Disease-Causing Allele-Specific Silencing by RNA Interference
Abstract
:Scientific Terms
| microRNA (miRNA) | 21~23-nucleotide-long small non-coding RNA that functions as a mediator in transcriptional and post-transcriptional regulation of gene expression. MiRNA, like siRNA, is incorporated into RISC and works. Over 2000 miRNA genes have been found in the human genome |
| “Seed” region | nucleotides at positions 2-8 relative to the 5'-end of miRNA. The region is considered to be a key determinant of target specificity |
| Short-hairpin RNA (shRNA) | RNA sequence that forms a hairpin turn and can be processed by Dicer, an RNase III enzyme, to siRNA |
| Allele | one of a number of alternative forms of the same gene |
| Single nucleotide polymorphism (SNP) | single nucleotide variation, whose frequency in a population is more than 1% |
| Induced pluripotent stem cell (iPSC) | pluripotent stem cell artificially derived from a non-pluripotent cell |
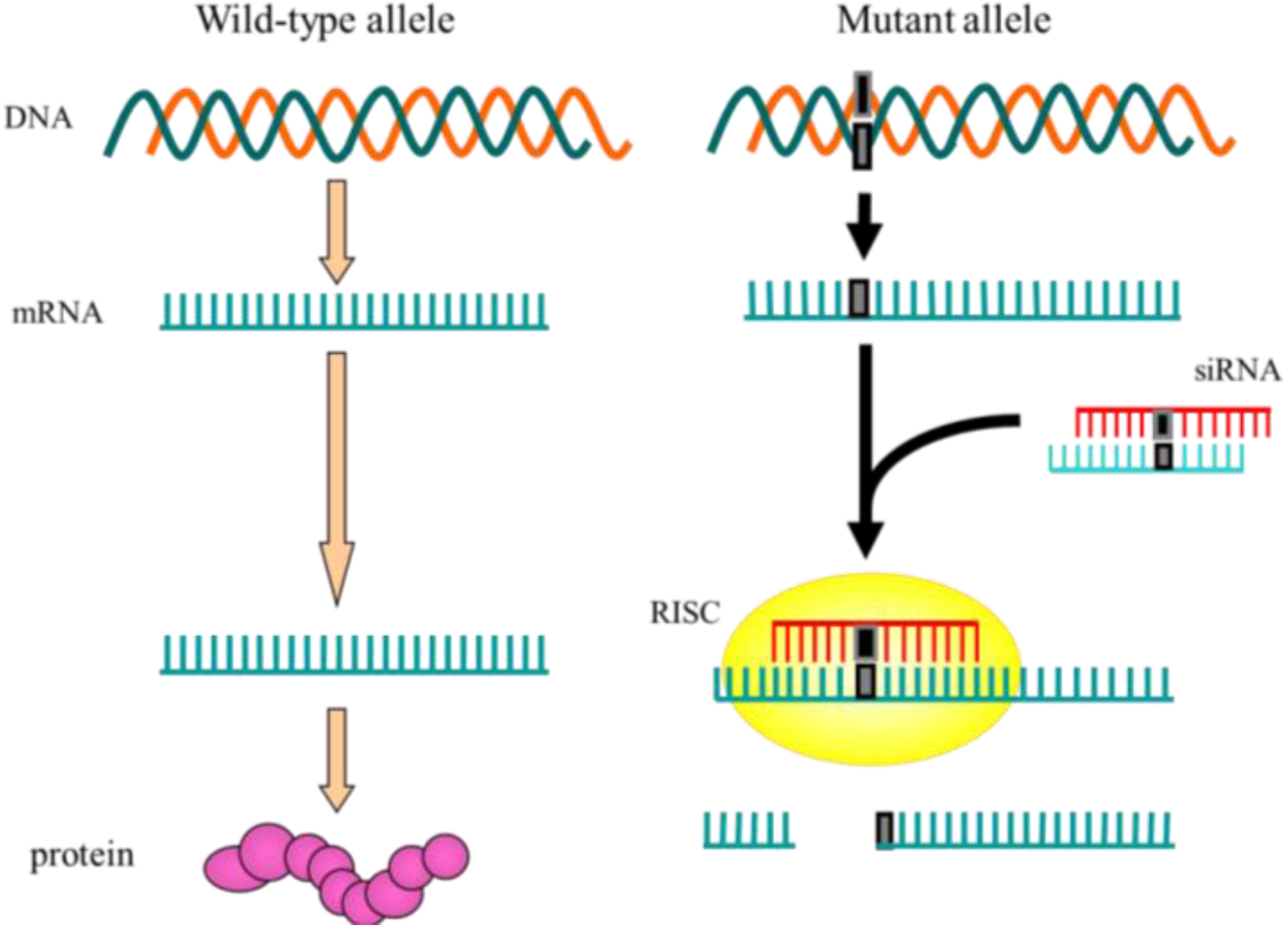
1. Introduction
2. Allele-Specific Silencing by RNAi As an Advanced Method for Therapeutic Use
| Disease | Target gene | Target variation | Inducer of RNAi | Competent siRNA(s) carrying mutation site(s) at the central position | Modification | References |
|---|---|---|---|---|---|---|
| Familial Alzheimer's disease | Amyloid precursor protein (APP) | K670N-M671L (Swedish mutant) | synthetic siRNA | yes | no | Miller VM. et al. (2004) [43] Ohnishi Y. et al. (2006) [44] Feng X. et al. (2006) [45] |
| Amyloid precursor protein (APP) | K670N-M671L (Swedish mutant) | synthetic siRNA | no | nucleotide mismatches | Ohnishi Y. et al. (2008) [46] | |
| Amyloid precursor protein (APP) | V717F (London mutant) | synthetic siRNA | yes | no | Ohnishi Y. et al. (2006) [44] | |
| Amyloid precursor protein (APP) | V717I (London mutant) | synthetic shRNA | yes | no | Feng X. et al. (2006) [45] | |
| Preseniline 1 (PSEN1) | L392V | synthetic siRNA | yes | 2-Thiouridine chemical modification | Sierant M. et al. (2011) [47] | |
| Amyotrophic lateral sclerosis (ALS) | Superoxide dismutase (SOD1) | G93A | shRNA expression vector | yes | no | Xia X. et al. (2006) [48] |
| Superoxide dismutase (SOD1) | G85R | synthetic siRNA | yes/no *1 | nucleotide mismatch | Schwarz DS. et al. (2006) [49] | |
| Slow channel congenital myasthenic syndrome (SCCMS) | Acetylcholine receptor (AChR) | aS226F | synthetic siRNA/shRNA | yes | no | Abdelgany A. et al. (2003) [50] |
| Frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) | Microtubule-associated protein TAU (MAPT) | V337M | synthetic siRNA | yes | nucleotide mismatch | Miller VM. et al. (2003,2004) [43,51] |
| Ehlers-Danlos syndrome (vEDS) | Procollagen type III (COL3A1) | G252V | synthetic siRNA | yes | no | Muller GA. et al. (2012) [52] |
| Sickle cell anemia | Hemoglobin-beta locus (HBB) | E6V | synthetic siRNA | yes | no | Dykxhoorn DM. et al. (2006) [53] |
| Familial amyloidotic polyneuropathy (FAP) | Transthyretin (TTR) | V30M | synthetic siRNA | yes | no | Kurosawa T. et al. (2005) [54] |
