RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation
Abstract
:1. Overview
2. Cap-Dependent Translation Initiation
3. IRES-Dependent Translation Initiation
4. IRES Classification
5. Advantages of Having an IRES
6. IRES-Transacting Factors
7. Most ITAFs Are Nucleus–Cytoplasmic Shuttling RNA-Binding Proteins
8. Impact of Post-Translational Modification of ITAFs on IRES Activity
9. ITAFs and IRES-Dependent Viral Tropism
10. What about Cellular IRESs?
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abrahao, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef] [Green Version]
- Stern-Ginossar, N.; Thompson, S.R.; Mathews, M.B.; Mohr, I. Translational Control in Virus-Infected Cells. Cold Spring Harb. Perspect. Biol. 2019, 11, a033001. [Google Scholar] [CrossRef] [Green Version]
- Jan, E.; Mohr, I.; Walsh, D. A Cap-to-Tail Guide to mRNA Translation Strategies in Virus-Infected Cells. Annu. Rev. Virol. 2016, 3, 283–307. [Google Scholar] [CrossRef]
- Walsh, D.; Mohr, I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011, 9, 860–875. [Google Scholar] [CrossRef]
- Schneider, R.J.; Mohr, I. Translation initiation and viral tricks. Trends Biochem. Sci. 2003, 28, 130–136. [Google Scholar] [CrossRef]
- Bushell, M.; Sarnow, P. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 2002, 158, 395–399. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, R.E. Translational control by viral proteinases. Virus Res. 2006, 119, 76–88. [Google Scholar] [CrossRef]
- Walsh, D.; Mathews, M.B.; Mohr, I. Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013, 5, a012351. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. The Organizing Principles of Eukaryotic Ribosome Recruitment. Annu. Rev. Biochem. 2019, 88, 307–335. [Google Scholar] [CrossRef]
- Weisser, M.; Ban, N. Extensions, Extra Factors, and Extreme Complexity: Ribosomal Structures Provide Insights into Eukaryotic Translation. Cold Spring Harb. Perspect. Biol. 2019, 11, a032367. [Google Scholar] [CrossRef]
- Merrick, W.C.; Pavitt, G.D. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb. Perspect. Biol. 2018, 10, a033092. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem. Sci. 2017, 42, 589–611. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Haimov, O.; Sinvani, H.; Dikstein, R. Cap-dependent, scanning-free translation initiation mechanisms. Biochim. Biophys. Acta 2015, 1849, 1313–1318. [Google Scholar] [CrossRef]
- Ryu, I.; Kim, Y.K. Translation initiation mediated by nuclear cap-binding protein complex. BMB Rep. 2017, 50, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.S.; Kranzusch, P.J.; Doudna, J.A.; Cate, J.H. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 2016, 536, 96–99. [Google Scholar] [CrossRef] [Green Version]
- De la Parra, C.; Ernlund, A.; Alard, A.; Ruggles, K.; Ueberheide, B.; Schneider, R.J. A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 2018, 9, 3068. [Google Scholar] [CrossRef] [Green Version]
- Soto-Rifo, R.; Rubilar, P.S.; Ohlmann, T. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res. 2013, 41, 6286–6299. [Google Scholar] [CrossRef]
- Elfakess, R.; Dikstein, R. A translation initiation element specific to mRNAs with very short 5'UTR that also regulates transcription. PLoS ONE 2008, 3, e3094. [Google Scholar] [CrossRef] [Green Version]
- Galloway, A.; Cowling, V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 270–279. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar] [CrossRef] [Green Version]
- Fakim, H.; Fabian, M.R. Communication Is Key: 5'-3' Interactions that Regulate mRNA Translation and Turnover. Adv. Exp. Med. Biol. 2019, 1203, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, A.L.; Pasquinelli, A.E. Tales of Detailed Poly(A) Tails. Trends Cell Biol. 2019, 29, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Alekhina, O.M.; Terenin, I.M.; Dmitriev, S.E.; Vassilenko, K.S. Functional Cyclization of Eukaryotic mRNAs. Int. J. Mol. Sci. 2020, 21, 1677. [Google Scholar] [CrossRef] [Green Version]
- Lomakin, I.B.; Steitz, T.A. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 2013, 500, 307–311. [Google Scholar] [CrossRef] [Green Version]
- Passmore, L.A.; Schmeing, T.M.; Maag, D.; Applefield, D.J.; Acker, M.G.; Algire, M.A.; Lorsch, J.R.; Ramakrishnan, V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 2007, 26, 41–50. [Google Scholar] [CrossRef]
- Parsyan, A.; Svitkin, Y.; Shahbazian, D.; Gkogkas, C.; Lasko, P.; Merrick, W.C.; Sonenberg, N. mRNA helicases: The tacticians of translational control. Nat. Rev. Mol. Cell Biol. 2011, 12, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Borukhov, S.I.; Hellen, C.U. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 1998, 394, 854–859. [Google Scholar] [CrossRef]
- Pestova, T.V.; Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002, 16, 2906–2922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valasek, L.; Nielsen, K.H.; Zhang, F.; Fekete, C.A.; Hinnebusch, A.G. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004, 24, 9437–9455. [Google Scholar] [CrossRef] [Green Version]
- Algire, M.A.; Maag, D.; Lorsch, J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell 2005, 20, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Llacer, J.L.; Hussain, T.; Marler, L.; Aitken, C.E.; Thakur, A.; Lorsch, J.R.; Hinnebusch, A.G.; Ramakrishnan, V. Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol. Cell 2015, 59, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Llacer, J.L.; Hussain, T.; Saini, A.K.; Nanda, J.S.; Kaur, S.; Gordiyenko, Y.; Kumar, R.; Hinnebusch, A.G.; Lorsch, J.R.; Ramakrishnan, V. Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start-codon recognition. Elife 2018, 7, e39273. [Google Scholar] [CrossRef]
- Pestova, T.V.; Lomakin, I.B.; Lee, J.H.; Choi, S.K.; Dever, T.E.; Hellen, C.U. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 2000, 403, 332–335. [Google Scholar] [CrossRef]
- Acker, M.G.; Shin, B.S.; Dever, T.E.; Lorsch, J.R. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J. Biol. Chem. 2006, 281, 8469–8475. [Google Scholar] [CrossRef] [Green Version]
- Fringer, J.M.; Acker, M.G.; Fekete, C.A.; Lorsch, J.R.; Dever, T.E. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell Biol. 2007, 27, 2384–2397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogorad, A.M.; Lin, K.Y.; Marintchev, A. eIF2B Mechanisms of Action and Regulation: A Thermodynamic View. Biochemistry 2018, 57, 1426–1435. [Google Scholar] [CrossRef]
- Dorner, A.J.; Semler, B.L.; Jackson, R.J.; Hanecak, R.; Duprey, E.; Wimmer, E. In Vitro translation of poliovirus RNA: Utilization of internal initiation sites in reticulocyte lysate. J. Virol. 1984, 50, 507–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.K.; Krausslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during In Vitro translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelletier, J.; Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hunt, S.L.; Reynolds, J.E.; Kaminski, A. Cap-dependent and cap-independent translation: Operational distinctions and mechanistic interpretations. Curr. Top. Microbiol. Immunol. 1995, 203, 1–29. [Google Scholar] [PubMed]
- Chen, C.Y.; Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995, 268, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Thompson, S.R. So you want to know if your message has an IRES? Wiley Interdiscip. Rev. RNA 2012, 3, 697–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, R.J.; Howell, M.T.; Kaminski, A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 1990, 15, 477–483. [Google Scholar] [CrossRef]
- Kuhn, R.; Luz, N.; Beck, E. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1990, 64, 4625–4631. [Google Scholar] [CrossRef] [Green Version]
- Glass, M.J.; Jia, X.Y.; Summers, D.F. Identification of the hepatitis A virus internal ribosome entry site: In vivo and In Vitro analysis of bicistronic RNAs containing the HAV 5' noncoding region. Virology 1993, 193, 842–852. [Google Scholar] [CrossRef]
- Hruby, D.E.; Roberts, W.K. Encephalomyocarditis virus RNA. II. Polyadenylic acid requirement for efficient translation. J. Virol. 1977, 23, 338–344. [Google Scholar] [CrossRef] [Green Version]
- Michel, Y.M.; Poncet, D.; Piron, M.; Kean, K.M.; Borman, A.M. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000, 275, 32268–32276. [Google Scholar] [CrossRef] [Green Version]
- Svitkin, Y.V.; Imataka, H.; Khaleghpour, K.; Kahvejian, A.; Liebig, H.D.; Sonenberg, N. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 2001, 7, 1743–1752. [Google Scholar]
- Paulous, S.; Malnou, C.E.; Michel, Y.M.; Kean, K.M.; Borman, A.M. Comparison of the capacity of different viral internal ribosome entry segments to direct translation initiation in poly(A)-dependent reticulocyte lysates. Nucleic Acids Res. 2003, 31, 722–733. [Google Scholar] [CrossRef] [Green Version]
- Bergamini, G.; Preiss, T.; Hentze, M.W. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 2000, 6, 1781–1790. [Google Scholar] [CrossRef] [Green Version]
- Svitkin, Y.V.; Costa-Mattioli, M.; Herdy, B.; Perreault, S.; Sonenberg, N. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP). RNA 2007, 13, 2330–2340. [Google Scholar] [CrossRef] [Green Version]
- Baird, S.D.; Turcotte, M.; Korneluk, R.G.; Holcik, M. Searching for IRES. Rna 2006, 12, 1755–1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Lastra, M.; Rivas, A.; Barria, M.I. Protein synthesis in eukaryotes: The growing biological relevance of cap-independent translation initiation. Biol. Res. 2005, 38, 121–146. [Google Scholar] [CrossRef] [Green Version]
- Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; Gritsenko, A.A.; Stern-Ginossar, N.; Yakhini, Z.; Weinberger, A.; Segal, E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016, 351. [Google Scholar] [CrossRef]
- Hong, J.J.; Wu, T.Y.; Chang, T.Y.; Chen, C.Y. Viral IRES prediction system—A web server for prediction of the IRES secondary structure in silico. PLoS ONE 2013, 8, e79288. [Google Scholar] [CrossRef]
- Peguero-Sanchez, E.; Pardo-Lopez, L.; Merino, E. IRES-dependent translated genes in fungi: Computational prediction, phylogenetic conservation and functional association. BMC Genom. 2015, 16, 1059. [Google Scholar] [CrossRef] [PubMed]
- Kolekar, P.; Pataskar, A.; Kulkarni-Kale, U.; Pal, J.; Kulkarni, A. IRESPred: Web Server for Prediction of Cellular and Viral Internal Ribosome Entry Site (IRES). Sci. Rep. 2016, 6, 27436. [Google Scholar] [CrossRef] [PubMed]
- Gritsenko, A.A.; Weingarten-Gabbay, S.; Elias-Kirma, S.; Nir, R.; de Ridder, D.; Segal, E. Sequence features of viral and human Internal Ribosome Entry Sites predictive of their activity. PLoS Comput. Biol. 2017, 13, e1005734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Gribskov, M. IRESpy: An XGBoost model for prediction of internal ribosome entry sites. BMC Bioinform. 2019, 20, 409. [Google Scholar] [CrossRef]
- Gupta, A.; Bansal, M. RNA-mediated translation regulation in viral genomes: Computational advances in the recognition of sequences and structures. Brief. Bioinform. 2020, 21, 1151–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.Y.; Hsieh, C.C.; Hong, J.J.; Chen, C.Y.; Tsai, Y.S. IRSS: A web-based tool for automatic layout and analysis of IRES secondary structure prediction and searching system in silico. BMC Bioinform. 2009, 10, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Mugavero, J.; Stauft, C.B.; Wimmer, E. Dengue and Zika Virus 5′ Untranslated Regions Harbor Internal Ribosomal Entry Site Functions. mBio 2019, 10, e00459-19. [Google Scholar] [CrossRef] [Green Version]
- Balvay, L.; Lopez Lastra, M.; Sargueil, B.; Darlix, J.L.; Ohlmann, T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007, 5, 128–140. [Google Scholar] [CrossRef]
- Barrera, A.; Olguin, V.; Vera-Otarola, J.; Lopez-Lastra, M. Cap-independent translation initiation of the unspliced RNA of retroviruses. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194583. [Google Scholar] [CrossRef] [PubMed]
