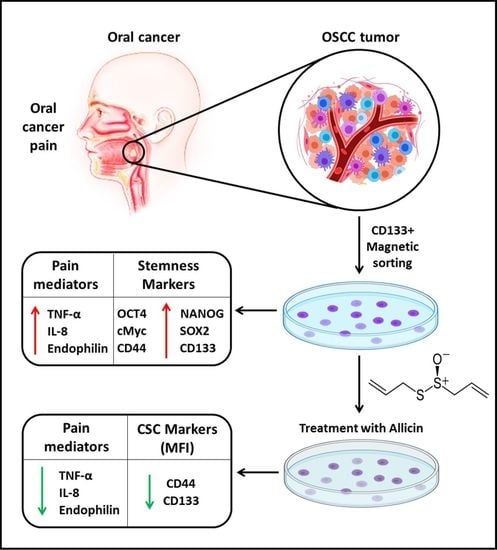
Allicin Could Potentially Alleviate Oral Cancer Pain by Inhibiting “Pain Mediators” TNF-alpha, IL-8, and Endothelin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Ethical Permissions
2.2. Preparation of the Single-Cell Suspension
2.3. Treatment Groups
2.4. 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
2.5. Magnetic Sorting of CD133+ Cells
2.6. RT-PCR for Stemness Markers and Gene Expression Analysis of TNF-alpha, IL-8, and Endothelin
2.7. ELISA for Analysis of Protein Levels of TNF-alpha, IL-8, and Endothelin
2.8. Statistical Analysis
3. Results
3.1. Sorted CD133+ Cells Show High Expression of Stemness and Pluripotency Markers
3.2. High Concentration of Allicin Does Not Affect Cell Viability Significantly
3.3. Allicin Downregulates Gene Expression of Pain Mediators in a Dose-Dependent Manner
3.4. Allicin Inhibits the Secretion of Pain Mediators at the Protein Level in a Dose-Dependent Manner
3.5. Allicin Significantly Reduces the Expression of Cancer Stem Cell Markers CD44 and CD133 in a Dose-Dependent Manner
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bagan, J.; Sarrion, G.; Jimenez, Y. Oral cancer: Clinical features. Oral Oncol. 2010, 46, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Morimata, J.; Otomaru, T.; Murase, M.; Haraguchi, M.; Sumita, Y.; Taniguchi, H. Investigation of factor affecting health-related quality of life in head and neck cancer patients. Gerodontology 2013, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, P.; Li, W. Comparison of orofacial pain of patients with different stages of precancer and oral cancer. Sci. Rep. 2017, 7, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, J.B.; Elad, S.; Eliav, E.; Jurevic, R.; Benoliel, R. Orofacial Pain in Cancer: Part II—Clinical Perspectives and Management. J. Dent. Res. 2007, 86, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.J.; Klasser, G.D.; Epstein, J.B. Cancer and Orofacial Pain. Oral Maxillofac. Surg. Clin. N. Am. 2008, 20, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Lam, D.K.; Dang, D.; Flynn, A.N.; Hardt, M.; Schmidt, B.L. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain 2015, 156, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benoliel, R.; Epstein, J.; Eliav, E.; Jurevic, R.; Elad, S. Orofacial Pain in Cancer: Part I—Mechanisms. J. Dent. Res. 2007, 86, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-C.; Liao, C.-T.; Chang, J.T.-C. Orofacial pain and predictors in oral squamous cell carcinoma patients receiving treatment. Oral Oncol. 2011, 47, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Viet, C.T.; Schmidt, B.L. Biologic Mechanisms of Oral Cancer Pain and Implications for Clinical Therapy. J. Dent. Res. 2012, 91, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Kamstra, J.I.; Jager-Wittenaar, H.; Dijkstra, P.U.; Huisman, P.M.; van Oort, R.P.; van der Laan, B.F.A.M.; Roodenburg, J.L.N. Oral symptoms and functional outcome related to oral and oropharyngeal cancer. Support. Care Cancer 2011, 19, 1327–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruparel, S.; Bendele, M.; Wallace, A.; Green, D. Released Lipids Regulate Transient Receptor Potential Channel (TRP)-Dependent Oral Cancer Pain. Mol. Pain 2015, 11, s12990015–s12990016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, J.; Yamazaki, Y.; Satoh, A.; Notani, K.; Kitagawa, Y. Pain