Evaluation of Microbiome Alterations Following Consumption of BIOHM, a Novel Probiotic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of BIOHM Probiotic
2.2. Participants
2.3. HMP Patient Comparison Selection
2.4. DNA Extraction
2.5. Bacterial 16S rRNA Gene or Pan Fungal ITS Amplicon Library Preparation
2.6. Next-Generation Sequencing, Classification, and Analysis
2.7. Statistical Analyses
3. Results
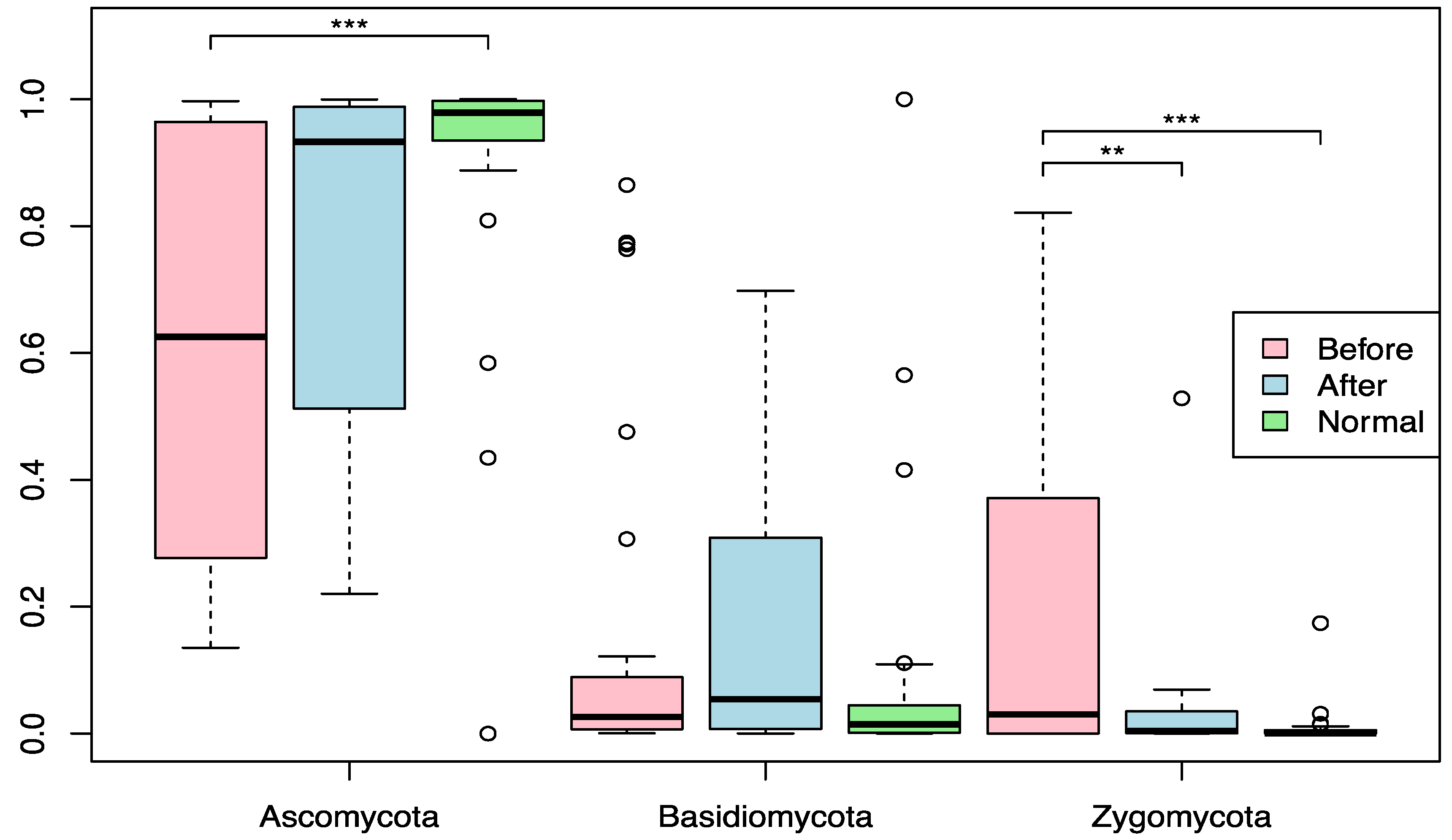
3.1. Effect of BIOHM on the Mycobiome Community
3.2. Effect of BIOHM on Candida Genus and Species Level
3.3. Effect of BIOHM on the Bacteriome Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hoarau, G.; Mukherjee, P.K.; Gower, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 2016, 7, e01250-16. [Google Scholar] [CrossRef] [Green Version]
- Hager, C.L.; Isham, N.; Schrom, K.P.; Chandra, J.; McCormick, T.; Miyagi, M.; Ghannoum, M.A. Effects of a Novel Probiotic Combination on Pathogenic Bacterial-Fungal Polymicrobial Biofilms. mBio 2019, 10, e00338-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guarner, F.; Khan, A.G.; Garisch, J.; Eliakim, R.; Gangl, A.; Thomson, A.; Krabshuis, J.; LeMair, A.; Kaufmann, P.; De Paula, J.A.; et al. World Gastroenterology Organisation Global Guidelines. J. Clin. Gastroenterol. 2012, 46, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Sartor, R.B. Probiotic therapy of intestinal inflammation and infections. Curr. Opin. Gastroenterol. 2005, 21, 44–50. [Google Scholar] [PubMed]
- Jakubczyk, D.; Leszczyńska, K.; Górska, S. The Effectiveness of Probiotics in the Treatment of Inflammatory Bowel Disease (IBD)—A Critical Review. Nutrients 2020, 12, 1973. [Google Scholar] [CrossRef] [PubMed]
- Jorjão, A.L.; De Oliveira, F.E.; Leão, M.V.P.; Carvalho, C.A.T.; Jorge, A.O.C.; Oliveira, L. Live and Heat-KilledLactobacillus rhamnosusATCC 7469 May Induce Modulatory Cytokines Profiles on Macrophages RAW 264.7. Sci. World J. 2015, 2015, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, K.A.; O’Hara, A.M.; Van Pijkeren, J.-P.; Douillard, F.P.; O’Toole, P. Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J. Med. Microbiol. 2009, 58, 996–1005. [Google Scholar] [CrossRef] [Green Version]
- Wickens, K.; Black, P.N.; Stanley, T.V.; Mitchell, E.; Fitzharris, P.; Tannock, G.W.; Purdie, G.; Crane, J. A differential effect of 2 probiotics in the prevention of eczema and atopy: A double-blind, randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 2008, 122, 788–794. [Google Scholar] [CrossRef] [PubMed]