- Tahiri-Alaoui, A.; Smith, L.P.; Baigent, S.; Kgosana, L.; Petherbridge, L.J.; Lambeth, L.S.; James, W.; Nair, V. Identification of an intercistronic internal ribosome entry site in a Marek's disease virus immediate-early gene. J. Virol. 2009, 83, 5846–5853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieleski, L.; Talbot, S.J. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J. Virol. 2001, 75, 1864–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isaksson, A.; Berggren, M.; Ricksten, A. Epstein-Barr virus U leader exon contains an internal ribosome entry site. Oncogene 2003, 22, 572–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiba, S.; Jamal, A.; Suzuki, N. First Evidence for Internal Ribosomal Entry Sites in Diverse Fungal Virus Genomes. mBio 2018, 9, e02350-17. [Google Scholar] [CrossRef] [Green Version]
- Jan, E. Divergent IRES elements in invertebrates. Virus Res. 2006, 119, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, N.; Uchiumi, T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009, 139, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Luria, N.; Smith, E.; Lachman, O.; Laskar, O.; Sela, N.; Dombrovsky, A. Isolation and characterization of a novel cripavirus, the first Dicistroviridae family member infecting the cotton mealybug Phenacoccus solenopsis. Arch. Virol. 2020, 165, 1987–1994. [Google Scholar] [CrossRef]
- Miras, M.; Miller, W.A.; Truniger, V.; Aranda, M.A. Non-canonical Translation in Plant RNA Viruses. Front. Plant Sci. 2017, 8, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.M.; Koh, D.C.; Liu, D. Identification of plant virus IRES. Methods Mol. Biol. 2008, 451, 125–133. [Google Scholar]
- Faye, M.D.; Holcik, M. The role of IRES trans-acting factors in carcinogenesis. Biochim. Biophys. Acta 2015, 1849, 887–897. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; Barna, M. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Spriggs, K.A.; Bushell, M.; Willis, A.E. Translational regulation of gene expression during conditions of cell stress. Mol. Cell 2010, 40, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Godet, A.C.; David, F.; Hantelys, F.; Tatin, F.; Lacazette, E.; Garmy-Susini, B.; Prats, A.C. IRES Trans-Acting Factors, Key Actors of the Stress Response. Int. J. Mol. Sci. 2019, 20, 924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwan, T.; Thompson, S.R. Noncanonical Translation Initiation in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2019, 11, a032672. [Google Scholar] [CrossRef] [PubMed]
- Horvilleur, E.; Wilson, L.A.; Bastide, A.; Pineiro, D.; Poyry, T.A.; Willis, A.E. Cap-Independent Translation in Hematological Malignancies. Front. Oncol. 2015, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Le Quesne, J.P.; Spriggs, K.A.; Bushell, M.; Willis, A.E. Dysregulation of protein synthesis and disease. J. Pathol. 2010, 220, 140–151. [Google Scholar] [CrossRef]
- Keiper, B.D. Cap-Independent mRNA Translation in Germ Cells. Int. J. Mol. Sci. 2019, 20, 173. [Google Scholar] [CrossRef] [Green Version]
- Macejak, D.G.; Sarnow, P. Translational regulation of the immunoglobulin heavy-chain binding protein mRNA. Enzyme 1990, 44, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Stoneley, M.; Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene 2004, 23, 3200–3207. [Google Scholar] [CrossRef] [Green Version]
- Bonnal, S.; Boutonnet, C.; Prado-Lourenco, L.; Vagner, S. IRESdb: The Internal Ribosome Entry Site database. Nucleic Acids Res. 2003, 31, 427–428. [Google Scholar] [CrossRef]
- Mokrejs, M.; Masek, T.; Vopalensky, V.; Hlubucek, P.; Delbos, P.; Pospisek, M. IRESite--a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res. 2010, 38, D131–D136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mokrejs, M.; Vopalensky, V.; Kolenaty, O.; Masek, T.; Feketova, Z.; Sekyrova, P.; Skaloudova, B.; Kriz, V.; Pospisek, M. IRESite: The database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res. 2006, 34, D125–D130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Li, Y.; Wang, C.; Zhang, H.; Zhang, H.; Jiang, B.; Guo, X.; Song, X. IRESbase: A Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites. Genom. Proteom. Bioinform. 2020, 18, 129–139. [Google Scholar] [CrossRef]
- Yang, T.H.; Wang, C.Y.; Tsai, H.C.; Liu, C.T. Human IRES Atlas: An integrative platform for studying IRES-driven translational regulation in humans. Database 2021, 2021, baab025. [Google Scholar] [CrossRef]
- Balvay, L.; Soto Rifo, R.; Ricci, E.P.; Decimo, D.; Ohlmann, T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta 2009, 1789, 542–557. [Google Scholar] [CrossRef]
- Deforges, J.; Locker, N.; Sargueil, B. mRNAs that specifically interact with eukaryotic ribosomal subunits. Biochimie 2015, 114, 48–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filbin, M.E.; Kieft, J.S. Toward a structural understanding of IRES RNA function. Curr. Opin. Struct. Biol. 2009, 19, 267–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mailliot, J.; Martin, F. Viral internal ribosomal entry sites: Four classes for one goal. Wiley Interdiscip. Rev. RNA 2018, 9, e1458. [Google Scholar] [CrossRef] [PubMed]
- Lukavsky, P.J. Structure and function of HCV IRES domains. Virus Res. 2009, 139, 166–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leslie, A.P. Translation initiation on picornavirus RNA. In Translational Control of Gene Expression; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2000; Volume 39. [Google Scholar]
- Kieft, J.S. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008, 33, 274–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niepmann, M. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta 2009, 1789, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Salas, E.; Fernandez-Miragall, O. Picornavirus IRES: Structure function relationship. Curr. Pharm. Des. 2004, 10, 3757–3767. [Google Scholar] [CrossRef]
- Niepmann, M.; Gerresheim, G.K. Hepatitis C Virus Translation Regulation. Int. J. Mol. Sci. 2020, 21, 2328. [Google Scholar] [CrossRef] [Green Version]
- Woo, P.C.; Lau, S.K.; Choi, G.K.; Huang, Y.; Teng, J.L.; Tsoi, H.W.; Tse, H.; Yeung, M.L.; Chan, K.H.; Jin, D.Y.; et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 2012, 86, 2797–2808. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.E.; Powell, M.J.; Hoover, S.E.; Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell Biol. 2000, 20, 4990–4999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolaway, K.E.; Lazaridis, K.; Belsham, G.J.; Carter, M.J.; Roberts, L.O. The 5' untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. J. Virol. 2001, 75, 10244–10249. [Google Scholar] [CrossRef] [Green Version]
- Shibuya, N.; Nakashima, N. Characterization of the 5′ internal ribosome entry site of Plautia stali intestine virus. J. Gen. Virol. 2006, 87, 3679–3686. [Google Scholar] [CrossRef] [PubMed]
- Gross, L.; Vicens, Q.; Einhorn, E.; Noireterre, A.; Schaeffer, L.; Kuhn, L.; Imler, J.L.; Eriani, G.; Meignin, C.; Martin, F. The IRES5'UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure. Nucleic Acids Res. 2017, 45, 8993–9004. [Google Scholar] [CrossRef]
- Roberts, L.O.; Groppelli, E. An atypical IRES within the 5' UTR of a dicistrovirus genome. Virus Res. 2009, 139, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Khong, A.; Bonderoff, J.M.; Spriggs, R.V.; Tammpere, E.; Kerr, C.H.; Jackson, T.J.; Willis, A.E.; Jan, E. Temporal Regulation of Distinct Internal Ribosome Entry Sites of the Dicistroviridae Cricket Paralysis Virus. Viruses 2016, 8, 25. [Google Scholar] [CrossRef] [Green Version]
- Majzoub, K.; Hafirassou, M.L.; Meignin, C.; Goto, A.; Marzi, S.; Fedorova, A.; Verdier, Y.; Vinh, J.; Hoffmann, J.A.; Martin, F.; et al. RACK1 controls IRES-mediated translation of viruses. Cell 2014, 159, 1086–1095. [Google Scholar] [CrossRef] [Green Version]
- Masoumi, A.; Hanzlik, T.N.; Christian, P.D. Functionality of the 5′- and intergenic IRES elements of cricket paralysis virus in a range of insect cell lines, and its relationship with viral activities. Virus Res. 2003, 94, 113–120. [Google Scholar] [CrossRef]
- Jan, E.; Sarnow, P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002, 324, 889–902. [Google Scholar] [CrossRef]
- Pestova, T.V.; Lomakin, I.B.; Hellen, C.U. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004, 5, 906–913. [Google Scholar] [CrossRef]
- Pfingsten, J.S.; Kieft, J.S. RNA structure-based ribosome recruitment: Lessons from the Dicistroviridae intergenic region IRESes. RNA 2008, 14, 1255–1263. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.E.; Pestova, T.V.; Hellen, C.U.; Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell 2000, 102, 511–520. [Google Scholar] [CrossRef] [Green Version]
- Jan, E.; Thompson, S.R.; Wilson, J.E.; Pestova, T.V.; Hellen, C.U.; Sarnow, P. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harb. Symp. Quant. Biol. 2001, 66, 285–292. [Google Scholar] [CrossRef]
- Abaeva, I.S.; Vicens, Q.; Bochler, A.; Soufari, H.; Simonetti, A.; Pestova, T.V.; Hashem, Y.; Hellen, C.U.T. The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism. Cell Rep. 2020, 33, 108476. [Google Scholar] [CrossRef]
- Boros, A.; Pankovics, P.; Simmonds, P.; Reuter, G. Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS ONE 2011, 6, e29145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Othman, Z.; Sulaiman, M.K.; Willcocks, M.M.; Ulryck, N.; Blackbourn, D.J.; Sargueil, B.; Roberts, L.O.; Locker, N. Functional analysis of Kaposi's sarcoma-associated herpesvirus vFLIP expression reveals a new mode of IRES-mediated translation. RNA 2014, 20, 1803–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundhoff, A.; Ganem, D. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2001, 75, 1857–1863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Low, W.; Harries, M.; Ye, H.; Du, M.Q.; Boshoff, C.; Collins, M. Internal ribosome entry site regulates translation of Kaposi's sarcoma-associated herpesvirus FLICE inhibitory protein. J. Virol. 2001, 75, 2938–2945. [Google Scholar] [CrossRef] [Green Version]
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 2011, 10, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Alwine, J.C. 19S late mRNAs of simian virus 40 have an internal ribosome entry site upstream of the virion structural protein 3 coding sequence. J. Virol. 2006, 80, 6553–6558. [Google Scholar] [CrossRef] [Green Version]
- Brasey, A.; Lopez-Lastra, M.; Ohlmann, T.; Beerens, N.; Berkhout, B.; Darlix, J.L.; Sonenberg, N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003, 77, 3939–3949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plank, T.D.; Whitehurst, J.T.; Kieft, J.S. Cell type specificity and structural determinants of IRES activity from the 5' leaders of different HIV-1 transcripts. Nucleic Acids Res. 2013, 41, 6698–6714. [Google Scholar] [CrossRef] [Green Version]
- Ohlmann, T.; Lopez-Lastra, M.; Darlix, J.L. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 2000, 275, 11899–11906. [Google Scholar] [CrossRef] [Green Version]
- Camerini, V.; Decimo, D.; Balvay, L.; Pistello, M.; Bendinelli, M.; Darlix, J.L.; Ohlmann, T. A dormant internal ribosome entry site controls translation of feline immunodeficiency virus. J. Virol. 2008, 82, 3574–3583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, C.B.; Shen, X.; Egan, M.A.; Pierson, T.C.; Walker, C.M.; Siliciano, R.F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001, 75, 181–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deforges, J.; de Breyne, S.; Ameur, M.; Ulryck, N.; Chamond, N.; Saaidi, A.; Ponty, Y.; Ohlmann, T.; Sargueil, B. Two ribosome recruitment sites direct multiple translation events within HIV1 Gag open reading frame. Nucleic Acids Res. 2017, 45, 7382–7400. [Google Scholar] [CrossRef]
