156 |
| 24 | Adhatoda vasica | 0.01106956 |
| 25 | Fagonia cretica | 0.01092521 |
| 26 | Berberies aristata | 0.01090131 |
| 27 | Berberis aristata | 0.0107864 |
| 28 | Bharangi—Clerodendrum | 0.01015191 |
| 29 | Syzygium aromaticum | 0.01012223 |
| 30 | Operculina turpethum | 0.01011386 |
| 31 | Terminalia bellcrica | 0.01009841 |
| 32 | Hollerrhena antidysentrica | 0.01003895 |
| No. | Medicinal Plant | Metabolite | Antimicrobial Property | Reference |
|---|---|---|---|---|
| 1 | Piper longum | isolates | Antibacterial | [44] |
| 2 | Piper nigrum | β-Caryophyllene (C15H24), limonene (C10H16) | Antibacterial, Antifungal | [25,26] |
| 3 | Zingiber officinale | Gingerol (C17H26O4) | Antibacterial | [27] |
| 4 | Myristica fragrans | Eugenol (C10H12O2) | Antibacterial, Antifungal | [28] |
| 5 | Plumbago zeylanica | plumbagin (C11H8O3) | Antibacterial | [29] |
| 6 | Trichosanthes dioica | isolates | Antibacterial | [30] |
| 7 | Cinnamomum zeylanicum | isolates | Antibacterial, Antifungal | [31] |
| 8 | Elettaria cardamomum | α-Pinene (C10H16), 1,8-Cineole (C10H18O) | Antibacterial, Antifungal | [32,33] |
| 9 | Inula racemosa | Alantolactone (C15H20O2) | Antibacterial | [34] |
| 10 | Curcuma longa | Curcumin (C21H20O6) | Antibacterial, Antioxidant | [35] |
| 11 | Piper chaba | isolates | Antibacterial, Antiviral | [36] |
| 12 | Carum carvi | carvone (C10H14O), limonene (C10H16) | Antibacterial | [26,37] |
| 13 | Terminalia chebula | isolates | Antibacterial, Antiviral | [38] |
| 14 | Aconitum heterophyllum | heterophylline (C22H26N2O4) | Antibacterial | [39] |
| 15 | Emblica officinalis | isolates, Ag NPs | Antibacterial, Antioxidant | [40,41] |
| 16 | Fagonia cretica | isolates | Antibacterial | [42] |
| 17 | Bharangi | isolates | Antibacterial | [43] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, P.; Nasution, A.K.; Ono, N.; Kanaya, S.; Altaf-Ul-Amin, M. Investigating Potential Anti-Bacterial Natural Products Based on Ayurvedic Formulae Using Supervised Network Analysis and Machine Learning Approaches. Pharmaceuticals 2025, 18, 192. https://doi.org/10.3390/ph18020192
Gao P, Nasution AK, Ono N, Kanaya S, Altaf-Ul-Amin M. Investigating Potential Anti-Bacterial Natural Products Based on Ayurvedic Formulae Using Supervised Network Analysis and Machine Learning Approaches. Pharmaceuticals. 2025; 18(2):192. https://doi.org/10.3390/ph18020192
Chicago/Turabian StyleGao, Pei, Ahmad Kamal Nasution, Naoaki Ono, Shigehiko Kanaya, and Md. Altaf-Ul-Amin. 2025. "Investigating Potential Anti-Bacterial Natural Products Based on Ayurvedic Formulae Using Supervised Network Analysis and Machine Learning Approaches" Pharmaceuticals 18, no. 2: 192. https://doi.org/10.3390/ph18020192
APA StyleGao, P., Nasution, A. K., Ono, N., Kanaya, S., & Altaf-Ul-Amin, M. (2025). Investigating Potential Anti-Bacterial Natural Products Based on Ayurvedic Formulae Using Supervised Network Analysis and Machine Learning Approaches. Pharmaceuticals, 18(2), 192. https://doi.org/10.3390/ph18020192