- De Seta, F.; Parazzini, F.; De Leo, R.; Banco, R.; Maso, G.; De Santo, D.; Sartore, A.; Stabile, G.; Inglese, S.; Tonon, M.; et al. Lactobacillus plantarum P17630 for preventing Candida vaginitis recurrence: A retrospective comparative study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 182, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.Y.; Cheah, Y.K.; Seow, H.F.; Sandai, D.; Than, L.T.L. Probiotic L actobacillus rhamnosus GR-1 and L actobacillus reuteri RC-14 exhibit strong antifungal effects against vulvovaginal candidiasis-causing C andida glabrata isolates. J. Appl. Microbiol. 2015, 118, 1180–1190. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Reid, G.; Challis, J.R.; Gloor, G.B.; Asztalos, E.; Money, D.; Seney, S.; Bocking, A.D. Effect of Oral Probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on the Vaginal Microbiota, Cytokines and Chemokines in Pregnant Women. Nutrients 2020, 12, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, R.; Wei, J.; Ma, D.; Jiang, L.; Dan, H.; Zhou, Y.; Ji, N.; Zeng, X.; Chen, Q. A meta-analysis of randomized trials assessing the effects of probiotic preparations on oral candidiasis in the elderly. Arch. Oral Biol. 2017, 83, 187–192. [Google Scholar] [CrossRef]
- Hayama, K.; Ishijima, S.; Ono, Y.; Izumo, T.; Ida, M.; Shibata, H.; Abe, S. Protective activity of S-PT84, a heat-killed preparation of Lactobacillus pentosus, against oral and gastric candidiasis in an experimental murine model. Nippon. Ishinkin Gakkai Zasshi 2014, 55, J123–J129. [Google Scholar]
- Zitvogel, L.; Galluzzi, L.; Viaud, S.; Vétizou, M.; Daillère, R.; Merad, M.; Kroemer, G. Cancer and the gut microbiota: An unexpected link. Sci. Transl. Med. 2015, 7, 271ps1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alard, J.; Peucelle, V.; Boutillier, D.; Breton, J.; Kuylle, S.; Pot, B.; Holowacz, S.; Grangette, C. New probiotic strains for inflammatory bowel disease management identified by combining in vitro and in vivo approaches. Benef. Microbes 2018, 9, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Dore, M.P.; Bibbò, S.; Fresi, G.; Bassotti, G.; Pes, G.M. Side Effects Associated with Probiotic Use in Adult Patients with Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Nutrients 2019, 11, 2913. [Google Scholar] [CrossRef] [Green Version]
- Dore, M.P.; Rocchi, C.; Longo, N.P.; Scanu, A.M.; Vidili, G.; Padedda, F.; Pes, G.M. Effect of Probiotic Use on Adverse Events in Adult Patients with Inflammatory Bowel Disease: A Retrospective Cohort Study. Probiotics Antimicrob. Proteins 2019, 12, 152–159. [Google Scholar] [CrossRef]