- Nicholson, M.G.; Rue, S.M.; Clements, J.E.; Barber, S.A. An internal ribosome entry site promotes translation of a novel SIV Pr55(Gag) isoform. Virology 2006, 349, 325–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locker, N.; Chamond, N.; Sargueil, B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011, 39, 2367–2377. [Google Scholar] [CrossRef] [PubMed]
- Herbreteau, C.H.; Weill, L.; Decimo, D.; Prevot, D.; Darlix, J.L.; Sargueil, B.; Ohlmann, T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 2005, 12, 1001–1007. [Google Scholar] [CrossRef]
- Weill, L.; James, L.; Ulryck, N.; Chamond, N.; Herbreteau, C.H.; Ohlmann, T.; Sargueil, B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010, 38, 1367–1381. [Google Scholar] [CrossRef] [Green Version]
- De Breyne, S.; Soto-Rifo, R.; Lopez-Lastra, M.; Ohlmann, T. Translation initiation is driven by different mechanisms on the HIV-1 and HIV-2 genomic RNAs. Virus Res. 2013, 171, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Abaeva, I.S.; Pestova, T.V.; Hellen, C.U. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi arva virus IRES. Nucleic Acids Res. 2016, 44, 2362–2377. [Google Scholar] [CrossRef]
- Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996, 16, 6859–6869. [Google Scholar] [CrossRef] [Green Version]
- Kolupaeva, V.G.; de Breyne, S.; Pestova, T.V.; Hellen, C.U. In Vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007, 430, 409–439. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.R.; Abaeva, I.S.; Pestova, T.V.; Hellen, C.U. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014, 33, 76–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pestova, T.V.; Shatsky, I.N.; Hellen, C.U. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996, 16, 6870–6878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whetter, L.E.; Day, S.P.; Elroy-Stein, O.; Brown, E.A.; Lemon, S.M. Low efficiency of the 5' nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J. Virol. 1994, 68, 5253–5263. [Google Scholar] [CrossRef] [Green Version]
- Borman, A.M.; Kean, K.M. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 1997, 237, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.A.; Zajac, A.J.; Lemon, S.M. In Vitro characterization of an internal ribosomal entry site (IRES) present within the 5' nontranslated region of hepatitis A virus RNA: Comparison with the IRES of encephalomyocarditis virus. J. Virol. 1994, 68, 1066–1074. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, A.M.; Jan, E.; Sarnow, P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 2006, 12, 894–902. [Google Scholar] [CrossRef] [Green Version]
- Fraser, C.S.; Doudna, J.A. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007, 5, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998, 12, 67–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, T.; Machida, K.; Iwasaki, W.; Shigeta, T.; Nishimoto, M.; Takahashi, M.; Sakamoto, A.; Yonemochi, M.; Harada, Y.; Shigematsu, H.; et al. HCV IRES Captures an Actively Translating 80S Ribosome. Mol. Cell 2019, 74, 1205–1214.e8. [Google Scholar] [CrossRef]
- Fitzgerald, K.D.; Semler, B.L. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta 2009, 1789, 518–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Miragall, O.; Lopez de Quinto, S.; Martinez-Salas, E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 2009, 139, 172–182. [Google Scholar] [CrossRef]
- Romero-Lopez, C.; Berzal-Herranz, A. The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle. Int. J. Mol. Sci. 2020, 21, 1479. [Google Scholar] [CrossRef] [Green Version]
- Honda, M.; Beard, M.R.; Ping, L.H.; Lemon, S.M. A phylogenetically conserved stem-loop structure at the 5' border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999, 73, 1165–1174. [Google Scholar] [CrossRef] [Green Version]
- Rijnbrand, R.; Yang, Y.; Beales, L.; Bodola, F.; Goettge, K.; Cohen, L.; Lanford, R.E.; Lemon, S.M.; Martin, A. A chimeric GB virus B with 5' nontranslated RNA sequence from hepatitis C virus causes hepatitis in tamarins. Hepatology 2005, 41, 986–994. [Google Scholar] [CrossRef]
- Fernandez, N.; Fernandez-Miragall, O.; Ramajo, J.; Garcia-Sacristan, A.; Bellora, N.; Eyras, E.; Briones, C.; Martinez-Salas, E. Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation. Nucleic Acids Res. 2011, 39, 8572–8585. [Google Scholar] [CrossRef] [PubMed]
- Bassili, G.; Tzima, E.; Song, Y.; Saleh, L.; Ochs, K.; Niepmann, M. Sequence and secondary structure requirements in a highly conserved element for foot-and-mouth disease virus internal ribosome entry site activity and eIF4G binding. J. Gen. Virol. 2004, 85, 2555–2565. [Google Scholar] [CrossRef]
- Fernandez-Miragall, O.; Martinez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 2003, 9, 1333–1344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, M.E.; Seamons, R.A.; Belsham, G.J. A selection system for functional internal ribosome entry site (IRES) elements: Analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 1999, 5, 1167–1179. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Salas, E.; Saiz, J.C.; Davila, M.; Belsham, G.J.; Domingo, E. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol. 1993, 67, 3748–3755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svitkin, Y.V.; Maslova, S.V.; Agol, V.I. The genomes of attenuated and virulent poliovirus strains differ in their In Vitro translation efficiencies. Virology 1985, 147, 243–252. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Pestova, T.V.; Maslova, S.V.; Agol, V.I. Point mutations modify the response of poliovirus RNA to a translation initiation factor: A comparison of neurovirulent and attenuated strains. Virology 1988, 166, 394–404. [Google Scholar] [CrossRef]
- Muzychenko, A.R.; Lipskaya, G.; Maslova, S.V.; Svitkin, Y.V.; Pilipenko, E.V.; Nottay, B.K.; Kew, O.M.; Agol, V.I. Coupled mutations in the 5'-untranslated region of the Sabin poliovirus strains during in vivo passages: Structural and functional implications. Virus Res. 1991, 21, 111–122. [Google Scholar] [CrossRef]
- Hertz, M.I.; Thompson, S.R. In vivo functional analysis of the Dicistroviridae intergenic region internal ribosome entry sites. Nucleic Acids Res. 2011, 39, 7276–7288. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, F.; Vallejos, M.; Walters, B.; Contreras, N.; Hertz, M.I.; Olivares, E.; Caceres, C.J.; Pino, K.; Letelier, A.; Thompson, S.R.; et al. Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J. 2016, 283, 2508–2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallejos, M.; Carvajal, F.; Pino, K.; Navarrete, C.; Ferres, M.; Huidobro-Toro, J.P.; Sargueil, B.; Lopez-Lastra, M. Functional and structural analysis of the internal ribosome entry site present in the mRNA of natural variants of the HIV-1. PLoS ONE 2012, 7, e35031. [Google Scholar] [CrossRef]
- Vallejos, M.; Deforges, J.; Plank, T.D.; Letelier, A.; Ramdohr, P.; Abraham, C.G.; Valiente-Echeverria, F.; Kieft, J.S.; Sargueil, B.; Lopez-Lastra, M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011, 39, 6186–6200. [Google Scholar] [CrossRef]
- Plank, T.D.; Whitehurst, J.T.; Cencic, R.; Pelletier, J.; Kieft, J.S. Internal translation initiation from HIV-1 transcripts is conferred by a common RNA structure. Translation 2014, 2, e27694. [Google Scholar] [CrossRef] [PubMed]
- Dorokhov, Y.L.; Skulachev, M.V.; Ivanov, P.A.; Zvereva, S.D.; Tjulkina, L.G.; Merits, A.; Gleba, Y.Y.; Hohn, T.; Atabekov, J.G. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA 2002, 99, 5301–5306. [Google Scholar] [CrossRef] [Green Version]
- Terenin, I.M.; Dmitriev, S.E.; Andreev, D.E.; Royall, E.; Belsham, G.J.; Roberts, L.O.; Shatsky, I.N. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol. Cell. Biol. 2005, 25, 7879–7888. [Google Scholar] [CrossRef] [Green Version]
- Embarc-Buh, A.; Francisco-Velilla, R.; Martinez-Salas, E. RNA-Binding Proteins at the Host-Pathogen Interface Targeting Viral Regulatory Elements. Viruses 2021, 13, 952. [Google Scholar] [CrossRef]
- Martinez-Salas, E.; Lozano, G.; Fernandez-Chamorro, J.; Francisco-Velilla, R.; Galan, A.; Diaz, R. RNA-binding proteins impacting on internal initiation of translation. Int. J. Mol. Sci. 2013, 14, 21705–21726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolupaeva, V.G.; Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 1998, 273, 18599–18604. [Google Scholar] [CrossRef] [Green Version]
- Lomakin, I.B.; Hellen, C.U.; Pestova, T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000, 20, 6019–6029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez de Quinto, S.; Martinez-Salas, E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 2000, 6, 1380–1392. [Google Scholar] [CrossRef] [Green Version]
- Chamond, N.; Deforges, J.; Ulryck, N.; Sargueil, B. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res. 2014, 42, 10373–10384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahan, N.; Wimmer, E.; Mueller, S. Polypyrimidine tract binding protein-1 (PTB1) is a determinant of the tissue and host tropism of a human rhinovirus/poliovirus chimera PV1(RIPO). PLoS ONE 2013, 8, e60791. [Google Scholar] [CrossRef] [Green Version]
- Asnani, M.; Kumar, P.; Hellen, C.U. Widespread distribution and structural diversity of Type IV IRESs in members of Picornaviridae. Virology 2015, 478, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Lozano, G.; Martinez-Salas, E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015, 12, 113–120. [Google Scholar] [CrossRef] [Green Version]
- Pisarev, A.V.; Chard, L.S.; Kaku, Y.; Johns, H.L.; Shatsky, I.N.; Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004, 78, 4487–4497. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, T.R.; Dhote, V.; Yu, Y.; Hellen, C.U. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J. Virol. 2012, 86, 1468–1486. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Alvarez, E.; Carrasco, L. The multifaceted poliovirus 2A protease: Regulation of gene expression by picornavirus proteases. J. Biomed. Biotechnol. 2011, 2011, 369648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiz, M.; Martinez-Salas, E. Uncovering targets of the Leader protease: Linking RNA-mediated pathways and antiviral defense. Wiley Interdiscip. Rev. RNA 2021, 12, e1645. [Google Scholar] [CrossRef] [PubMed]
- Prevot, D.; Darlix, J.L.; Ohlmann, T. Conducting the initiation of protein synthesis: The role of eIF4G. Biol. Cell 2003, 95, 141–156. [Google Scholar] [CrossRef]
- Ziegler, E.; Borman, A.M.; Deliat, F.G.; Liebig, H.D.; Jugovic, D.; Kean, K.M.; Skern, T.; Kuechler, E. Picornavirus 2A proteinase-mediated stimulation of internal initiation of translation is dependent on enzymatic activity and the cleavage products of cellular proteins. Virology 1995, 213, 549–557. [Google Scholar] [CrossRef]
- Ziegler, E.; Borman, A.M.; Kirchweger, R.; Skern, T.; Kean, K.M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 1995, 69, 3465–3474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohlmann, T.; Rau, M.; Pain, V.M.; Morley, S.J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. Embo J. 1996, 15, 1371–1382. [Google Scholar] [CrossRef]
- Aumayr, M.; Schrempf, A.; Uzulmez, O.; Olek, K.M.; Skern, T. Interaction of 2A proteinase of human rhinovirus genetic group A with eIF4E is required for eIF4G cleavage during infection. Virology 2017, 511, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Avanzino, B.C.; Fuchs, G.; Fraser, C.S. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc. Natl. Acad. Sci. USA 2017, 114, 9611–9616. [Google Scholar] [CrossRef] [Green Version]