| Fibrodysplasia ossificans progressiva (FOP) | Activin A receptor type I (ACVR1) | R206H, G356D | synthetic siRNA | yes | nucleotide mismatch | Takahashi M. et al. (2012) [55] |
| Activin A receptor type I (ACVR1) | R206H | synthetic siRNA | yes | no | kaplan J. et al. (2012) [56] | |
| Tumors | Phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) | 1633G -> A 3140A -> G | synthetic siRNA | yes | no | Huang H. et al. (2009) [57] |
| Spinocerebellar ataxia type 1 (SCA1) | Ataxin-1 (ATXN1) | flanking region of expanded CAG repeat | shRNA expression vector | N/A *2 | no | Xia H. et al. (2004) [58] |
| Machado-Joseph disease/spinocerebellar ataxia type 3 (MJD/SCA3) | ATAXIN3/MJD1 | SNPs linked to expanded CAG repeat | synthetic siRNA / shRNA expression vector | yes | no | Miller VM. et al. (2003) [51] Alves S. et al. (2008) [59] Nobrega C. et al. (2013) [60] |
| Spinocerebellar ataxia type 7 (SCA7) | Ataxin-7 (ATXN7) | SNP linked to expanded CAG repeat | shRNA expression vector | no | no | Scholefield J. et al. (2009) [61] |
| Parkinson's disease | Leucine-rich repeat kinase 2 (LRRK2) | R1441G, R1441C | shRNA expression vector | yes | no | de Ynigo-Mojado L. et al. (2011) [62] |
| Leucine-rich repeat kinase 2 (LRRK2) | G20195S | shRNA expression vector | no | no | Sibley CR. et al. (2011) [63] | |
| alpha-synuclein | A30P | shRNA expression vector | no | nucleotide mismatch | Sibley CR. et al. (2011) [63] | |
| Huntington disease | Huntingtin (HTT) | SNPs linked to expanded CAG repeat | synthetic siRNA | yes/no *1 | nucleotide mismatch | Pfister EL. et al. (2009) [64] Takahashi M. et al. (2010) [65] |
3. Assessment of Allele-Specific RNAi
4. siRNAs and shRNAs Conferring Allele-Specific Silencing
5. Enhancement of Allele-Specific Silencing by Improved siRNA Duplexes
4. Summary
Acknowledgments
Conflict of Interest
References
- Sharp, P.A. RNAi and double-strand RNA. Genes Dev. 1999, 13, 139–41. [Google Scholar]
- Bosher, J.M.; Labouesse, M. RNA interference: genetic wand and genetic watchdog. Nat. Cell. Biol. 2000, 2, E31–E36. [Google Scholar] [CrossRef]
- Vaucheret, H.; Beclin, C.; Fagard, M. Post-transcriptional gene silencing in plants. J. Cell. Sci. 2001, 114, 3083–3091. [Google Scholar]
- Cerutti, H. RNA interference: Traveling in the cell and gaining functions? Trends Genet. 2003, 19, 39–46. [Google Scholar]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar]
- Hammond, S.M.; Bernstein, E.; Beach, D.; Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000, 404, 293–296. [Google Scholar]
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 2000, 101, 25–33. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar]
- Nykanen, A.; Haley, B.; Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001, 107, 309–321. [Google Scholar] [CrossRef]
