Proteomic Characterization of Colorectal Cancer Tissue from Patients Identifies Novel Putative Protein Biomarkers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Biopsies from Patients with CRC
2.2. Two-dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) and Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS) for Protein Identification
2.3. Western Blotting
3. Results
3.1. Patient Characteristics
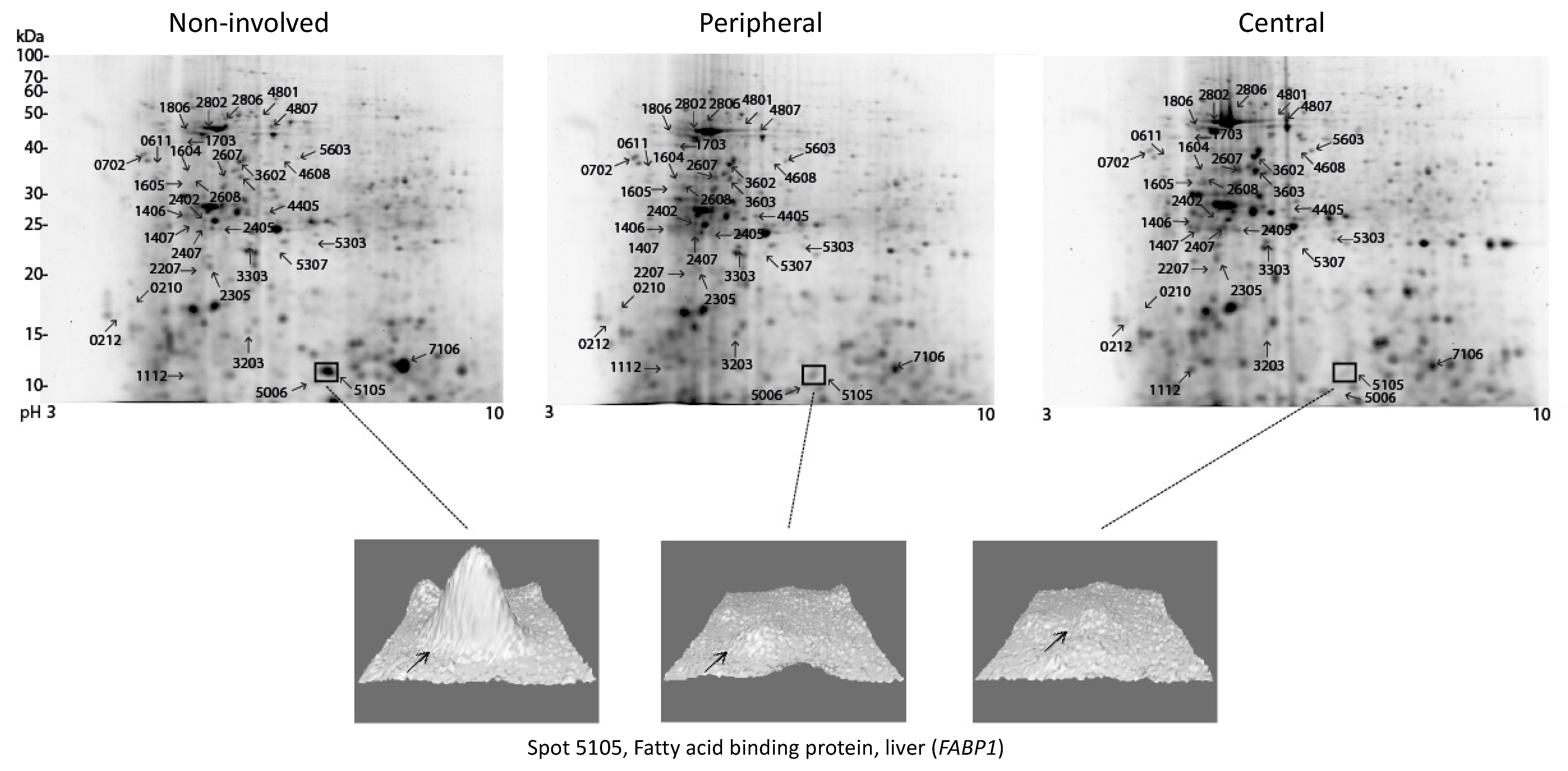
3.2. Protein Expression Patterns in the Central versus the Peripheral Part of the Tumor
3.3. Proteomic Comparison of Various Parts of the Tumor with Non-involved Colorectal Tissue
3.4. Protein Expression Patterns in High TNM Stage versus Low TNM Stage
3.5. Immunological Evaluation of Identified Differentially Expressed Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nambiar, P.R.; Gupta, R.R.; Misra, V. An Omics based survey of human colon cancer. Mutat. Res. 2010, 693, 3–18. [Google Scholar] [CrossRef]
- Lindhorst, P.H.; Hummon, A.B. Proteomics of Colorectal Cancer: Tumors, Organoids, and Cell Cultures—A Minireview. Front. Mol. Biosci. 2020, 7, 604492. [Google Scholar] [CrossRef]
- Ludvigsen, M.; Thorlacius-Ussing, L.; Vorum, H.; Moyer, M.P.; Stender, M.T.; Thorlacius-Ussing, O.; Honoré, B. Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 3466. [Google Scholar] [CrossRef]
- Moyer, M.P.; Manzano, L.A.; Merriman, R.L.; Stauffer, J.S.; Tanzer, L.R. NCM460, a normal human colon mucosal epithelial cell line. In Vitro Cell. Dev. Biol. Anim. 1996, 32, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Leibovitz, A.; Stinson, J.C.; McCombs, W.B., III; McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar]
- Brattain, M.G.; Fine, W.D.; Khaled, F.M.; Thompson, J.; Brattain, D.E. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981, 41, 1751–1756. [Google Scholar] [PubMed]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknæs, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Sugihara, Y.; Taniguchi, H.; Kushima, R.; Tsuda, H.; Kubota, D.; Ichikawa, H.; Fujita, S.; Kondo, T. Laser microdissection and two-dimensional difference gel electrophoresis reveal proteomic intra-tumor heterogeneity in colorectal cancer. J. Proteom. 2012, 78, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.C.; Lim, Y.M. Proteome Heterogeneity in Colorectal Cancer. Proteomics 2018, 18, 1700169. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Chen, J.; Pande, M.; Lynch, P.M.; Frazier, M.L. A pilot study comparing protein expression in different segments of the normal colon and rectum and in normal colon versus adenoma in patients with Lynch syndrome. J. Cancer Res. Clin. Oncol 2013, 139, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Gajadhar, A.S.; Johnson, H.; Slebos, R.J.; Shaddox, K.; Wiles, K.; Washington, M.K.; Herline, A.J.; Levine, D.A.; Liebler, D.; White, F.M. Phosphotyrosine Signaling Analysis in Human Tumors Is Confounded by Systemic Ischemia-Driven Artifacts and Intra-Specimen Heterogeneity. Cancer Res. 2015, 75, 1495–1503. [Google Scholar] [CrossRef] [Green Version]