- Sukarieh, R.; Sonenberg, N.; Pelletier, J. Nuclear assortment of eIF4E coincides with shut-off of host protein synthesis upon poliovirus infection. J. Gen. Virol. 2010, 91, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.C.; Yu, S.L.; Chen, J.J.; Chang, S.Y.; Yan, B.S.; Hong, Q.S.; Singh, S.; Kao, C.L.; Chen, H.Y.; Su, K.Y.; et al. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 2011, 9, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Borman, A.M.; Le Mercier, P.; Girard, M.; Kean, K.M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997, 25, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Garcia, L.; Angulo, J.; Ramos, H.; Barrera, A.; Pino, K.; Vera-Otarola, J.; Lopez-Lastra, M. The internal ribosome entry site of the Dengue virus mRNA is active when cap-dependent translation initiation is inhibited. J. Virol. 2020, 95, e01998-20. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, E.; Castello, A.; Carrasco, L.; Izquierdo, J.M. Poliovirus 2A protease triggers a selective nucleo-cytoplasmic redistribution of splicing factors to regulate alternative pre-mRNA splicing. PLoS ONE 2013, 8, e73723. [Google Scholar] [CrossRef] [Green Version]
- Watters, K.; Inankur, B.; Gardiner, J.C.; Warrick, J.; Sherer, N.M.; Yin, J.; Palmenberg, A.C. Differential Disruption of Nucleocytoplasmic Trafficking Pathways by Rhinovirus 2A Proteases. J. Virol. 2017, 91, e02472-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, M.E.; Kumar, A.; Wells, J.A.; Hobman, T.C.; Julien, O.; Hardy, J.A. The Unique Cofactor Region of Zika Virus NS2B-NS3 Protease Facilitates Cleavage of Key Host Proteins. ACS Chem. Biol. 2018, 13, 2398–2405. [Google Scholar] [CrossRef]
- Alvarez, E.; Menendez-Arias, L.; Carrasco, L. The eukaryotic translation initiation factor 4GI is cleaved by different retroviral proteases. J. Virol. 2003, 77, 12392–12400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perales, C.; Carrasco, L.; Ventoso, I. Cleavage of eIF4G by HIV-1 protease: Effects on translation. FEBS Lett. 2003, 533, 89–94. [Google Scholar] [CrossRef] [Green Version]
- Ventoso, I.; Blanco, R.; Perales, C.; Carrasco, L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. USA 2001, 98, 12966–12971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohlmann, T.; Prevot, D.; Decimo, D.; Roux, F.; Garin, J.; Morley, S.J.; Darlix, J.L. In Vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002, 318, 9–20. [Google Scholar] [CrossRef]
- Castello, A.; Franco, D.; Moral-Lopez, P.; Berlanga, J.J.; Alvarez, E.; Wimmer, E.; Carrasco, L. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS ONE 2009, 4, e7997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Breyne, S.; Ohlmann, T. Focus on Translation Initiation of the HIV-1 mRNAs. Int. J. Mol. Sci. 2018, 20, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gingras, A.C.; Svitkin, Y.; Belsham, G.J.; Pause, A.; Sonenberg, N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. USA 1996, 93, 5578–5583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haghighat, A.; Mader, S.; Pause, A.; Sonenberg, N. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995, 14, 5701–5709. [Google Scholar] [CrossRef]
- Marcotrigiano, J.; Gingras, A.C.; Sonenberg, N.; Burley, S.K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 1999, 3, 707–716. [Google Scholar] [CrossRef]
- Richter, J.D.; Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005, 433, 477–480. [Google Scholar] [CrossRef]
- Edgil, D.; Polacek, C.; Harris, E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 2006, 80, 2976–2986. [Google Scholar] [CrossRef] [Green Version]
- Aviner, R.; Li, K.H.; Frydman, J.; Andino, R. Cotranslational prolyl hydroxylation is essential for flavivirus biogenesis. Nature 2021, 596, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.; Magg, V.; Uch, F.; Mutz, P.; Klein, P.; Haneke, K.; Lohmann, V.; Bartenschlager, R.; Fackler, O.T.; Locker, N.; et al. Flavivirus Infection Uncouples Translation Suppression from Cellular Stress Responses. mBio 2017, 8, e02150-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.R.; Kuo, S.H.; Lin, C.Y.; Fu, P.J.; Lin, Y.S.; Yeh, T.M.; Liu, H.S. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both In Vitro and in vivo. Sci. Rep. 2018, 8, 489. [Google Scholar] [CrossRef]
- Harding, H.P.; Novoa, I.; Zhang, Y.; Zeng, H.; Wek, R.; Schapira, M.; Ron, D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000, 6, 1099–1108. [Google Scholar] [CrossRef]
- Hou, J.N.; Chen, T.H.; Chiang, Y.H.; Peng, J.Y.; Yang, T.H.; Cheng, C.C.; Sofiyatun, E.; Chiu, C.H.; Chiang-Ni, C.; Chen, W.J. PERK Signal-Modulated Protein Translation Promotes the Survivability of Dengue 2 Virus-Infected Mosquito Cells and Extends Viral Replication. Viruses 2017, 9, 262. [Google Scholar] [CrossRef] [Green Version]
- Goh, W.C.; Rogel, M.E.; Kinsey, C.M.; Michael, S.F.; Fultz, P.N.; Nowak, M.A.; Hahn, B.H.; Emerman, M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: A mechanism for selection of Vpr in vivo. Nat. Med. 1998, 4, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Yilmaz, A.; Marsh, K.; Cochrane, A.; Boris-Lawrie, K. Thriving under stress: Selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012, 8, e1002612. [Google Scholar] [CrossRef]
- Gendron, K.; Ferbeyre, G.; Heveker, N.; Brakier-Gingras, L. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011, 39, 902–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monette, A.; Ajamian, L.; Lopez-Lastra, M.; Mouland, A.J. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: Implications for HIV-1 gene expression. J. Biol. Chem. 2009, 284, 31350–31362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amorim, R.; Costa, S.M.; Cavaleiro, N.P.; da Silva, E.E.; da Costa, L.J. HIV-1 transcripts use IRES-initiation under conditions where Cap-dependent translation is restricted by poliovirus 2A protease. PLoS ONE 2014, 9, e88619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monette, A.; Valiente-Echeverria, F.; Rivero, M.; Cohen, E.A.; Lopez-Lastra, M.; Mouland, A.J. Dual mechanisms of translation initiation of the full-length HIV-1 mRNA contribute to gag synthesis. PLoS ONE 2013, 8, e68108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohlmann, T.; Mengardi, C.; Lopez-Lastra, M. Translation initiation of the HIV-1 mRNA. Translation 2014, 2, e960242. [Google Scholar] [CrossRef] [PubMed]
- Borman, A.; Howell, M.T.; Patton, J.G.; Jackson, R.J. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 1993, 74 Pt 9, 1775–1788. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Ehrenfeld, E. Translation of poliovirus RNA in vitro: Changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 1979, 97, 396–405. [Google Scholar] [CrossRef]
- Glass, M.J.; Summers, D.F. Identification of a trans-acting activity from liver that stimulates hepatitis A virus translation in vitro. Virology 1993, 193, 1047–1050. [Google Scholar] [CrossRef]
- Toyoda, H.; Koide, N.; Kamiyama, M.; Tobita, K.; Mizumoto, K.; Imura, N. Host factors required for internal initiation of translation on poliovirus RNA. Arch. Virol. 1994, 138, 1–15. [Google Scholar] [CrossRef]
- Gamarnik, A.V.; Andino, R. Replication of poliovirus in Xenopus oocytes requires two human factors. EMBO J. 1996, 15, 5988–5998. [Google Scholar] [CrossRef]
- Belsham, G.J.; Sonenberg, N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996, 60, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Belsham, G.J.; Sonenberg, N. Picornavirus RNA translation: Roles for cellular proteins. Trends Microbiol. 2000, 8, 330–335. [Google Scholar] [CrossRef]
- Cobbold, L.C.; Spriggs, K.A.; Haines, S.J.; Dobbyn, H.C.; Hayes, C.; De Moor, C.H.; Lilley, K.S.; Bushell, M.; Willis, A.E. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell Biol. 2008, 28, 40–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivas-Aravena, A.; Ramdohr, P.; Vallejos, M.; Valiente-Echeverria, F.; Dormoy-Raclet, V.; Rodriguez, F.; Pino, K.; Holzmann, C.; Huidobro-Toro, J.P.; Gallouzi, I.E.; et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology 2009, 392, 178–185. [Google Scholar] [CrossRef] [Green Version]
- Barrera, A.; Ramos, H.; Vera-Otarola, J.; Fernandez-Garcia, L.; Angulo, J.; Olguin, V.; Pino, K.; Mouland, A.J.; Lopez-Lastra, M. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res. 2020, 48, 10479–10499. [Google Scholar] [CrossRef] [PubMed]
- Hellen, C.U.; Witherell, G.W.; Schmid, M.; Shin, S.H.; Pestova, T.V.; Gil, A.; Wimmer, E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 1993, 90, 7642–7646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminski, A.; Hunt, S.L.; Patton, J.G.; Jackson, R.J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1995, 1, 924–938. [Google Scholar]
- Meerovitch, K.; Svitkin, Y.V.; Lee, H.S.; Lejbkowicz, F.; Kenan, D.J.; Chan, E.K.; Agol, V.I.; Keene, J.D.; Sonenberg, N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993, 67, 3798–3807. [Google Scholar] [CrossRef] [Green Version]
- Svitkin, Y.V.; Meerovitch, K.; Lee, H.S.; Dholakia, J.N.; Kenan, D.J.; Agol, V.I.; Sonenberg, N. Internal translation initiation on poliovirus RNA: Further characterization of La function in poliovirus translation in vitro. J. Virol. 1994, 68, 1544–1550. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.K.; Jang, S.K. La protein is required for efficient translation driven by encephalomyocarditis virus internal ribosomal entry site. J. Gen. Virol. 1999, 80 Pt 12, 3159–3166. [Google Scholar] [CrossRef]
- Blyn, L.B.; Swiderek, K.M.; Richards, O.; Stahl, D.C.; Semler, B.L.; Ehrenfeld, E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: Identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 1996, 93, 11115–11120. [Google Scholar] [CrossRef] [Green Version]
- Blyn, L.B.; Towner, J.S.; Semler, B.L.; Ehrenfeld, E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997, 71, 6243–6246. [Google Scholar] [CrossRef] [Green Version]
- Gamarnik, A.V.; Andino, R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 1997, 3, 882–892. [Google Scholar]
- Pilipenko, E.V.; Pestova, T.V.; Kolupaeva, V.G.; Khitrina, E.V.; Poperechnaya, A.N.; Agol, V.I.; Hellen, C.U. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000, 14, 2028–2045. [Google Scholar] [CrossRef] [PubMed]
- Monie, T.P.; Perrin, A.J.; Birtley, J.R.; Sweeney, T.R.; Karakasiliotis, I.; Chaudhry, Y.; Roberts, L.O.; Matthews, S.; Goodfellow, I.G.; Curry, S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007, 26, 3936–3944. [Google Scholar] [CrossRef] [Green Version]
- Hunt, S.L.; Hsuan, J.J.; Totty, N.; Jackson, R.J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999, 13, 437–448. [Google Scholar] [CrossRef] [Green Version]
- Boussadia, O.; Niepmann, M.; Creancier, L.; Prats, A.C.; Dautry, F.; Jacquemin-Sablon, H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003, 77, 3353–3359. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.C.; Jackson, R.J. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 2004, 85, 2279–2287. [Google Scholar] [CrossRef]
- Huang, P.N.; Lin, J.Y.; Locker, N.; Kung, Y.A.; Hung, C.T.; Lin, J.Y.; Huang, H.I.; Li, M.L.; Shih, S.R. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 2011, 39, 9633–9648. [Google Scholar] [CrossRef] [Green Version]
- Dave, P.; George, B.; Sharma, D.K.; Das, S. Polypyrimidine tract-binding protein (PTB) and PTB-associated splicing factor in CVB3 infection: An ITAF for an ITAF. Nucleic Acids Res. 2017, 45, 9068–9084. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.Y.; Brewer, G.; Li, M.L. HuR and Ago2 Bind the Internal Ribosome Entry Site of Enterovirus 71 and Promote Virus Translation and Replication. PLoS ONE 2015, 10, e0140291. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Song, L.; Cong, H.; Tien, P. Nuclear Protein Sam68 Interacts with the Enterovirus 71 Internal Ribosome Entry Site and Positively Regulates Viral Protein Translation. J. Virol. 2015, 89, 10031–10043. [Google Scholar] [CrossRef] [Green Version]
- Dong, Q.; Men, R.; Dan, X.; Chen, Y.; Li, H.; Chen, G.; Zee, B.; Wang, M.H.T.; He, M.L. Hsc70 regulates the IRES activity and serves as an antiviral target of enterovirus A71 infection. Antivir. Res. 2018, 150, 39–46. [Google Scholar] [CrossRef]
- Su, Y.S.; Hsieh, P.Y.; Li, J.S.; Pao, Y.H.; Chen, C.J.; Hwang, L.H. The Heat Shock Protein 70 Family of Chaperones Regulates All Phases of the Enterovirus A71 Life Cycle. Front. Microbiol. 2020, 11, 1656. [Google Scholar] [CrossRef]