- Martinez, J.; Patkaniowska, A.; Urlaub, H.; Luhrmann, R.; Tuschl, T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 2002, 110, 563–574. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvagner, G.; Haley, B.; Zamore, P.D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathway. Mol. Cell. 2002, 10, 537–548. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004, 15, 185–97. [Google Scholar] [CrossRef]
- Haley, B.; Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct Mol. Biol. 2004, 11, 599–606. [Google Scholar] [CrossRef]
- Rivas, F.V.; Tolia, N.H.; Song, J.J.; Aragon, J.P.; Liu, J.; Hannon, G.J.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349. [Google Scholar]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef]
- Rand, T.A.; Ginalski, K.; Grishin, N.V.; Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. USA 2004, 101, 14385–14389. [Google Scholar] [CrossRef]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef]
- Svoboda, P.; Stein, P.; Hayashi, H.; Schultz, R.M. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 2000, 127, 4147–4156. [Google Scholar]
- Wianny, F.; Zernicka-Goetz, M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell. Biol. 2000, 2, 70–75. [Google Scholar] [CrossRef]
- Svoboda, P.; Stein, P.; Schultz, R.M. RNAi in mouse oocytes and preimplantation embryos: Effectiveness of hairpin dsRNA. Biochem. Biophys. Res. Commun. 2001, 287, 1099–1104. [Google Scholar]
- Player, M.R.; Torrence, P.F. The 2–5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol. Ther. 1998, 78, 55–113. [Google Scholar] [CrossRef]
- Clemens, M.J.; Elia, A. The double-stranded RNA-dependent protein kinase PKR: Structure and function. J. Interferon. Cytokine Res. 1997, 17, 503–524. [Google Scholar] [CrossRef]
- Gale, M., Jr.; Katze, M.G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 1998, 78, 29–46. [Google Scholar] [CrossRef]
- Caplen, N.J.; Fleenor, J.; Fire, A.; Morgan, R.A. dsRNA-mediated gene silencing in cultured Drosophila cells: A tissue culture model for the analysis of RNA interference. Gene 2000, 252, 95–105. [Google Scholar] [CrossRef]
- Yang, S.; Tutton, S.; Pierce, E.; Yoon, K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 2001, 21, 7807–7816. [Google Scholar] [CrossRef]
- Billy, E.; Brondani, V.; Zhang, H.; Muller, U.; Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 14428–14433. [Google Scholar]
- Ui-Tei, K.; Zenno, S.; Miyata, Y.; Saigo, K. Sensitive assay of RNA interference in Drosophila and Chinese hamster cultured cells using firefly luciferase gene as target. FEBS Lett. 2000, 479, 79–82. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar]
- McManus, M.T.; Petersen, C.P.; Haines, B.B.; Chen, J.; Sharp, P.A. Gene silencing using micro-RNA designed hairpins. RNA 2002, 8, 842–850. [Google Scholar] [CrossRef]
- Yu, J.Y.; DeRuiter, S.L.; Turner, D.L. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 2002, 99, 6047–6052. [Google Scholar]
- Paddison, P.J.; Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef]
- Zhou, H.; Xia, X.G.; Xu, Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005, 33, e62. [Google Scholar] [CrossRef]