is associated with an endophytic cancer growth pattern in patients with oral squamous cell carcinoma before treatment. Odontology 2010, 98, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, V.; Nepal, A.; Olaisen, C.; Bachke, S.; Hira, J.; Søgaard, C.; Røst, L.; Misund, K.; Andreassen, T.; Melø, T.; et al. Anti-Cancer Potential of Homemade Fresh Garlic Extract Is Related to Increased Endoplasmic Reticulum Stress. Nutrients 2018, 10, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Yang, D. Allicin suppresses the migration and invasion in cervical cancer cells mainly by inhibiting NRF2. Exp. Ther. Med. 2019, 17, 1523–1528. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, K.G.; von Zeidler, S.V.; Lamas, A.Z.; de Podestá, J.R.V.; Sena, A.; Souza, E.D.; Lenzi, J.; Lemos, E.M.; Gouvea, S.A.; Bissoli, N.S. Relationship of inflammatory markers and pain in patients with head and neck cancer prior to anticancer therapy. Braz. J. Med. Biol. Res. 2014, 47, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.; Cahill, C.M. TNF-α and neuropathic pain—A review. J. Neuroinflamm. 2010, 7, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, P.K.; Chandra, G.; Bogra, J.; Gupta, R.; Kumar, V.; Hussain, S.R.; Jain, A.; Mahdi, A.A.; Ahmad, M.K. Association of Genetic Polymorphism in the Interleukin-8 Gene with Risk of Oral Cancer and Its Correlation with Pain. Biochem. Genet. 2016, 54, 95–106. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, G.A.G.; Hinsley, E.E.; Hunter, K.; Lambert, D.W. The endothelin axis in head and neck cancer: A promising therapeutic opportunity? J. Oral Pathol. Med. 2014, 43, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Kopruszinski, C.M.; dos Reis, R.C.; Gambeta, E.; Acco, A.; Rae, G.A.; King, T.; Chichorro, J.G. Blockade of endothelin receptors reduces tumor-induced ongoing pain and evoked hypersensitivity in a rat model of facial carcinoma induced pain. Eur. J. Pharmacol. 2018, 818, 132–140. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| TNF-alpha | 5′-CCG ATG GGT TGT ACC TTG TC-3′ | 5′-GGGCTG GGT AGA GAA TGG AT-3′ |
| IL-8 | 5′-GTG CAG TTT TGC CAA GGA GT-3′ | 5′-TTA TGA ATT CTC AGC CCT CTT CAA-3′ |
| Endothelin 1 (ET-1) | 5′-CTT TGA GGG ACC TGA AGC TG-3′ | 5′-AGT TCT TTT CCT GCT TGG CA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamir, A.H.; Patil, S. Allicin Could Potentially Alleviate Oral Cancer Pain by Inhibiting “Pain Mediators” TNF-alpha, IL-8, and Endothelin. Curr. Issues Mol. Biol. 2021, 43, 187-196. https://doi.org/10.3390/cimb43010016
Alamir AH, Patil S. Allicin Could Potentially Alleviate Oral Cancer Pain by Inhibiting “Pain Mediators” TNF-alpha, IL-8, and Endothelin. Current Issues in Molecular Biology. 2021; 43(1):187-196. https://doi.org/10.3390/cimb43010016
Chicago/Turabian StyleAlamir, Abdulwahab H., and Shankargouda Patil. 2021. "Allicin Could Potentially Alleviate Oral Cancer Pain by Inhibiting “Pain Mediators” TNF-alpha, IL-8, and Endothelin" Current Issues in Molecular Biology 43, no. 1: 187-196. https://doi.org/10.3390/cimb43010016
APA StyleAlamir, A. H., & Patil, S. (2021). Allicin Could Potentially Alleviate Oral Cancer Pain by Inhibiting “Pain Mediators” TNF-alpha, IL-8, and Endothelin. Current Issues in Molecular Biology, 43(1), 187-196. https://doi.org/10.3390/cimb43010016