- Su, Y.S.; Hwang, L.H.; Chen, C.J. Heat Shock Protein A6, a Novel HSP70, Is Induced During Enterovirus A71 Infection to Facilitate Internal Ribosomal Entry Site-Mediated Translation. Front. Microbiol. 2021, 12, 664955. [Google Scholar] [CrossRef] [PubMed]
- Witherell, G.W.; Wimmer, E. Encephalomyocarditis virus internal ribosomal entry site RNA-protein interactions. J. Virol. 1994, 68, 3183–3192. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Eisenring, I.A.; Cajero-Juarez, M.; del Angel, R.M. Cell proteins bind to a linear polypyrimidine-rich sequence within the 5′-untranslated region of rhinovirus 14 RNA. J. Virol. 1995, 69, 6819–6824. [Google Scholar] [CrossRef] [Green Version]
- Bedard, K.M.; Daijogo, S.; Semler, B.L. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007, 26, 459–467. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, K.D.; Semler, B.L. Re-localization of cellular protein SRp20 during poliovirus infection: Bridging a viral IRES to the host cell translation apparatus. PLoS Pathog. 2011, 7, e1002127. [Google Scholar] [CrossRef] [Green Version]
- Hunt, S.L.; Jackson, R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. Rna 1999, 5, 344–359. [Google Scholar] [CrossRef]
- Lin, J.Y.; Li, M.L.; Shih, S.R. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 2009, 37, 47–59. [Google Scholar] [CrossRef]
- Chen, L.L.; Kung, Y.A.; Weng, K.F.; Lin, J.Y.; Horng, J.T.; Shih, S.R. Enterovirus 71 infection cleaves a negative regulator for viral internal ribosomal entry site-driven translation. J. Virol. 2013, 87, 3828–3838. [Google Scholar] [CrossRef] [Green Version]
- Cathcart, A.L.; Rozovics, J.M.; Semler, B.L. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J. Virol. 2013, 87, 10423–10434. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.Y.; Li, M.L.; Brewer, G. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS ONE 2014, 9, e103827. [Google Scholar] [CrossRef] [Green Version]
- Ullmer, W.; Semler, B.L. Direct and Indirect Effects on Viral Translation and RNA Replication Are Required for AUF1 Restriction of Enterovirus Infections in Human Cells. mBio 2018, 9, e01669-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Yang, D.; Sun, C.; Wang, H.; Zhao, B.; Zhou, G.; Yu, L. hnRNP K Is a Novel Internal Ribosomal Entry Site-Transacting Factor That Negatively Regulates Foot-and-Mouth Disease Virus Translation and Replication and Is Antagonized by Viral 3C Protease. J. Virol. 2020, 94, e00803-20. [Google Scholar] [CrossRef]
- Pacheco, A.; Lopez de Quinto, S.; Ramajo, J.; Fernandez, N.; Martinez-Salas, E. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 2009, 37, 582–590. [Google Scholar] [CrossRef]
- Merrill, M.K.; Gromeier, M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 2006, 80, 6936–6942. [Google Scholar] [CrossRef] [Green Version]
- Kafasla, P.; Morgner, N.; Poyry, T.A.; Curry, S.; Robinson, C.V.; Jackson, R.J. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol. Cell 2009, 34, 556–568. [Google Scholar] [CrossRef] [Green Version]
- Conte, M.R.; Grune, T.; Ghuman, J.; Kelly, G.; Ladas, A.; Matthews, S.; Curry, S. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 2000, 19, 3132–3141. [Google Scholar] [CrossRef]
- Maris, C.; Dominguez, C.; Allain, F.H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, J.; Ye, F.; Wang, G.; Han, W.; Wei, Z.; Yin, B.; Yuan, J.; Qiang, B.; Peng, X. Polypyrimidine Tract-Binding Protein Regulates Enterovirus 71 Translation Through Interaction with the Internal Ribosomal Entry Site. Virol. Sin. 2019, 34, 66–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kafasla, P.; Morgner, N.; Robinson, C.V.; Jackson, R.J. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010, 29, 3710–3722. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.E.; Hardin, C.C.; Lemon, S.M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5'-nontranslated RNA of hepatitis A virus. J. Biol. Chem. 1996, 271, 14134–14142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, B.; Bhattacharyya, S.; Das, S. Polypyrimidine tract-binding protein interacts with coxsackievirus B3 RNA and influences its translation. J. Gen. Virol. 2010, 91, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Siddiqui, A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 1995, 69, 6367–6375. [Google Scholar] [CrossRef] [Green Version]
- Caceres, C.J.; Contreras, N.; Angulo, J.; Vera-Otarola, J.; Pino-Ajenjo, C.; Llorian, M.; Ameur, M.; Lisboa, F.; Pino, K.; Lowy, F.; et al. Polypyrimidine tract-binding protein binds to the 5′ untranslated region of the mouse mammary tumor virus mRNA and stimulates cap-independent translation initiation. FEBS J. 2016, 283, 1880–1901. [Google Scholar] [CrossRef] [Green Version]
- Izumi, R.E.; Valdez, B.; Banerjee, R.; Srivastava, M.; Dasgupta, A. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 2001, 76, 17–29. [Google Scholar] [CrossRef]
- Han, S.; Wang, X.; Guan, J.; Wu, J.; Zhang, Y.; Li, P.; Liu, Z.; Abdullah, S.W.; Zhang, Z.; Jin, Y.; et al. Nucleolin Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus by Supporting the Assembly of Translation Initiation Complexes. J. Virol. 2021, 95, e0023821. [Google Scholar] [CrossRef]
- Paek, K.Y.; Kim, C.S.; Park, S.M.; Kim, J.H.; Jang, S.K. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008, 82, 12082–12093. [Google Scholar] [CrossRef] [Green Version]
- Costa-Mattioli, M.; Svitkin, Y.; Sonenberg, N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004, 24, 6861–6870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, P.; Lim, T.; Yuan, J.; Zhang, M.; Chau, D.; McManus, B.; Yang, D. Specific interaction of HeLa cell proteins with coxsackievirus B3 3'UTR: La autoantigen binds the 3′ and 5′ UTR independently of the poly(A) tail. Cell. Microbiol. 2007, 9, 1705–1715. [Google Scholar] [CrossRef]
- Cordes, S.; Kusov, Y.; Heise, T.; Gauss-Muller, V. La autoantigen suppresses IRES-dependent translation of the hepatitis A virus. Biochem. Biophys. Res. Commun. 2008, 368, 1014–1019. [Google Scholar] [CrossRef]
- Hahm, B.; Kim, Y.K.; Kim, J.H.; Kim, T.Y.; Jang, S.K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 1998, 72, 8782–8788. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Paek, K.Y.; Ha, S.H.; Cho, S.; Choi, K.; Kim, C.S.; Ryu, S.H.; Jang, S.K. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 2004, 24, 7878–7890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.M.; Paek, K.Y.; Hong, K.Y.; Jang, C.J.; Cho, S.; Park, J.H.; Kim, J.H.; Jan, E.; Jang, S.K. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res. 2011, 39, 7791–7802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dave, P.; George, B.; Balakrishnan, S.; Sharma, D.K.; Raheja, H.; Dixit, N.M.; Das, S. Strand-specific affinity of host factor hnRNP C1/C2 guides positive to negative-strand ratio in Coxsackievirus B3 infection. RNA Biol. 2019, 16, 1286–1299. [Google Scholar] [CrossRef]
- Shih, S.R.; Stollar, V.; Li, M.L. Host factors in enterovirus 71 replication. J. Virol. 2011, 85, 9658–9666. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Razanau, A.; Feng, D.; Lobo, V.G.; Xie, J. Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res. 2012, 40, 8059–8071. [Google Scholar] [CrossRef] [Green Version]
- Lewis, H.A.; Musunuru, K.; Jensen, K.B.; Edo, C.; Chen, H.; Darnell, R.B.; Burley, S.K. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 2000, 100, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Masaki, T.; Shimakami, T.; Lemon, S.M. hnRNP L and NF90 interact with hepatitis C virus 5'-terminal untranslated RNA and promote efficient replication. J. Virol. 2014, 88, 7199–7209. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Dong, X.; Li, Y.; Zhang, Q.; Kim, C.; Song, Y.; Kang, L.; Liu, Y.; Wu, K.; Wu, J. PolyC-binding protein 1 interacts with 5′-untranslated region of enterovirus 71 RNA in membrane-associated complex to facilitate viral replication. PLoS ONE 2014, 9, e87491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, K.; Kim, J.H.; Li, X.; Paek, K.Y.; Ha, S.H.; Ryu, S.H.; Wimmer, E.; Jang, S.K. Identification of cellular proteins enhancing activities of internal ribosomal entry sites by competition with oligodeoxynucleotides. Nucleic Acids Res. 2004, 32, 1308–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graff, J.; Cha, J.; Blyn, L.B.; Ehrenfeld, E. Interaction of poly(rC) binding protein 2 with the 5' noncoding region of hepatitis A virus RNA and its effects on translation. J. Virol. 1998, 72, 9668–9675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Seitz, S.; Kusov, Y.; Zell, R.; Gauss-Muller, V. RNA interaction and cleavage of poly(C)-binding protein 2 by hepatitis A virus protease. Biochem. Biophys. Res. Commun. 2007, 364, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Dixit, U.; Pandey, A.K.; Mishra, P.; Sengupta, A.; Pandey, V.N. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res. 2016, 44, 5271–5287. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Barraud, P. Functions of double-stranded RNA-binding domains in nucleocytoplasmic transport. RNA Biol. 2014, 11, 1226–1232. [Google Scholar] [CrossRef] [Green Version]
- Leulliot, N.; Quevillon-Cheruel, S.; Graille, M.; van Tilbeurgh, H.; Leeper, T.C.; Godin, K.S.; Edwards, T.E.; Sigurdsson, S.T.; Rozenkrants, N.; Nagel, R.J.; et al. A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J. 2004, 23, 2468–2477. [Google Scholar] [CrossRef] [Green Version]
- Ramos, A.; Grunert, S.; Adams, J.; Micklem, D.R.; Proctor, M.R.; Freund, S.; Bycroft, M.; St Johnston, D.; Varani, G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000, 19, 997–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.M.; Ou, B.T.; Chen, C.Y.; Chan, H.H.; Chen, C.J.; Wang, R.Y. Staufen1 Protein Participates Positively in the Viral RNA Replication of Enterovirus 71. Viruses 2019, 11, 142. [Google Scholar] [CrossRef] [Green Version]
- Ramos, H.; Monette, A.; Niu, M.; Barrera, A.; Lopez-Ulloa, B.; Fuentes, Y.; Guizar, P.; Pino, K.; DesGroseillers, L.; Mouland, A.J.; et al. The double-stranded RNA-binding protein, Staufen1, is an IRES-transacting factor regulating HIV-1 cap-independent translation initiation. Nucleic Acids Res. 2022, 50, 411–429. [Google Scholar] [CrossRef]
- Korf, M.; Jarczak, D.; Beger, C.; Manns, M.P.; Kruger, M. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J. Hepatol. 2005, 43, 225–234. [Google Scholar] [CrossRef]
- Mihailovich, M.; Militti, C.; Gabaldon, T.; Gebauer, F. Eukaryotic cold shock domain proteins: Highly versatile regulators of gene expression. Bioessays 2010, 32, 109–118. [Google Scholar] [CrossRef]
- Caceres, C.J.; Angulo, J.; Contreras, N.; Pino, K.; Vera-Otarola, J.; Lopez-Lastra, M. Targeting deoxyhypusine hydroxylase activity impairs cap-independent translation initiation driven by the 5'untranslated region of the HIV-1, HTLV-1, and MMTV mRNAs. Antivir. Res. 2016, 134, 192–206. [Google Scholar] [CrossRef]
- Liu, J.; Henao-Mejia, J.; Liu, H.; Zhao, Y.; He, J.J. Translational regulation of HIV-1 replication by HIV-1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. J. Neuroimmune Pharmacol. 2011, 6, 308–321. [Google Scholar] [CrossRef]
- Jarvelin, A.I.; Noerenberg, M.; Davis, I.; Castello, A. The new (dis)order in RNA regulation. Cell Commun. Signal. 2016, 14, 9. [Google Scholar] [CrossRef] [Green Version]
- Birney, E.; Kumar, S.; Krainer, A.R. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993, 21, 5803–5816. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, E.; Zhang, X.; Ceman, S. Arginines of the RGG box regulate FMRP association with polyribosomes and mRNA. Hum. Mol. Genet. 2010, 19, 1314–1323. [Google Scholar] [CrossRef] [Green Version]
- Tolbert, M.; Morgan, C.E.; Pollum, M.; Crespo-Hernandez, C.E.; Li, M.L.; Brewer, G.; Tolbert, B.S. HnRNP A1 Alters the Structure of a Conserved Enterovirus IRES Domain to Stimulate Viral Translation. J. Mol. Biol. 2017, 429, 2841–2858. [Google Scholar] [CrossRef]