- Ludvigsen, M.; Pedersen, M.B.; Lauridsen, K.L.; Poulsen, T.; Hamilton-Dutoit, S.J.; Besenbacher, S.; Bendix, K.; Møller, M.B.; Nørgaard, P.; D’Amore, F.; et al. Proteomic profiling identifies outcome-predictive markers in patients with peripheral T-cell lymphoma, not otherwise specified. Blood Adv. 2018, 2, 2533–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Kamper, P.; Ludvigsen, M.; Bendix, K.; Hamilton-Dutoit, S.; Rabinovich, G.A.; Møller, M.B.; Nyengaard, J.R.; Honoré, B.; D’Amore, F. Proteomic analysis identifies galectin-1 as a predictive biomarker for relapsed/refractory disease in classical Hodgkin lymphoma. Blood 2011, 117, 6638–6649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Du, S.; Yu, J.; Yang, X.; Yang, C.; Zhou, D.; Wang, Q.; Qin, S.; Yan, X.; He, L.; et al. Common housekeeping proteins are upregulated in colorectal adenocarcinoma and hepatocellular carcinoma, making the total protein a better housekeeper. Oncotarget 2016, 7, 66679–66688. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Luo, H.; Wang, R.; Wu, W.W.; Phue, J.-N.; Shen, R.-F.; Juhl, H.; Wu, L.; Alterovitz, W.-L.; Simonyan, V.; et al. Novel reference genes in colorectal cancer identify a distinct subset of high stage tumors and their associated histologically normal colonic tissues. BMC Med. Genet. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Honoré, B.; Vorum, H.; Baandrup, U. hnRNPs H, H′ and F behave differently with respect to posttranslational cleavage and subcellular localization. FEBS Lett. 1999, 456, 274–280. [Google Scholar] [CrossRef]
- Smith, L.M.; The Consortium for Top Down Proteomics; Kelleher, N.L. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [Green Version]
- Stulík, J.; Koupilová, K.; Österreicher, J.; Knížek, J.; Macela, A.; Bureš, J.; Jandík, P.; Langr, F.; Dědič, K.; Jungblut, P.R. Protein abundance alterations in matched sets of macroscopically normal colon mucosa and colorectal carcinoma. Electrophoresis 1999, 20, 3638–3646. [Google Scholar] [CrossRef]
- Melle, C.; Ernst, G.; Schimmel, B.; Bleul, A.; Mothes, H.; Kaufmann, R.; Settmacher, U.; Von Eggeling, F. Different expression of calgizzarin (S100A11) in normal colonic epithelium, adenoma and colorectal carcinoma. Int. J. Oncol. 2006, 28, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Melle, C.; Ernst, G.; Schimmel, B.; Bleul, A.; von Eggeling, F. Colon-Derived Liver Metastasis, Colorectal Carcinoma, and Hepatocellular Carcinoma Can Be Discriminated by the Ca2+-Binding Proteins S100A6 and S100A11. PLoS ONE 2008, 3, e3767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, M.-L.; Zhu, X.-J.; Liu, J.; Shi, P.-C.; Kang, Y.