- Stegmeier, F.; Hu, G.; Rickles, R.J.; Hannon, G.J.; Elledge, S.J. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 2005, 102, 13212–13217. [Google Scholar]
- Hohjoh, H. RNA interference (RNA(i)) induction with various types of synthetic oligonucleotide duplexes in cultured human cells. FEBS Lett. 2002, 521, 195–199. [Google Scholar] [CrossRef]
- Ui-Tei, K.; Naito, Y.; Takahashi, F.; Haraguchi, T.; Ohki-Hamazaki, H.; Juni, A.; Ueda, R.; Saigo, K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004, 32, 936–948. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Rodriguez-Lebron, E.; Paulson, H.L. Allele-specific RNA interference for neurological disease. Gene Ther. 2006, 13, 576–581. [Google Scholar] [CrossRef]
- Miller, V.M.; Gouvion, C.M.; Davidson, B.L.; Paulson, H.L. Targeting Alzheimer’s disease genes with RNA interference: An efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004, 32, 661–668. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Tokunaga, K.; Kaneko, K.; Hohjoh, H. Assessment of allele-specific gene silencing by RNA interference with mutant and wild-type reporter alleles. J. RNAi Gene Silencing 2006, 2, 154–160. [Google Scholar]
- Feng, X.; Zhao, P.; He, Y.; Zuo, Z. Allele-specific silencing of Alzheimer’s disease genes: The amyloid precursor protein genes with Swedish or London mutations. Gene 2006, 371, 68–74. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Tamura, Y.; Yoshida, M.; Tokunaga, K.; Hohjoh, H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS One 2008, 3, e2248. [Google Scholar]
- Sierant, M.; Paduszynska, A.; Kazmierczak-Baranska, J.; Nacmias, B.; Sorbi, S.; Bagnoli, S.; Sochacka, E.; Nawrot, B. Specific silencing of L392V PSEN1 mutant allele by RNA Interference. Int. J. Alzheimers Dis. 2011, 2011, 809218. [Google Scholar]
- Xia, X.; Zhou, H.; Huang, Y.; Xu, Z. Allele-specific RNAi selectively silences mutant SOD1 and achieves significant therapeutic benefit in vivo. Neurobiol. Dis. 2006, 23, 578–586. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Ding, H.; Kennington, L.; Moore, J.T.; Schelter, J.; Burchard, J.; Linsley, P.S.; Aronin, N.; Xu, Z.; Zamore, P.D. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006, 2, e140. [Google Scholar] [CrossRef]
- Abdelgany, A.; Wood, M.; Beeson, D. Allele-specific silencing of a pathogenic mutant acetylcholine receptor subunit by RNA interference. Hum. Mol. Genet. 2003, 12, 2637–2644. [Google Scholar] [CrossRef]
- Miller, V.M.; Xia, H.; Marrs, G.L.; Gouvion, C.M.; Lee, G.; Davidson, B.L.; Paulson, H.L. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. USA 2003, 100, 7195–7200. [Google Scholar]
- Muller, G.A.; Hansen, U.; Xu, Z.; Griswold, B.; Talan, M.I.; McDonnell, N.B.; Briest, W. Allele-specific siRNA knockdown as a personalized treatment strategy for vascular Ehlers-Danlos syndrome in human fibroblasts. FASEB J. 2012, 26, 668–677. [Google Scholar] [CrossRef]