- Schirle, N.T.; MacRae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003, 10, 1026–1032. [Google Scholar] [CrossRef]
- Chahal, J.; Gebert, L.F.R.; Gan, H.H.; Camacho, E.; Gunsalus, K.C.; MacRae, I.J.; Sagan, S.M. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5' terminus. Nucleic Acids Res. 2019, 47, 5307–5324. [Google Scholar] [CrossRef] [Green Version]
- Gebert, L.F.R.; Law, M.; MacRae, I.J. A structured RNA motif locks Argonaute2:miR-122 onto the 5' end of the HCV genome. Nat. Commun. 2021, 12, 6836. [Google Scholar] [CrossRef]
- Bauer, D.; Merz, D.R.; Pelz, B.; Theisen, K.E.; Yacyshyn, G.; Mokranjac, D.; Dima, R.I.; Rief, M.; Zoldak, G. Nucleotides regulate the mechanical hierarchy between subdomains of the nucleotide binding domain of the Hsp70 chaperone DnaK. Proc. Natl. Acad. Sci. USA 2015, 112, 10389–10394. [Google Scholar] [CrossRef] [Green Version]
- Ali, N.; Siddiqui, A. The La antigen binds 5' noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. USA 1997, 94, 2249–2254. [Google Scholar] [CrossRef] [Green Version]
- Ali, N.; Pruijn, G.J.; Kenan, D.J.; Keene, J.D.; Siddiqui, A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000, 275, 27531–27540. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Li, W.; Noble, W.S.; Payan, D.; Anderson, D.C. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 2004, 3, 949–957. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, A.; Reigadas, S.; Martinez-Salas, E. Riboproteomic analysis of polypeptides interacting with the internal ribosome-entry site element of foot-and-mouth disease viral RNA. Proteomics 2008, 8, 4782–4790. [Google Scholar] [CrossRef]
- Gosert, R.; Chang, K.H.; Rijnbrand, R.; Yi, M.; Sangar, D.V.; Lemon, S.M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol. Cell. Biol. 2000, 20, 1583–1595. [Google Scholar] [CrossRef] [Green Version]
- Anwar, A.; Ali, N.; Tanveer, R.; Siddiqui, A. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 2000, 275, 34231–34235. [Google Scholar] [CrossRef] [Green Version]
- Brocard, M.; Paulous, S.; Komarova, A.V.; Deveaux, V.; Kean, K.M. Evidence that PTB does not stimulate HCV IRES-driven translation. Virus Genes 2007, 35, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Domitrovich, A.M.; Diebel, K.W.; Ali, N.; Sarker, S.; Siddiqui, A. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 2005, 335, 72–86. [Google Scholar] [CrossRef] [Green Version]
- Tischendorf, J.J.; Beger, C.; Korf, M.; Manns, M.P.; Kruger, M. Polypyrimidine tract-binding protein (PTB) inhibits Hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation, but does not affect HCV replication. Arch. Virol. 2004, 149, 1955–1970. [Google Scholar] [CrossRef]
- Sp Ngberg, K.; Schwartz, S. Poly(C)-binding protein interacts with the hepatitis C virus 5′ untranslated region. J. Gen. Virol. 1999, 80 Pt 6, 1371–1376. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.S.; Seol, S.K.; Song, O.K.; Park, J.H.; Jang, S.K. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J. Virol. 2007, 81, 3852–3865. [Google Scholar] [CrossRef] [Green Version]
- Belsham, G.J.; Sonenberg, N.; Svitkin, Y.V. The role of the La autoantigen in internal initiation. Curr. Top. Microbiol. Immunol. 1995, 203, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Francisco-Velilla, R.; Azman, E.B.; Martinez-Salas, E. Impact of RNA-Protein Interaction Modes on Translation Control: The Versatile Multidomain Protein Gemin5. Bioessays 2019, 41, e1800241. [Google Scholar] [CrossRef] [PubMed]
- Niepmann, M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 1996, 388, 39–42. [Google Scholar] [CrossRef] [Green Version]
- Kaminski, A.; Jackson, R.J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 1998, 4, 626–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kafasla, P.; Mickleburgh, I.; Llorian, M.; Coelho, M.; Gooding, C.; Cherny, D.; Joshi, A.; Kotik-Kogan, O.; Curry, S.; Eperon, I.C.; et al. Defining the roles and interactions of PTB. Biochem. Soc. Trans. 2012, 40, 815–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawicka, K.; Bushell, M.; Spriggs, K.A.; Willis, A.E. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008, 36, 641–647. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, T.; Ichikawa, M.; Arai, J.; Tateiwa, H.; Fu, L.; Higuchi, K.; Yoshimura, N. Molecular cloning and characterization of a new neuron-specific homologue of rat polypyrimidine tract binding protein. J. Biochem. 2000, 128, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Markovtsov, V.; Nikolic, J.M.; Goldman, J.A.; Turck, C.W.; Chou, M.Y.; Black, D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000, 20, 7463–7479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polydorides, A.D.; Okano, H.J.; Yang, Y.Y.; Stefani, G.; Darnell, R.B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. USA 2000, 97, 6350–6355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, H.; Tsukahara, K.; Kanaoka, Y.; Jinno, S.; Okayama, H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999, 19, 3829–3841. [Google Scholar] [CrossRef] [Green Version]
- Makeyev, A.V.; Liebhaber, S.A. Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics 2000, 67, 301–316. [Google Scholar] [CrossRef]
- Makeyev, A.V.; Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef] [Green Version]
- Thandapani, P.; O'Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castello, A.; Fischer, B.; Frese, C.K.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Comprehensive Identification of RNA-Binding Domains in Human Cells. Mol. Cell 2016, 63, 696–710. [Google Scholar] [CrossRef] [Green Version]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell. Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Garcia-Moreno, M.; Noerenberg, M.; Ni, S.; Jarvelin, A.I.; Gonzalez-Almela, E.; Lenz, C.E.; Bach-Pages, M.; Cox, V.; Avolio, R.; Davis, T.; et al. System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Mol. Cell 2019, 74, 196–211.e11. [Google Scholar] [CrossRef] [Green Version]
- Flather, D.; Semler, B.L. Picornaviruses and nuclear functions: Targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015, 6, 594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, R.E.; Frye, M. Noncanonical functions of the serine-arginine-rich splicing factor (SR) family of proteins in development and disease. Bioessays 2021, 43, e2000242. [Google Scholar] [CrossRef] [PubMed]
- Wegener, M.; Muller-McNicoll, M. View from an mRNP: The Roles of SR Proteins in Assembly, Maturation and Turnover. Adv. Exp. Med. Biol. 2019, 1203, 83–112. [Google Scholar] [CrossRef]
- Dreyfuss, G.; Matunis, M.J.; Pinol-Roma, S.; Burd, C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993, 62, 289–321. [Google Scholar] [CrossRef] [PubMed]
- Gorlach, M.; Burd, C.G.; Portman, D.S.; Dreyfuss, G. The hnRNP proteins. Mol. Biol. Rep. 1993, 18, 73–78. [Google Scholar] [CrossRef]
- Kim, V.N.; Dreyfuss, G. Nuclear mRNA binding proteins couple pre-mRNA splicing and post-splicing events. Mol. Cells 2001, 12, 1–10. [Google Scholar] [PubMed]
- Dreyfuss, G.; Kim, V.N.; Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002, 3, 195–205. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [Green Version]
- Busch, A.; Hertel, K.J. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA 2012, 3, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Nakielny, S.; Dreyfuss, G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell. Biol. 1996, 134, 1365–1373. [Google Scholar] [CrossRef]
- Pinol-Roma, S.; Dreyfuss, G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 1992, 355, 730–732. [Google Scholar] [CrossRef] [PubMed]
- Amero, S.A.; Raychaudhuri, G.; Cass, C.L.; van Venrooij, W.J.; Habets, W.J.; Krainer, A.R.; Beyer, A.L. Independent deposition of heterogeneous nuclear ribonucleoproteins and small nuclear ribonucleoprotein particles at sites of transcription. Proc. Natl. Acad. Sci. USA 1992, 89, 8409–8413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matunis, E.L.; Matunis, M.J.; Dreyfuss, G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J. Cell Biol. 1993, 121, 219–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinol-Roma, S.; Swanson, M.S.; Gall, J.G.; Dreyfuss, G. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell Biol. 1989, 109, 2575–2587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mili, S.; Shu, H.J.; Zhao, Y.; Pinol-Roma, S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: Candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 2001, 21, 7307–7319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamma, H.; Portman, D.S.; Dreyfuss, G. Cell type-specific expression of hnRNP proteins. Exp. Cell Res. 1995, 221, 187–196. [Google Scholar] [CrossRef]
- Singh, R.; Valcarcel, J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005, 12, 645–653. [Google Scholar] [CrossRef]
- Lizcano-Perret, B.; Michiels, T. Nucleocytoplasmic Trafficking Perturbation Induced by Picornaviruses. Viruses 2021, 13, 1210. [Google Scholar] [CrossRef]
- Yarbrough, M.L.; Mata, M.A.; Sakthivel, R.; Fontoura, B.M. Viral subversion of nucleocytoplasmic trafficking. Traffic 2014, 15, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Le Sage, V.; Mouland, A.J. Viral subversion of the nuclear pore complex. Viruses 2013, 5, 2019–2042. [Google Scholar] [CrossRef] [PubMed]
- Ghildyal, R.; Jordan, B.; Li, D.; Dagher, H.; Bardin, P.G.; Gern, J.E.; Jans, D.A. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J. Virol. 2009, 83, 7349–7352. [Google Scholar] [CrossRef] [Green Version]
- Walker, E.J.; Younessi, P.; Fulcher, A.J.; McCuaig, R.; Thomas, B.J.; Bardin, P.G.; Jans, D.A.; Ghildyal, R. Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells. PLoS ONE 2013, 8, e71316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, K.D.; Chase, A.J.; Cathcart, A.L.; Tran, G.P.; Semler, B.L. Viral proteinase requirements for the nucleocytoplasmic relocalization of cellular splicing factor SRp20 during picornavirus infections. J. Virol. 2013, 87, 2390–2400. [Google Scholar] [CrossRef] [Green Version]
- Belov, G.A.; Lidsky, P.V.; Mikitas, O.V.; Egger, D.; Lukyanov, K.A.; Bienz, K.; Agol, V.I. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 2004, 78, 10166–10177. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Bi, J.; Liu, J.; Liu, X.; Wu, X.; Jiang, P.; Yoo, D.; Zhang, Y.; Wu, J.; Wan, R.; et al. 3Cpro of foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2014, 88, 4908–4920. [Google Scholar] [CrossRef] [Green Version]
- Lidsky, P.V.; Hato, S.; Bardina, M.V.; Aminev, A.G.; Palmenberg, A.C.; Sheval, E.V.; Polyakov, V.Y.; van Kuppeveld, F.J.; Agol, V.I. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 2006, 80, 2705–2717. [Google Scholar] [CrossRef] [Green Version]
- Gustin, K.E.; Sarnow, P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001, 20, 240–249. [Google Scholar] [CrossRef] [Green Version]
- McBride, A.E.; Schlegel, A.; Kirkegaard, K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. USA 1996, 93, 2296–2301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spurrell, J.C.; Wiehler, S.; Zaheer, R.S.; Sanders, S.P.; Proud, D. Human airway epithelial cells produce IP-10 (CXCL10) In Vitro and in vivo upon rhinovirus infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 289, L85–L95. [Google Scholar] [CrossRef] [Green Version]
- Rozovics, J.M.; Chase, A.J.; Cathcart, A.L.; Chou, W.; Gershon, P.D.; Palusa, S.; Wilusz, J.; Semler, B.L. Picornavirus modification of a host mRNA decay protein. mBio 2012, 3, e00431-12. [Google Scholar] [CrossRef] [Green Version]
- Cammas, A.; Pileur, F.; Bonnal, S.; Lewis, S.M.; Leveque, N.; Holcik, M.; Vagner, S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell. 2007, 18, 5048–5059. [Google Scholar] [CrossRef] [Green Version]
- Waggoner, S.; Sarnow, P. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 1998, 72, 6699–6709. [Google Scholar] [CrossRef] [Green Version]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. (Eds.) Retroviruses; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997. [Google Scholar]