-L.; Lin, Z.; Cao, Y.-P. An Integrated Bioinformatic Analysis of the S100 Gene Family for the Prognosis of Colorectal Cancer. BioMed Res. Int. 2020, 2020, 4746929. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Wang, F.; Ji, Q.; Geng, H.; Yan, X.; Wang, L.; Tie, W.; Liu, X.; Thorne, R.F.; Liu, G.; et al. Proteome Analyses Reveal S100A11, S100P, and RBM25 Are Tumor Biomarkers in Colorectal Cancer. Proteom. Clin. Appl. 2020, 15, e2000056. [Google Scholar] [CrossRef]
- Moravkova, P.; Kohoutova, D.; Vavrova, J.; Bures, J. Serum S100A6, S100A8, S100A9 and S100A11 proteins in colorectal neoplasia: Results of a single centre prospective study. Scand. J. Clin. Lab. Investig. 2019, 80, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, L.C.; Dundas, S.R.; Curran, S.; I Murray, G. Liver fatty acid binding protein expression in colorectal neoplasia. Br. J. Cancer 2004, 90, 1955–1960. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.-L.; Pan, L.-L.; Huang, T.; Wang, J.-H. The transcriptome difference between colorectal tumor and normal tissues revealed by single-cell sequencing. J. Cancer 2019, 10, 5883–5890. [Google Scholar] [CrossRef]
- Honoré, B.; Baandrup, U.; Vorum, H. Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res. 2004, 294, 199–209. [Google Scholar] [CrossRef]
- Takahashi, K.; Fujiya, M.; Konishi, H.; Murakami, Y.; Iwama, T.; Sasaki, T.; Kunogi, T.; Sakatani, A.; Ando, K.; Ueno, N.; et al. Heterogenous Nuclear Ribonucleoprotein H1 Promotes Colorectal Cancer Progression through the Stabilization of mRNA of Sphingosine-1-Phosphate Lyase 1. Int. J. Mol. Sci. 2020, 21, 4514. [Google Scholar] [CrossRef]
- Xu, C.; Xie, N.; Su, Y.; Sun, Z.; Liang, Y.; Zhang, N.; Liu, D.; Jia, S.; Xing, X.; Han, L.; et al. HnRNP F/H associate with hTERC and telomerase holoenzyme to modulate telomerase function and promote cell proliferation. Cell Death Differ. 2019, 27, 1998–2013. [Google Scholar] [CrossRef]
- Hall, E.R.; Cottam, G.L. Isozymes of pyruvate kinase in vertebrates: Their physical, chemical, kinetic and immunological properties. Int. J. Biochem. 1978, 9, 785–794. [Google Scholar] [CrossRef]
- Kanno, H.; Fujii, H.; Miwa, S. Structural analysis of human pyruvate kinase L-gene and identification of the promoter activity in erythroid cells. Biochem. Biophys. Res. Commun. 1992, 188, 516–523. [Google Scholar] [CrossRef]
- Takenaka, M.; Noguchi, T.; Sadahiro, S.; Hirai, H.; Yamada, K.; Matsuda, T.; Imai, E.; Tanaka, T. Isolation and characterization of the human pyruvate kinase M gene. Eur. J. Biochem. 1991, 198, 101–106. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, Y.-S.; Kang, S.W.; Kim, S.; Kim, T.-Y.; Lee, S.-H.; Hwang, S.W.; Kim, J.; Kim, E.N.; Ju, J.-S.; et al. Loss of PKM2 in Lgr5+ intestinal stem cells promotes colitis-associated colorectal cancer. Sci. Rep. 2019, 9, 6212. [Google Scholar] [CrossRef] [Green Version]
