Comparative Analysis of the Transcriptomes of Persisting and Abscised Fruitlets: Insights into Plant Hormone and Carbohydrate Metabolism Regulated Self-Thinning of Pecan Fruitlets during the Early Stage
Abstract
:1. Introduction
2. Experimental Section
2.1. Plant Materials, Fruit Development, and Drop Dynamics Analysis
2.2. RNA Isolation and cDNA Library Preparation and Sequencing
2.3. Analysis of Differential Gene Expression
2.4. Plant Hormone Content Analysis
3. Results
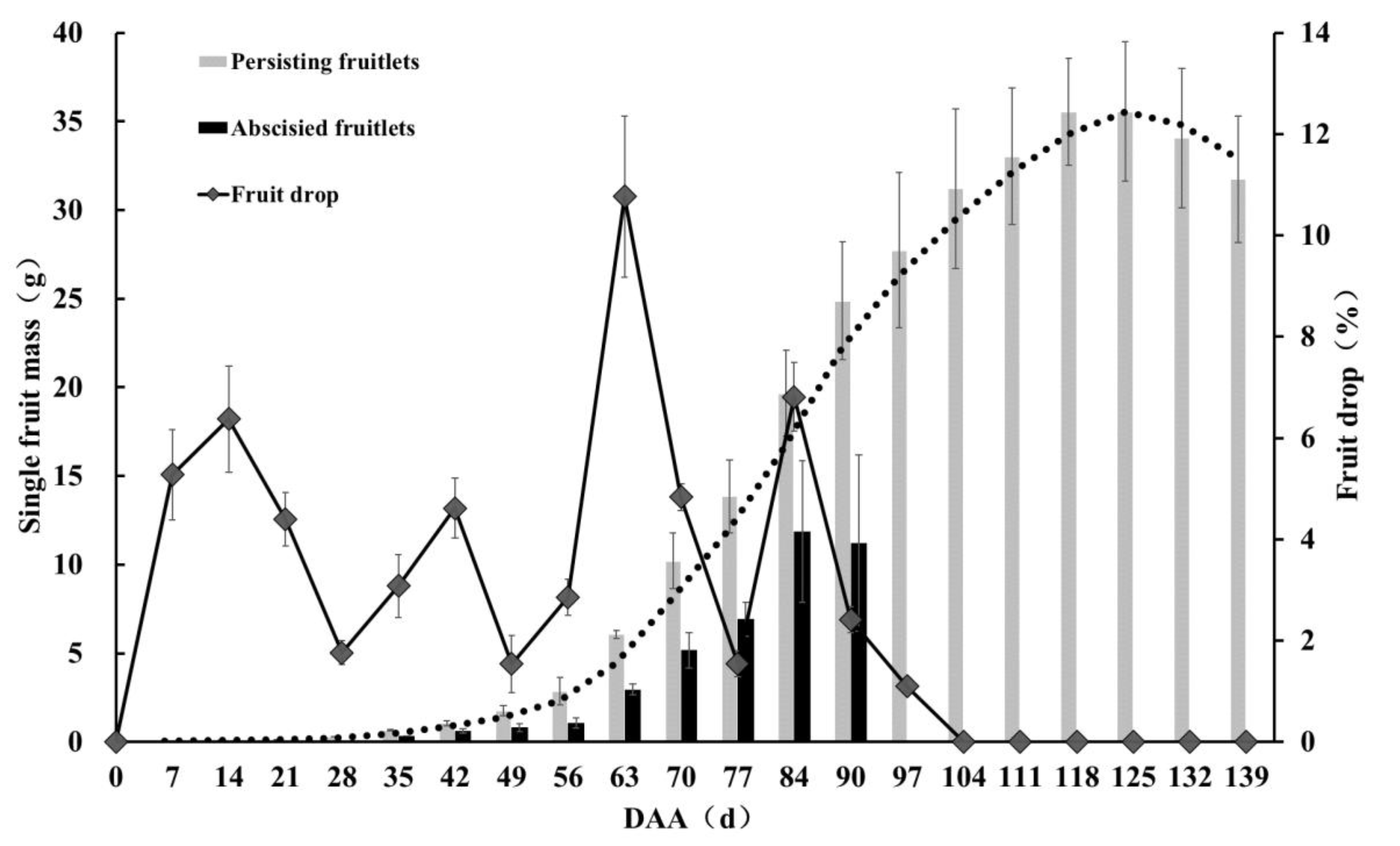
3.1. Analysis of Pecan Fruit Drop Dynamics
3.2. RNA Sequencing of Transcriptomes of Persisting and Abscised Fruitlets and Mapping of RNA Sequences to Reference Genome
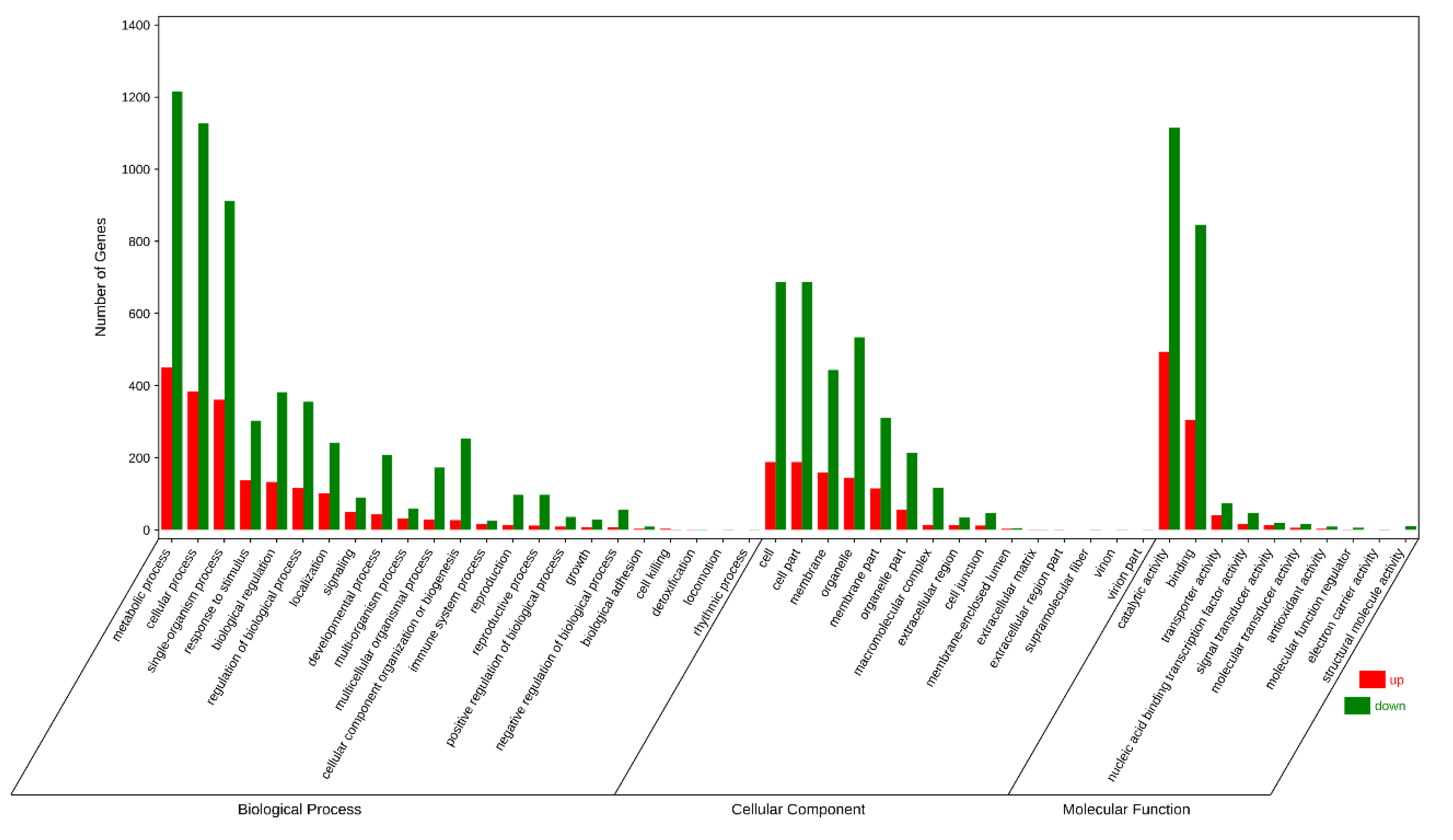
3.3. Differentially Expressed Gene Analysis
3.4. Identification of Differentially Expressed Proteins Involved in Plant Hormone Signal Transduction
3.5. Key Identified Differentially Expressed Proteins Involved in Starch and Sucrose Metabolism and Galactose Metabolism
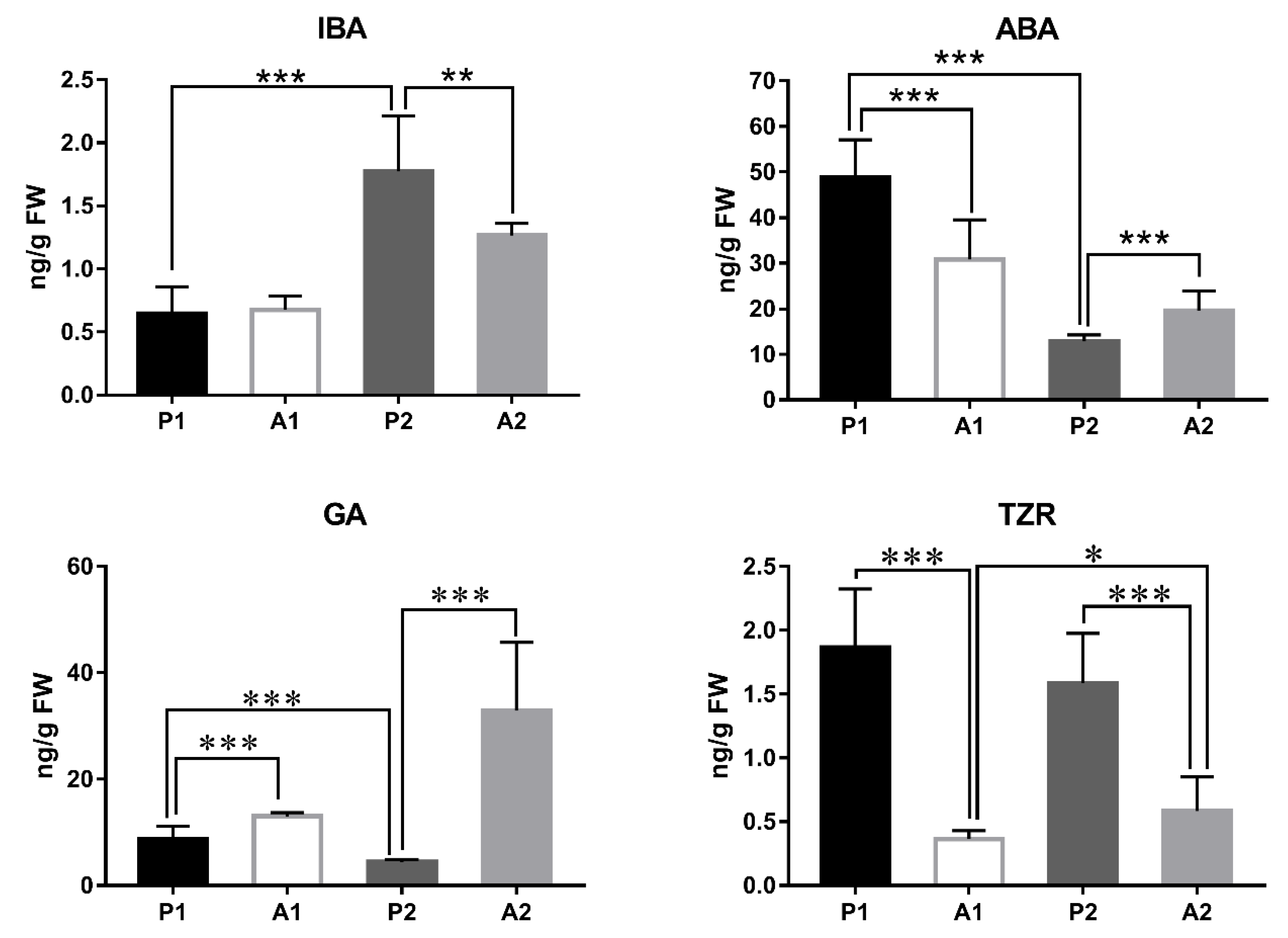
3.6. Plant Hormone Content Analysis
3.7. Response of Transcription Factors in the Comparison of A1 vs. P1
4. Discussion
4.1. Increased IBA Content and Decreased ABA and GA Might Promote Fruit Development in Pecan