- Cochrane, A.W.; McNally, M.T.; Mouland, A.J. The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology 2006, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Messer, L.I.; Levin, J.G.; Chattopadhyay, S.K. Metabolism of viral RNA in murine leukemia virus-infected cells; evidence for differential stability of viral message and virion precursor RNA. J. Virol. 1981, 40, 683–690. [Google Scholar] [CrossRef] [Green Version]
- Butsch, M.; Boris-Lawrie, K. Destiny of unspliced retroviral RNA: Ribosome and/or virion? J. Virol. 2002, 76, 3089–3094. [Google Scholar] [CrossRef] [Green Version]
- Pinol-Roma, S.; Choi, Y.D.; Matunis, M.J.; Dreyfuss, G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988, 2, 215–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, S.M.; Holcik, M. For IRES trans-acting factors, it is all about location. Oncogene 2008, 27, 1033–1035. [Google Scholar] [CrossRef]
- Cho, H.; Lee, H.C.; Jang, S.K.; Kim, Y.K. Iron increases translation initiation directed by internal ribosome entry site of hepatitis C virus. Virus Genes 2008, 37, 154–160. [Google Scholar] [CrossRef]
- Pollard, V.W.; Malim, M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [Google Scholar] [CrossRef] [PubMed]
- Truman, C.T.; Jarvelin, A.; Davis, I.; Castello, A. HIV Rev-isited. Open Biol. 2020, 10, 200320. [Google Scholar] [CrossRef]
- Kumar, R.; Mehta, D.; Mishra, N.; Nayak, D.; Sunil, S. Role of Host-Mediated Post-Translational Modifications (PTMs) in RNA Virus Pathogenesis. Int. J. Mol. Sci. 2020, 22, 323. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Gao, F.H. The Molecular Mechanisms and the Role of hnRNP K Protein Post- Translational Modification in DNA Damage Repair. Curr. Med. Chem. 2017, 24, 614–621. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, W.; Han, Q.; Wang, Y.; Li, C.; Zhang, P.; Xu, H. Post-translational modification control of RNA-binding protein hnRNPK function. Open Biol. 2019, 9, 180239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, G.; Dhar, S.; Bedford, M.T. PRMT5 regulates IRES-dependent translation via methylation of hnRNP A1. Nucleic Acids Res. 2017, 45, 4359–4369. [Google Scholar] [CrossRef] [Green Version]
- Jo, O.D.; Martin, J.; Bernath, A.; Masri, J.; Lichtenstein, A.; Gera, J. Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin D1 and c-myc internal ribosome entry site function through Akt signaling. J. Biol. Chem. 2008, 283, 23274–23287. [Google Scholar] [CrossRef] [Green Version]
- Roy, R.; Durie, D.; Li, H.; Liu, B.Q.; Skehel, J.M.; Mauri, F.; Cuorvo, L.V.; Barbareschi, M.; Guo, L.; Holcik, M.; et al. hnRNPA1 couples nuclear export and translation of specific mRNAs downstream of FGF-2/S6K2 signalling. Nucleic Acids Res. 2014, 42, 12483–12497. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.L.; Lewis, S.M. Methylarginines within the RGG-Motif Region of hnRNP A1 Affect Its IRES Trans-Acting Factor Activity and Are Required for hnRNP A1 Stress Granule Localization and Formation. J. Mol. Biol. 2017, 429, 295–307. [Google Scholar] [CrossRef]
- Martin, J.; Masri, J.; Cloninger, C.; Holmes, B.; Artinian, N.; Funk, A.; Ruegg, T.; Anderson, L.; Bashir, T.; Bernath, A.; et al. Phosphomimetic substitution of heterogeneous nuclear ribonucleoprotein A1 at serine 199 abolishes AKT-dependent internal ribosome entry site-transacting factor (ITAF) function via effects on strand annealing and results in mammalian target of rapamycin complex 1 (mTORC1) inhibitor sensitivity. J. Biol. Chem. 2011, 286, 16402–16413. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Luo, M.; Chang, G.; Ren, W.; Wu, K.; Li, X.; Shen, J.; Zhao, X.; Hu, Y. Phosphorylation of Ser6 in hnRNPA1 by S6K2 regulates glucose metabolism and cell growth in colorectal cancer. Oncol Lett. 2017, 14, 7323–7331. [Google Scholar] [CrossRef] [Green Version]
- Velazquez-Cruz, A.; Banos-Jaime, B.; Diaz-Quintana, A.; De la Rosa, M.A.; Diaz-Moreno, I. Post-translational Control of RNA-Binding Proteins and Disease-Related Dysregulation. Front. Mol. Biosci. 2021, 8, 658852. [Google Scholar] [CrossRef] [PubMed]
- Kung, Y.A.; Hung, C.T.; Chien, K.Y.; Shih, S.R. Control of the negative IRES trans-acting factor KHSRP by ubiquitination. Nucleic Acids Res. 2017, 45, 271–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caceres, C.J.; Angulo, J.; Lowy, F.; Contreras, N.; Walters, B.; Olivares, E.; Allouche, D.; Merviel, A.; Pino, K.; Sargueil, B.; et al. Non-canonical translation initiation of the spliced mRNA encoding the human T-cell leukemia virus type 1 basic leucine zipper protein. Nucleic Acids Res. 2018, 46, 11030–11047. [Google Scholar] [CrossRef]
- Valiente-Echeverria, F.; Vallejos, M.; Monette, A.; Pino, K.; Letelier, A.; Huidobro-Toro, J.P.; Mouland, A.J.; Lopez-Lastra, M. A cis-acting element present within the Gag open reading frame negatively impacts on the activity of the HIV-1 IRES. PLoS ONE 2013, 8, e56962. [Google Scholar] [CrossRef]
- Holland, J.J. Receptor affinities as major determinants of enterovirus tissue tropisms in humans. Virology 1961, 15, 312–326. [Google Scholar] [CrossRef]
- Komar, A.A.; Hatzoglou, M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011, 10, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Evans, D.M.; Dunn, G.; Minor, P.D.; Schild, G.C.; Cann, A.J.; Stanway, G.; Almond, J.W.; Currey, K.; Maizel, J.V., Jr. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 1985, 314, 548–550. [Google Scholar] [CrossRef]
- Kauder, S.E.; Racaniello, V.R. Poliovirus tropism and attenuation are determined after internal ribosome entry. J. Clin. Investig. 2004, 113, 1743–1753. [Google Scholar] [CrossRef]
- Yanagiya, A.; Ohka, S.; Hashida, N.; Okamura, M.; Taya, C.; Kamoshita, N.; Iwasaki, K.; Sasaki, Y.; Yonekawa, H.; Nomoto, A. Tissue-specific replicating capacity of a chimeric poliovirus that carries the internal ribosome entry site of hepatitis C virus in a new mouse model transgenic for the human poliovirus receptor. J. Virol. 2003, 77, 10479–10487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilipenko, E.V.; Viktorova, E.G.; Guest, S.T.; Agol, V.I.; Roos, R.P. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 2001, 20, 6899–6908. [Google Scholar] [CrossRef]
- Guest, S.; Pilipenko, E.; Sharma, K.; Chumakov, K.; Roos, R.P. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol. 2004, 78, 11097–11107. [Google Scholar] [CrossRef] [Green Version]
- Domingo, E.; Garcia-Crespo, C.; Perales, C. Historical Perspective on the Discovery of the Quasispecies Concept. Annu. Rev. Virol. 2021, 8, 51–72. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Perales, C. Quasispecies and virus. Eur. Biophys. J. 2018, 47, 443–457. [Google Scholar] [CrossRef]
- Day, S.P.; Murphy, P.; Brown, E.A.; Lemon, S.M. Mutations within the 5' nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J. Virol. 1992, 66, 6533–6540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schultz, D.E.; Honda, M.; Whetter, L.E.; McKnight, K.L.; Lemon, S.M. Mutations within the 5' nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J. Virol. 1996, 70, 1041–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barria, M.I.; Gonzalez, A.; Vera-Otarola, J.; Leon, U.; Vollrath, V.; Marsac, D.; Monasterio, O.; Perez-Acle, T.; Soza, A.; Lopez-Lastra, M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 2009, 37, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Forton, D.M.; Karayiannis, P.; Mahmud, N.; Taylor-Robinson, S.D.; Thomas, H.C. Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants. J. Virol. 2004, 78, 5170–5183. [Google Scholar] [CrossRef] [Green Version]
- Laporte, J.; Malet, I.; Andrieu, T.; Thibault, V.; Toulme, J.J.; Wychowski, C.; Pawlotsky, J.M.; Huraux, J.M.; Agut, H.; Cahour, A. Comparative analysis of translation efficiencies of hepatitis C virus 5′ untranslated regions among intraindividual quasispecies present in chronic infection: Opposite behaviors depending on cell type. J. Virol. 2000, 74, 10827–10833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerat, H.; Shimizu, Y.K.; Lemon, S.M. Cell type-specific enhancement of hepatitis C virus internal ribosome entry site-directed translation due to 5' nontranslated region substitutions selected during passage of virus in lymphoblastoid cells. J. Virol. 2000, 74, 7024–7031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercuri, L.; Thomson, E.C.; Hughes, J.; Karayiannis, P. Quasispecies Changes with Distinctive Point Mutations in the Hepatitis C Virus Internal Ribosome Entry Site (IRES) Derived from PBMCs and Plasma. Adv. Virol. 2018, 2018, 4835252. [Google Scholar] [CrossRef]
- Piron, M.; Beguiristain, N.; Nadal, A.; Martinez-Salas, E.; Gomez, J. Characterizing the function and structural organization of the 5' tRNA-like motif within the hepatitis C virus quasispecies. Nucleic Acids Res. 2005, 33, 1487–1502. [Google Scholar] [CrossRef] [Green Version]
- Van Leeuwen, H.C.; Reusken, C.; Roeten, M.; Dalebout, T.J.; Riezu-Boj, J.I.; Ruiz, J.; Spaan, W.J.M. Evolution of naturally occurring 5′ non-translated region variants of hepatitis C virus genotype 1b in selectable replicons. J. Gen. Virol. 2004, 85, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Vopalensky, V.; Khawaja, A.; Roznovsky, L.; Mrazek, J.; Masek, T.; Pospisek, M. Characterization of Hepatitis C Virus IRES Quasispecies—From the Individual to the Pool. Front. Microbiol. 2018, 9, 731. [Google Scholar] [CrossRef]
- Floden, E.W.; Khawaja, A.; Vopalensky, V.; Pospisek, M. HCVIVdb: The hepatitis-C IRES variation database. BMC Microbiol. 2016, 16, 187. [Google Scholar] [CrossRef] [Green Version]
- Daude, C.; Decimo, D.; Trabaud, M.A.; Andre, P.; Ohlmann, T.; de Breyne, S. HIV-1 sequences isolated from patients promote expression of shorter isoforms of the Gag polyprotein. Arch. Virol. 2016, 161, 3495–3507. [Google Scholar] [CrossRef]
- Alexander, L.; Lu, H.H.; Wimmer, E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: Genetic hybrids and the expression of a foreign gene. Proc. Natl. Acad. Sci. USA 1994, 91, 1406–1410. [Google Scholar] [CrossRef] [Green Version]
- Gromeier, M.; Alexander, L.; Wimmer, E. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 1996, 93, 2370–2375. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Yang, D.; Gao, R.; Liang, T.; Wang, H.; Zhou, G.; Yu, L. Modification of the internal ribosome entry site element impairs the growth of foot-and-mouth disease virus in porcine-derived cells. J. Gen. Virol. 2016, 97, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.H.; Wimmer, E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 1996, 93, 1412–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reusken, C.; Dalebout, T.J.; Eerligh, P.; Bredenbeek, P.J.; Spaan, W.J.M. Analysis of hepatitis C virus/classical swine fever virus chimeric 5'NTRs: Sequences within the hepatitis C virus IRES are required for viral RNA replication. J. Gen. Virol. 2003, 84, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Gromeier, M.; Bossert, B.; Arita, M.; Nomoto, A.; Wimmer, E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 1999, 73, 958–964. [Google Scholar] [CrossRef] [Green Version]
- Ohlmann, T.; Jackson, R.J. The properties of chimeric picornavirus IRESes show that discrimination between internal translation initiation sites is influenced by the identity of the IRES and not just the context of the AUG codon. RNA 1999, 5, 764–778. [Google Scholar] [CrossRef] [Green Version]
- Merrill, M.K.; Dobrikova, E.Y.; Gromeier, M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein 76. J. Virol. 2006, 80, 3147–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromeier, M.; Lachmann, S.; Rosenfeld, M.R.; Gutin, P.H.; Wimmer, E. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA 2000, 97, 6803–6808. [Google Scholar] [CrossRef] [Green Version]