- Dykxhoorn, D.M.; Schlehuber, L.D.; London, I.M.; Lieberman, J. Determinants of specific RNA interference-mediated silencing of human beta-globin alleles differing by a single nucleotide polymorphism. Proc. Natl. Acad. Sci. USA 2006, 103, 5953–5958. [Google Scholar] [CrossRef]
- Kurosawa, T.; Igarashi, S.; Nishizawa, M.; Onodera, O. Selective silencing of a mutant transthyretin allele by small interfering RNAs. Biochem. Biophys. Res. Commun. 2005, 337, 1012–1018. [Google Scholar] [CrossRef]
- Takahashi, M.; Katagiri, T.; Furuya, H.; Hohjoh, H. Disease-causing allele-specific silencing against the ALK2 mutants, R206H and G356D, in fibrodysplasia ossificans progressiva. Gene Ther. 2012, 19, 781–785. [Google Scholar] [CrossRef]
- Kaplan, J.; Kaplan, F.S.; Shore, E.M. Restoration of normal BMP signaling levels and osteogenic differentiation in FOP mesenchymal progenitor cells by mutant allele-specific targeting. Gene Ther. 2012, 19, 786–790. [Google Scholar] [CrossRef]
- Huang, H.; Qiao, R.; Zhao, D.; Zhang, T.; Li, Y.; Yi, F.; Lai, F.; Hong, J.; Ding, X.; Yang, Z.; et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009, 37, 7560–7569. [Google Scholar]
- Xia, H.; Mao, Q.; Eliason, S.L.; Harper, S.Q.; Martins, I.H.; Orr, H.T.; Paulson, H.L.; Yang, L.; Kotin, R.M.; Davidson, B.L. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med. 2004, 10, 816–820. [Google Scholar] [CrossRef]
- Alves, S.; Nascimento-Ferreira, I.; Auregan, G.; Hassig, R.; Dufour, N.; Brouillet, E.; Pedroso de Lima, M.C.; Hantraye, P.; Pereira de Almeida, L.; Deglon, N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One 2008, 3, e3341. [Google Scholar] [CrossRef]
- Nobrega, C.; Nascimento-Ferreira, I.; Onofre, I.; Albuquerque, D.; Hirai, H.; Deglon, N.; de Almeida, L.P. Silencing mutant ataxin-3 rescues motor deficits and neuropathology in machado-joseph disease transgenic mice. PLoS One 2013, 8, e52396. [Google Scholar]
- Scholefield, J.; Greenberg, L.J.; Weinberg, M.S.; Arbuthnot, P.B.; Abdelgany, A.; Wood, M.J. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS One 2009, 4, e7232. [Google Scholar]
- De Ynigo-Mojado, L.; Martin-Ruiz, I.; Sutherland, J.D. Efficient allele-specific targeting of LRRK2 R1441 mutations mediated by RNAi. PLoS One 2011, 6, e21352. [Google Scholar] [CrossRef]
- Sibley, C.R.; Wood, M.J. Identification of allele-specific RNAi effectors targeting genetic forms of Parkinson's disease. PLoS One 2011, 6, e26194. [Google Scholar] [CrossRef]
- Pfister, E.L.; Kennington, L.; Straubhaar, J.; Wagh, S.; Liu, W.; DiFiglia, M.; Landwehrmeyer, B.; Vonsattel, J.P.; Zamore, P.D.; Aronin, N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol. 2009, 19, 774–778. [Google Scholar] [CrossRef]
- Takahashi, M.; Watanabe, S.; Murata, M.; Furuya, H.; Kanazawa, I.; Wada, K.; Hohjoh, H. Tailor-made RNAi knockdown against triplet repeat disease-causing alleles. Proc. Natl. Acad. Sci. USA 2010, 107, 21731–21736. [Google Scholar]