4.2. Plant Hormones Seem to Play a Key Role during the Abscission Progress in Pecan
4.3. Reduced Sugar Supply in Abscised Fruitlets Is One Reason for Fruitlet Abscission in Pecan
4.4. NAC Transcription Factors Participate in Fruit Development or Abscission Process
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Estornell, L.H.; Agusti, J.; Merelo, P.; Talon, M.; Tadeo, F.R. Elucidating mechanisms underlying organ abscission. Plant Sci. 2013, 199–200, 48–60. [Google Scholar] [CrossRef]
- Patharkar, O.R.; Walker, J.C. Advances in abscission signaling. J. Exp. Bot. 2018, 69, 733–740. [Google Scholar] [CrossRef]
- Eccher, G.; Begheldo, M.; Boschetti, A.; Ruperti, B.; Botton, A. Roles of ethylene production and ethylene receptor expression in regulating apple fruitlet abscission. Plant Physiol. 2015, 169, 125–137. [Google Scholar] [CrossRef] [Green Version]
- Giulia, E.; Alessandro, B.; Mariano, D.; Andrea, B.; Benedetto, R.; Angelo, R. Early induction of apple fruitlet abscission is characterized by an increase of both isoprene emission and abscisic acid content. Plant Physiol. 2013, 161, 1952–1969. [Google Scholar] [CrossRef] [Green Version]