- Sadahiro, A.; Fukao, A.; Kosaka, M.; Funakami, Y.; Takizawa, N.; Takeuchi, O.; Duncan, K.E.; Fujiwara, T. Translation of Hepatitis A Virus IRES Is Upregulated by a Hepatic Cell-Specific Factor. Front. Genet. 2018, 9, 307. [Google Scholar] [CrossRef]
- Carter, J.R.; Fraser, T.S.; Fraser, M.J. Examining the relative activity of several dicistrovirus intergenic internal ribosome entry site elements in uninfected insect and mammalian cell lines. J. Gen. Virol. 2008, 89, 3150–3155. [Google Scholar] [CrossRef]
- Urwin, P.; Yi, L.; Martin, H.; Atkinson, H.; Gilmartin, P.M. Functional characterization of the EMCV IRES in plants. Plant J. 2000, 24, 583–589. [Google Scholar] [CrossRef] [Green Version]
- Thompson, S.R.; Gulyas, K.D.; Sarnow, P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. USA 2001, 98, 12972–12977. [Google Scholar] [CrossRef] [Green Version]
- Colussi, T.M.; Costantino, D.A.; Zhu, J.; Donohue, J.P.; Korostelev, A.A.; Jaafar, Z.A.; Plank, T.D.; Noller, H.F.; Kieft, J.S. Initiation of translation in bacteria by a structured eukaryotic IRES RNA. Nature 2015, 519, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Skulachev, M.V.; Ivanov, P.A.; Karpova, O.V.; Korpela, T.; Rodionova, N.P.; Dorokhov, Y.L.; Atabekov, J.G. Internal initiation of translation directed by the 5'-untranslated region of the tobamovirus subgenomic RNA I(2). Virology 1999, 263, 139–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, J.; Nakashima, N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999, 73, 1219–1226. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.J. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 2013, 5, a011569. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, K.A.; Bushell, M.; Mitchell, S.A.; Willis, A.E. Internal ribosome entry segment-mediated translation during apoptosis: The role of IRES-trans-acting factors. Cell Death Differ. 2005, 12, 585–591. [Google Scholar] [CrossRef] [Green Version]
- Spriggs, K.A.; Stoneley, M.; Bushell, M.; Willis, A.E. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 2008, 100, 27–38. [Google Scholar] [CrossRef] [PubMed]
- King, H.A.; Cobbold, L.C.; Willis, A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010, 38, 1581–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komar, A.A.; Hatzoglou, M. Exploring Internal Ribosome Entry Sites as Therapeutic Targets. Front. Oncol. 2015, 5, 233. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Holcik, M. Strong eukaryotic IRESs have weak secondary structure. PLoS ONE 2009, 4, e4136. [Google Scholar] [CrossRef]
- Chappell, S.A.; Edelman, G.M.; Mauro, V.P. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. USA 2000, 97, 1536–1541. [Google Scholar] [CrossRef] [Green Version]
- Chappell, S.A.; LeQuesne, J.P.; Paulin, F.E.; deSchoolmeester, M.L.; Stoneley, M.; Soutar, R.L.; Ralston, S.H.; Helfrich, M.H.; Willis, A.E. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: A novel mechanism of oncogene de-regulation. Oncogene 2000, 19, 4437–4440. [Google Scholar] [CrossRef] [Green Version]
- Owens, G.C.; Chappell, S.A.; Mauro, V.P.; Edelman, G.M. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc. Natl. Acad. Sci. USA 2001, 98, 1471–1476. [Google Scholar] [CrossRef] [Green Version]
- Le Quesne, J.P.; Stoneley, M.; Fraser, G.A.; Willis, A.E. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 2001, 310, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.M.; Leong, L.E.; Hoang, L.T.; Wang, P.H.; Gutman, G.A.; Semler, B.L. Structurally distinct elements mediate internal ribosome entry within the 5′-noncoding region of a voltage-gated potassium channel mRNA. J. Biol. Chem. 2004, 279, 47419–47430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holcik, M.; Gordon, B.W.; Korneluk, R.G. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003, 23, 280–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiroki, K.; Ohsawa, C.; Sugi, N.; Wakiyama, M.; Miura, K.; Watanabe, M.; Suzuki, Y.; Sugano, S. Internal ribosome entry site-mediated translation of Smad5 in vivo: Requirement for a nuclear event. Nucleic Acids Res. 2002, 30, 2851–2861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoneley, M.; Subkhankulova, T.; Le Quesne, J.P.; Coldwell, M.J.; Jopling, C.L.; Belsham, G.J.; Willis, A.E. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000, 28, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.K.; Cheng, R.; Demeter, J.; Chen, J.; Weingarten-Gabbay, S.; Jiang, L.; Snyder, M.P.; Weissman, J.S.; Segal, E.; Jackson, P.K.; et al. Structured elements drive extensive circular RNA translation. Mol. Cell 2021, 81, 4300–4318.e13. [Google Scholar] [CrossRef]
- Prats, A.C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the Key for Translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [Green Version]

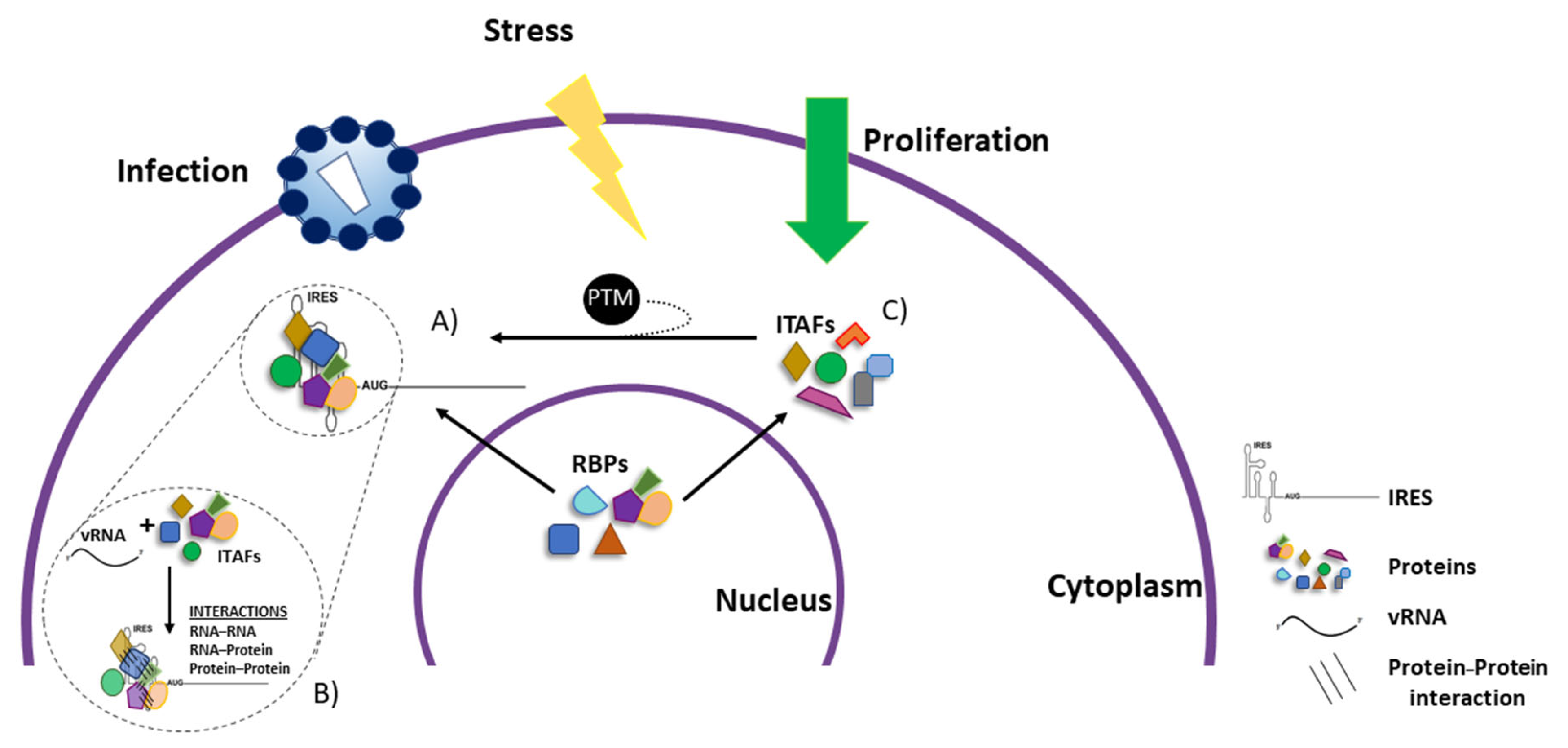


| ITAF | Viral IRES (Type of Modulation) | Binding Region | Main RNA Binding Domain (RBD) | Domain Description/Structure | Type of RNA that is Bound |
|---|---|---|---|---|---|
| hnRNPI (PTB) | Picornavirus EMCV (promotes) [259] | IRES [259] | RNA-recognition motif (RRM) | RRM is involved in different processes of gene expression, it has two consensus sequences RNP1 (Lys/Arg-Gly-Phe/Tyr-Gly/Ala-Phe/Tyr-Val/Ile/ Leu-X-Phe/Tyr), and RNP2 (Ile/Val/Leu-Phe/Tyr-Ile/Val/Leu-XAsn-Leu), where X can be any amino acid [260,261]. The RRM has an αβ sandwich fold. The RNP1 and RNP2 motifs are positioned in the central strands of the β sheet [260,261]. | ssRNA [261,262]. |
| Picornavirus EV71 (promotes) [263] | 5′ UTR [263] | ||||
| Picornavirus PV (promotes) [264] | IRES (Domain V) [264] | ||||
| Picornavirus HRV (promotes) [250] | IRES [174] | ||||
| Picornavirus HAV (promotes) [265] | 5′ UTR [265] | ||||
| Picornavirus CVB3 (promotes) [266] | 3′ and 5′ UTR [266] | ||||
| Flavivirus HCV (promotes) [267] | IRES [267] | ||||
| Retrovirus MMTV (promotes) [268] | 5′ UTR [268] | ||||
| Nucleolin | Picornavirus PV(promotes) [269] | IRES [269] | |||
| Picornavirus HRV (promotes) [269] | Unknown | ||||
| Picornavirus FMDV (promotes) [270] | Unknown | ||||
| hnRNPD (AUF1) | Picornavirus PV, HRV (reduces) [253] | 5′ UTR [253] | |||
| Picornavirus EV71 (reduces) [254] | 5′ UTR (stem loop II) [254] | ||||
| Flavivirus HCV (promotes) [271] | 5′ UTR [271] | ||||
| La | Picornavirus PV (promotes) [272] | 5′ UTR [272] | |||
| Picornavirus EMCV (promotes) [230] | IRES [230] | ||||
| Picornavirus CVB3 (promotes) [273] | 3′ and 5′ UTR [273] | ||||
| Picornavirus HAV (reduces) [274] | 5′ UTR [274] | ||||
| Flavivirus HCV (promotes) [272] | 5′ UTR [272] | ||||
| PSF | Picornavirus CVB3 (promotes) [240] | IRES [240] | |||
| SRp20 | Picornavirus PV (promotes) [248,249] | stem-loop IV [248,249] | |||
| hnRNP L | Flavivirus HCV (promotes) [275] | IRES [275] | |||
| hnRNP Q | Flavivirus HCV (promotes) [276,277] | downstream of the initiation codon [276,277] | |||
| hnRNP C1/C2 | Picornavirus CVB3 (reduces) [278] | IRES (Stem-Loop V) [278] | |||
| hnRNP K | Picornavirus EV71 (promotes) [279] | 5′ UTR (cloverleaf) [279] | K-homology domain (KH) | Involved in gene expression processes. Comprises about 70 residues with a hydrophobic cleft, formed by a Gly-X-X-Gly segment and a variable loop. KH domains are present in multiples copies [262,280,281]. Two or three alpha-helices clustered on the surface of antiparallel beta-sheets [262]. | ssRNA [262]. |
| Flavivirus HCV (promotes) [282] | 5′ UTR [282] | ||||
| Picornavirus FMDV (reduces) [256] | IRES (II, III and IV domains) [256] | ||||
| FBP-2 | Picornavirus EV71 (reduces) [251] | IRES [251] | |||
| hnRNPE1(PCBP1) | Picornavirus EV71 (promotes) [283] | 5′ UTR [283] | |||
| Picornavirus PV (promotes) [284] | 5′ UTR (cloverleaf) [284] | ||||
| Picornavirus HRV (promotes) [284] | IRES [284] | ||||
| hnRNPE2(PCBP2) | Picornavirus PV (promotes) [232,233,284] | 5′ UTR (cloverleaf) [284] | |||
| Picornavirus HRV (promotes) [284] | IRES [284] | ||||
| Picornavirus HAV (promotes) [285,286] | 5′ UTR (pyrimidine-rich tract) [285,286] | ||||
| Flavivirus HCV (promotes) [284] | IRES [284] | ||||
| Sam68 | Picornavirus EV71 (promotes) [239] | 5′ UTR (Stem-loops IV and V) [239] | |||
| Staufen 1 | Flavivirus HCV (promotes) [287] | 3′UTR (variable-stem-loop region) and IRES (domain IIId) [287] | Double-stranded RBD (dsRBD) | Domain of 70–90 amino acids, involved in RNA maturation [262,288,289,290]. Conserved topology α1-L1-β1-L2-β2-L3-b3-L4-α2, where L specifies a loop [262] | dsRNA [262,288,289,290]. |
| Picornavirus EV71 (promotes) [291] | 5′ UTR [291] | ||||
| Retrovirus HIV-1 (promotes) [292] | 5′ UTR [292] | ||||
| HuR | Picornavirus EV71 (promotes) [241] | Stem-loop II [241] | |||
| Flavivirus HCV (promotes) [224,293] | Unknown | ||||
| Retrovirus HIV-1 (reduces) [224] | Does not bind the IRES [224] | ||||
| Gemin 5 | Picornavirus FMDV (reduces) [257] | IRES [257] | |||
| Flavivirus HCV (reduces) [257] | IRES [257] | ||||
| DRBP76 | Picornavirus HRV 2 (reduces in neuronal cells) [258] | IRES (sldV/VI) [258] | |||
| Ebp1 | Picornavirus FMDV (promotes) [234] | IRES (Domain I) [234] | |||
| Unr | Picornavirus HRV (promotes) [237] | 5′ UTR [236] | Cold shock domain (CSD) | CSD is part of catalytic centers by binding different biomolecules. Participates in various processes of gene expression regulation [294]. Five-stranded antiparallel beta-barrel structure [294] | ssRNA [294] |
| Picornavirus PV (promotes) [237] | Unknown | ||||
| Picornavirus HRV-2 (promotes) [238] | 5′ UTR [238] | ||||
| eIF5A | Retrovirus (HIV-1, HTLV-1, MMTV) (promotes) [295,296] | Unknown | |||
| hnRNP A1 | Retrovirus HIV-1 (promotes) [213] | 5′ UTR [225] | RGG or GAR boxes | RBPs containing these repeats function in RNA metabolism processes such as transcription, splicing of pre-mRNA, localization, and post-translational modification [297]. Gly-rich regions are interspersed with aromatic and Arg residues and are present in disorder protein regions [298] | G-quadruplexes [299] |
| Retrovirus HTLV-1 (promotes) [225] | Unknown | ||||
| Retrovirus MMTV (promotes) [225] | Unknown | ||||
| Picornavirus EV71 (promotes) [300] | Stem-loop II and VI [300] | ||||
| Ago2 | Picornavirus EV71 (promotes) [241] | EV71 IRES (stem-loop II) [241]. | P-element-induced whimpy testes (PIWI) domain, a middle (MID) domain, and a PIWI/Argonaute/Zwille (PAZ) domain [301]. | Required in gene silencing by the RNA-induced silencing complex (RISC) [301,302]. Bi-lobed architecture composed of four globular domains (N-terminal(N), PIWI, MID, and PAZ connected through two structured linker domains [301]. | ssRNA [301,302]. |
| Flavivirus HCV (promotes) [303] | Stabilizes mir-122/HCV RNA interaction [303,304]. | ||||
| HSPA6 | Picornavirus EV-A71 (promotes) [245] | Unknown | Nucleotide-binding domain (NBD), also known as the ATPase domain. | Found in Hsp70 family proteins with ATP-regulated chaperone function [305]. Two lobes connected by α-helix in C-terminal [305]. | unknown |
| HSPA8 | Picornavirus EV-A71 (promotes) [243] | Does not bind to the 5′-UTR [243] | |||
| HSPA1 and HSPA9 | Picornavirus EV-A71 (promotes) [244] | Unknown |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ulloa, B.; Fuentes, Y.; Pizarro-Ortega, M.S.; López-Lastra, M. RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation. Viruses 2022, 14, 188. https://doi.org/10.3390/v14020188
López-Ulloa B, Fuentes Y, Pizarro-Ortega MS, López-Lastra M. RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation. Viruses. 2022; 14(2):188. https://doi.org/10.3390/v14020188
Chicago/Turabian StyleLópez-Ulloa, Brenda, Yazmín Fuentes, Magdalena S. Pizarro-Ortega, and Marcelo López-Lastra. 2022. "RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation" Viruses 14, no. 2: 188. https://doi.org/10.3390/v14020188
APA StyleLópez-Ulloa, B., Fuentes, Y., Pizarro-Ortega, M. S., & López-Lastra, M. (2022). RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation. Viruses, 14(2), 188. https://doi.org/10.3390/v14020188