- Fatmawati, N.N.D.; Gotoh, K.; Mayura, I.P.B.; Nocianitri, K.A.; Suwardana, G.N.R.; Komalasari, N.L.G.Y.; Ramona, Y.; Sakaguchi, M.; Matsushita, O.; Sujaya, I.N. Enhancement of intestinal epithelial barrier function by Weissella confusa F213 and Lactobacillus rhamnosus FBB81 probiotic candidates in an in vitro model of hydrogen peroxide-induced inflammatory bowel disease. BMC Res. Notes 2020, 13, 1–7. [Google Scholar] [CrossRef]
- Ghavami, S.B.; Yadegar, A.; Aghdaei, H.A.; Sorrentino, D.; Farmani, M.; Mir, A.S.; Azimirad, M.; Balaii, H.; Shahrokh, S.; Zali, M.R. Immunomodulation and Generation of Tolerogenic Dendritic Cells by Probiotic Bacteria in Patients with Inflammatory Bowel Disease. Int. J. Mol. Sci. 2020, 21, 6266. [Google Scholar] [CrossRef]
- Kumar, M.; Hemalatha, R.; Nagpal, R.; Singh, B.; Parasannanavar, D.; Verma, V.; Kumar, A.; Marotta, F.; Catanzaro, R.; Cuffari, B.; et al. Probiotic Approaches for Targeting Inflammatory Bowel Disease: An Update on Advances and Opportunities in Managing the Disease. Int. J. Probiotics Prebiotics 2016, 11, 99–116. [Google Scholar]
- Sato, N.; Yuzawa, M.; Aminul, I.; Tomokiyo, M.; Albarracin, L.; Garcia-Castillo, V.; Ideka-Ohtsubo, W.; Iwabuchi, N.; Xiao, J.-Z.; Garcia-Cancino, A.; et al. Evaluation of Porcine Intestinal Epitheliocytes as an In vitro Immunoassay System for the Selection of Probiotic Bifidobacteria to Alleviate Inflammatory Bowel Disease. Probiotics Antimicrob. Proteins 2021, 13, 824–836. [Google Scholar] [CrossRef]
- White, R.; Atherly, T.; Guard, B.; Rossi, G.; Wang, C.; Mosher, C.; Webb, C.; Hill, S.; Ackermann, M.; Sciabarra, P.; et al. Randomized, controlled trial evaluating the effect of multi-strain probiotic on the mucosal microbiota in canine idiopathic inflammatory bowel disease. Gut Microbes 2017, 8, 451–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craigen, B.; Dashiff, A.; Kadouri, D.E. The Use of Commercially Available Alpha-Amylase Compounds to Inhibit and Remove Staphylococcus aureus Biofilms. Open Microbiol. J. 2011, 5, 21–31. [Google Scholar]
- Kalpana, B.J.; Aarthy, S.; Pandian, S.K. Antibiofilm Activity of α-Amylase from Bacillus subtilis S8-18 Against Biofilm Forming Human Bacterial Pathogens. Appl. Biochem. Biotechnol. 2012, 167, 1778–1794. [Google Scholar] [CrossRef]
- Vaikundamoorthy, R.; Rajendran, R.; Selvaraju, A.; Moorthy, K.; Perumal, S. Development of thermostable amylase enzyme from Bacillus cereus for potential antibiofilm activity. Bioorganic Chem. 2018, 77, 494–506. [Google Scholar] [CrossRef]