- Botton, A.; Eccher, G.; Forcato, C.; Ferrarini, A.; Begheldo, M.; Zermiani, M.; Moscatello, S.; Battistelli, A.; Velasco, R.; Ramina, R.A.J.P.P. Signaling pathways mediating the induction of apple fruitlet abscission. Plant Physiol. 2011, 155, 185–208. [Google Scholar] [CrossRef] [Green Version]
- Celton, J.-M.; Dheilly, E.; Guillou, M.-C.; Simonneau, F.; Juchaux, M.; Costes, E.; Laurens, F.; Renou, J.-P. Additional amphivasal bundles in pedicel pith exacerbate central fruit dominance and induce self-thinning of lateral fruitlets in apple. Plant Physiol. 2014, 164, 1930–1951. [Google Scholar] [CrossRef] [Green Version]
- Sundaresan, S.; Philosoph-Hadas, S.; Riov, J.; Mugasimangalam, R.; Kuravadi, N.A.; Kochanek, B.; Salim, S.; Tucker, M.L.; Meir, S. De novo transcriptome sequencing and development of abscission zone-specific microarray as a new molecular tool for analysis of tomato organ abscission. Front. Plant Sci. 2015, 6, 1258. [Google Scholar] [CrossRef] [Green Version]
- Sundaresan, S.; Philosoph-Hadas, S.; Ma, C.; Jiang, C.Z.; Riov, J.; Mugasimangalam, R.; Kochanek, B.; Salim, S.; Reid, M.S.; Meir, S. The tomato hybrid proline-rich protein regulates the abscission zone competence to respond to ethylene signals. Hortic. Res. 2018, 5, 28. [Google Scholar] [CrossRef] [Green Version]