- Howie, A.; Forrester, L.; Glancey, M.; Schlager, J.; Beckett, G.; Hayes, J.; Wolf, C.; Powis, G. Glutathione S-transferase and glutathione peroxidase expression in normal and tumour human tissues. Carcinogenesis 1990, 11, 451–458. [Google Scholar] [CrossRef]
- Kodate, C.; Fukushi, A.; Narita, T.; Kudo, H.; Soma, Y.; Sato, K. Human placental form of glutathione S-transferase (GST-pi) as a new immunohistochemical marker for human colonic carcinoma. Jpn. J. Cancer Res. 1986, 77, 226–229. [Google Scholar]
- Ranganathan, S.; Tew, K.D. Immunohistochemical localization of glutathione S-transferases alpha, mu, and pi in normal tissue and carcinomas from human colon. Carcinogenesis 1991, 12, 2383–2387. [Google Scholar] [CrossRef]
- Bulus, H.; Oguztuzun, S.; Simsek, G.G.; Kılıç, M.; Ada, A.O.; Göl, S.; Kocdogan, A.K.; Kaygın, P.; Bozer, B.; Iscan, M. Expression of CYP and GST in human normal and colon tumor tissues. Biotech. Histochem. 2018, 94, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hengstler, J.G.; Böttger, T.; Tanner, B.; Dietrich, B.; Henrich, M.; Knapstein, P.G.; Junginger, T.; Oesch, F. Resistance factors in colon cancer tissue and the adjacent normal colon tissue: Glutathione S-transferases α and π, glutathione and aldehyde dehydrogenase. Cancer Lett. 1998, 128, 105–112. [Google Scholar] [CrossRef]
- Nasrabadi, P.N.; Nayeri, Z.; Gharib, E.; Salmanipour, R.; Masoomi, F.; Mahjoubi, F.; Zomorodipour, A. Establishment of a CALU, AURKA, and MCM2 gene panel for discrimination of metastasis from primary colon and lung cancers. PLoS ONE 2020, 15, e0233717. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Q.; Fan, Y.; He, X. Reticulocalbin 2 correlates with recurrence and prognosis in colorectal cancer. Am. J. Cancer Res. 2017, 7, 2169–2179. [Google Scholar] [PubMed]
- Komatsu, K.; Andoh, A.; Ishiguro, S.; Suzuki, N.; Hunai, H.; Kobune-Fujiwara, Y.; Kameyama, M.; Miyoshi, J.; Akedo, H.; Nakamura, H. Increased expression of S100A6 (Calcyclin), a calcium-binding protein of the S100 family, in human colorectal adenocarcinomas. Clin. Cancer Res. 2000, 6, 172–177. [Google Scholar] [PubMed]


| Patient ID No. | Sex | Age by the Time of Diagnosis | Localization | T | N | M | TNM-Stage | Rt | Recurrence | Death Due to CRC |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | M | 63 | Colon | 4 | 2 | 1 | High | No | NA | Yes |
| 13 | M | 73 | Colon | 3 | 1 | 1 | High | No | NA | Yes |
| 17 | M | 58 | Colon | 4 | 2 | 0 | High | No | Yes | Yes |
| 20 | F | 75 | Rectum | 2 | 0 | 0 | Low | Yes | No | - |
| 25 | M | 65 | Colon | 3 | 0 | 0 | Low | No | Yes | - |
| 40 | M | 69 | Colon | 2 | 0 | 0 | Low | No | No | - |
| 46 | M | 76 | Colon | 3 | 0 | 0 | Low | No | Yes | - |
| 48 | F | 80 | Colon | 3 | 0 | 1 | High | No | NA | Yes |
| 78 | F | 54 | Colon | 4 | 1 | 1 | High | No | NA | Yes |
| 97 | M | 79 | Colon | 3 | 0 | 0 | Low | No | No | - |