- Hager, C.L.; Ghannoum, M.A. The mycobiome: Role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig. Liver Dis. 2017, 49, 1171–1176. [Google Scholar] [CrossRef]
- Dressman, J.B.; Berardi, R.R.; Dermentzoglou, L.C.; Russell, T.L.; Schmaltz, S.P.; Barnett, J.L.; Jarvenpaa, K.M. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm. Res. 1990, 7, 756–761. [Google Scholar] [CrossRef]
- Bezkorovainy, A. Probiotics: Determinants of survival and growth in the gut. Am. J. Clin. Nutr. 2001, 73, 399s–405s. [Google Scholar] [CrossRef]
- Ghannoum, M.; Ghannoum, A.; Long, L.; Sun, P.L.; Isham, N. BIOHM Probiotics Retain Viability in Low pH Environments Simulating the Digestive Environment. J. Probiotics Heal. 2019, 07, 1–4. [Google Scholar]
- Andrews, G. The slow food story. Sound. 2006, 31, 88–102. [Google Scholar] [CrossRef]
- The 2017 NIH-Wide Microbiome Workshop Writing Team. 2017 NIH-wide workshop report on “The Human Microbiome: Emerging Themes at the Horizon of the 21st Century”. Microbiome 2019, 7, 32. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinform. 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.; DeCoffe, D.; Molcan, E.; Gibson, D.L. Diet-Induced Dysbiosis of the Intestinal Microbiota and the Effects on Immunity and Disease. Nutrients 2012, 4, 1095–1119. [Google Scholar] [CrossRef] [Green Version]
- Jumpertz, R.; Le, D.S.; Turnbaugh, P.; Trinidad, C.; Bogardus, C.; Gordon, J.I.; Krakoff, J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 2011, 94, 58–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deepak, P.; Loftus, E.V., Jr. Ustekinumab in treatment of Crohn’s disease: Design, development, and potential place in therapy. Drug Des. Dev. Ther. 2016, 10, 3685–3698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allez, M.; Karmiris, K.; Louis, E.; Van Assche, G.; Ben-Horin, S.; Klein, A.; Van Der Woude, J.; Baert, F.; Eliakim, R.; Katsanos, K.; et al. Report of the ECCO pathogenesis workshop on anti-TNF therapy failures in inflammatory bowel diseases: Definitions, frequency and pharmacological aspects. J. Crohn’s Coliti 2010, 4, 355–366. [Google Scholar] [CrossRef]
- Abraham, B.; Quigley, E.M.M. Antibiotics and probiotics in inflammatory bowel disease: When to use them? Front. Gastroenterol. 2020, 11, 62–69. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Rainer, B.M.; Thompson, K.G.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Kang, S.; Chien, A.L. Impact of lifestyle and demographics on the gut microbiota of acne patients and the response to minocycline. J. Dermatol. Treat. 2020, 32, 1–2. [Google Scholar]