- Hohjoh, H. Allele-specific silencing by RNA interference. Methods Mol. Biol. 2010, 623, 67–79. [Google Scholar] [CrossRef]
- Rodriguez-Lebron, E.; Gouvion, C.M.; Moore, S.A.; Davidson, B.L.; Paulson, H.L. Allele-specific RNAi mitigates phenotypic progression in a transgenic model of Alzheimer's disease. Mol. Ther. 2009, 17, 1563–1573. [Google Scholar] [CrossRef]
- Towne, C.; Raoul, C.; Schneider, B.L.; Aebischer, P. Systemic AAV6 delivery mediating RNA interference against SOD1: neuromuscular transduction does not alter disease progression in fALS mice. Mol. Ther. 2008, 16, 1018–1025. [Google Scholar] [CrossRef]
- Hamasaki, M.; Hashizume, Y.; Yamada, Y.; Katayama, T.; Hohjoh, H.; Fusaki, N.; Nakashima, Y.; Furuya, H.; Haga, N.; Takami, Y.; Era, T. Pathogenic mutation of ALK2 inhibits induced pluripotent stem cell reprogramming and maintenance: mechanisms of reprogramming and strategy for drug identification. Stem Cells 2012, 30, 2437–2449. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Tokunaga, K.; Hohjoh, H. Influence of assembly of siRNA elements into RNA-induced silencing complex by fork-siRNA duplex carrying nucleotide mismatches at the 3'- or 5'-end of the sense-stranded siRNA element. Biochem. Biophys. Res. Commun. 2005, 329, 516–521. [Google Scholar] [CrossRef]
- Hohjoh, H. Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett. 2004, 557, 193–198. [Google Scholar] [CrossRef]
- Chiu, Y.L.; Rana, T.M. siRNA function in RNAi: A chemical modification analysis. RNA 2003, 9, 1034–1048. [Google Scholar] [CrossRef]
- Czauderna, F.; Fechtner, M.; Dames, S.; Aygun, H.; Klippel, A.; Pronk, G.J.; Giese, K.; Kaufmann, J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003, 31, 2705–2716. [Google Scholar]
- Choung, S.; Kim, Y.J.; Kim, S.; Park, H.O.; Choi, Y.C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res. Commun. 2006, 342, 919–927. [Google Scholar] [CrossRef]
- Elmen, J.; Thonberg, H.; Ljungberg, K.; Frieden, M.; Westergaard, M.; Xu, Y.; Wahren, B.; Liang, Z.; Orum, H.; Koch, T.; Wahlestedt, C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005, 33, 439–447. [Google Scholar] [CrossRef]
- Dowler, T.; Bergeron, D.; Tedeschi, A.L.; Paquet, L.; Ferrari, N.; Damha, M.J. Improvements in siRNA properties mediated by 2'-deoxy-2'-fluoro-beta-D-arabinonucleic acid (FANA). Nucleic Acids Res. 2006, 34, 1669–1675. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hohjoh, H. Disease-Causing Allele-Specific Silencing by RNA Interference. Pharmaceuticals 2013, 6, 522-535. https://doi.org/10.3390/ph6040522
Hohjoh H. Disease-Causing Allele-Specific Silencing by RNA Interference. Pharmaceuticals. 2013; 6(4):522-535. https://doi.org/10.3390/ph6040522
Chicago/Turabian StyleHohjoh, Hirohiko. 2013. "Disease-Causing Allele-Specific Silencing by RNA Interference" Pharmaceuticals 6, no. 4: 522-535. https://doi.org/10.3390/ph6040522
APA StyleHohjoh, H. (2013). Disease-Causing Allele-Specific Silencing by RNA Interference. Pharmaceuticals, 6(4), 522-535. https://doi.org/10.3390/ph6040522