| Fold Change | |||||||
|---|---|---|---|---|---|---|---|
| Spot No. | Protein ID (Gene) | C/N | P/N | P/C | TNMH/TNML | ||
| P | C | N | |||||
| 0210 | MYL6 | 0.44 | |||||
| 0212 | TUBB2A fr * | 3.20 | 3.50 | ||||
| 0611 | SERPINA1 fr HSP90B1 fr or HSP90AB1 fr | 2.43 | |||||
| 0702 | SERPINA1 fr | 0.53 | |||||
| 1112 | TXN | 3.19 | 2.84 | ||||
| 1406 | ALB fr | 2.07 | |||||
| 1407 | ACTB fr; ENO1 fr; ALB fr; FGG fr | 3.72 | 3.37 | ||||
| 1604 | ATP5F1B fr | 0.50 | |||||
| 1605 | ACTB fr | 3.73 | 5.27 | ||||
| 1703 | HSP8A fr | 0.35 | |||||
| 1806 | HSP8A fr; VIM fr HSPA5 fr; gltX fr | 3.04 | 3.36 | ||||
| 2207 | NI | 0.32 | |||||
| 2305 | NI | 0.21 | |||||
| 2402 | NI | 2.04 | |||||
| 2405 | HNRNPF fr | 1.68 | 1.85 | ||||
| 2407 | HSPA5 fr; ALB fr. | 2.22 | |||||
| 2607 | TUBA1A fr; Actin fr | 2.08 | 2.38 | ||||
| 2608 | betaTub56D fr | 2.81 | |||||
| 2802 | actb | 0.32 | |||||
| 2806 | NI | 0.28 | |||||
| 3203 | NI | 0.28 | |||||
| 3303 | GSTP1; PRDX2; CRKL fr; EZR fr | 2.85 | 2.62 | ||||
| 3602 | Tubulin fr; Actin | 2.35 | |||||
| 3603 | Actin fr ACTN1 fr PDIA6 fr Tubulin fr NCL fr SRSF1 | 0.45 | |||||
| 4405 | PKM fr PRDX4; ATP5F1B fr; | 2.07 | 1.85 | ||||
| 4608 | TCP1 fr; PKM fr | 2.29 | |||||
| 4801 | LUM | 0.17 | |||||
| 4807 | NI | 0.21 | |||||
| 5006 | S100A11 | 5.50 | |||||
| 5105 | FABP1 | 0.29 | 0.23 | ||||
| 5303 | HNRNPH1/HNRNPH2 fr. RAB11A/RAB11B | 3.04 | |||||
| 5307 | HNRNPH1/HNRNPH2 fr; COL1A2 fr | 3.13 | 2.67 | ||||
| 5603 | PKM fr | 2.03 | |||||
| 7106 | HBB | 0.17 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludvigsen, M.; Thorlacius-Ussing, L.; Vorum, H.; Stender, M.T.; Thorlacius-Ussing, O.; Honoré, B. Proteomic Characterization of Colorectal Cancer Tissue from Patients Identifies Novel Putative Protein Biomarkers. Curr. Issues Mol. Biol. 2021, 43, 1043-1056. https://doi.org/10.3390/cimb43020074
Ludvigsen M, Thorlacius-Ussing L, Vorum H, Stender MT, Thorlacius-Ussing O, Honoré B. Proteomic Characterization of Colorectal Cancer Tissue from Patients Identifies Novel Putative Protein Biomarkers. Current Issues in Molecular Biology. 2021; 43(2):1043-1056. https://doi.org/10.3390/cimb43020074
Chicago/Turabian StyleLudvigsen, Maja, Louise Thorlacius-Ussing, Henrik Vorum, Mogens Tornby Stender, Ole Thorlacius-Ussing, and Bent Honoré. 2021. "Proteomic Characterization of Colorectal Cancer Tissue from Patients Identifies Novel Putative Protein Biomarkers" Current Issues in Molecular Biology 43, no. 2: 1043-1056. https://doi.org/10.3390/cimb43020074
APA StyleLudvigsen, M., Thorlacius-Ussing, L., Vorum, H., Stender, M. T., Thorlacius-Ussing, O., & Honoré, B. (2021). Proteomic Characterization of Colorectal Cancer Tissue from Patients Identifies Novel Putative Protein Biomarkers. Current Issues in Molecular Biology, 43(2), 1043-1056. https://doi.org/10.3390/cimb43020074