- Cerikcioglu, N.; Ilki, A.; Bilgen, H.; Ozek, E.; Metin, F.; Kalacs, S. The relationships between candidemia and candidal colonization and virulence factors of the colonizing strains in preterm infants. Turk. J. Pediatr. 2004, 46, 245–250. [Google Scholar]
- Coronado-Castellote, L.; Jiménez-Soriano, Y. Clinical and microbiological diagnosis of oral candidiasis. J. Clin. Exp. Dent. 2013, 5, e279–e286. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Álvarez, J.A.; Pérez-García, L.A.; Flores-Carreón, A.; Mora-Montes, H.M. The immune response against Candida spp. and Sporothrix schenckii. Revista Iberoamericana de Micología 2014, 31, 62–66. [Google Scholar] [CrossRef]
- Samonis, G.; Falagas, M.E.; Lionakis, S.; Ntaoukakis, M.; Kofteridis, D.P.; Ntalas, I.; Maraki, S. Saccharomyces boulardiiandCandida albicansexperimental colonization of the murine gut. Med. Mycol. 2011, 49, 395–399. [Google Scholar] [CrossRef] [Green Version]
- Miyazima, T.Y.; Ishikawa, K.H.; Mayer, M.; Saad, S.; Nakamae, A. Cheese supplemented with probiotics reduced theCandidalevels in denture wearers-RCT. Oral Dis. 2017, 23, 919–925. [Google Scholar] [CrossRef]
- Ribeiro, F.; De Barros, P.; Rossoni, R.; Junqueira, J.; Jorge, A. Lactobacillus rhamnosusinhibitsCandida albicansvirulence factorsin vitroand modulates immune system inGalleria mellonella. J. Appl. Microbiol. 2016, 122, 201–211. [Google Scholar] [CrossRef]
- Rossoni, R.; Fuchs, B.B.; de Barros, P.P.; Velloso, M.D.S.; Jorge, A.O.C.; Junqueira, J.C.; Mylonakis, E. Lactobacillus paracasei modulates the immune system of Galleria mellonella and protects against Candida albicans infection. PLOS ONE 2017, 12, e0173332. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghannoum, M.A.; McCormick, T.S.; Retuerto, M.; Bebek, G.; Cousineau, S.; Hartman, L.; Barth, C.; Schrom, K. Evaluation of Microbiome Alterations Following Consumption of BIOHM, a Novel Probiotic. Curr. Issues Mol. Biol. 2021, 43, 2135-2146. https://doi.org/10.3390/cimb43030148
Ghannoum MA, McCormick TS, Retuerto M, Bebek G, Cousineau S, Hartman L, Barth C, Schrom K. Evaluation of Microbiome Alterations Following Consumption of BIOHM, a Novel Probiotic. Current Issues in Molecular Biology. 2021; 43(3):2135-2146. https://doi.org/10.3390/cimb43030148
Chicago/Turabian StyleGhannoum, Mahmoud A., Thomas S. McCormick, Mauricio Retuerto, Gurkan Bebek, Susan Cousineau, Lynn Hartman, Charles Barth, and Kory Schrom. 2021. "Evaluation of Microbiome Alterations Following Consumption of BIOHM, a Novel Probiotic" Current Issues in Molecular Biology 43, no. 3: 2135-2146. https://doi.org/10.3390/cimb43030148