- de Jong, M.; Mariani, C.; Vriezen, W.H. The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 2009, 60, 1523–1532. [Google Scholar] [CrossRef] [Green Version]
- Nitsch, L.M.; Oplaat, C.; Feron, R.; Ma, Q.; Wolters-Arts, M.; Hedden, P.; Mariani, C.; Vriezen, W.H. Abscisic acid levels in tomato ovaries are regulated by LeNCED1 and SlCYP707A1. Planta 2009, 229, 1335–1346. [Google Scholar] [CrossRef] [Green Version]
- Vriezen, W.H.; Feron, R.; Maretto, F.; Keijman, J.; Mariani, C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 2008, 177, 60–76. [Google Scholar] [CrossRef]
- Cin, V.D.; Danesin, M.; Boschetti, A.; Dorigoni, A.; Ramina, A. Ethylene biosynthesis and perception in apple fruitlet abscission (Malus domestica L. Borck). J. Exp. Bot. 2005, 56, 2995–3005. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, N.; Serrano, A.; Abello, C.; Arce, A.; Espinoza, C.; Gouthu, S.; Deluc, L.; Arce-Johnson, P. Regulation of polar auxin transport in grapevine fruitlets (Vitis vinifera L.) and the proposed role of auxin homeostasis during fruit abscission. BMC Plant Biol. 2016, 16, 234. [Google Scholar] [CrossRef] [Green Version]
- Rook, F.; Corke, F.; Baier, M.; Holman, R.; May, A.G.; Bevan, M.W. Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J. 2006, 46, 1045–1058. [Google Scholar] [CrossRef]
- Rorat, T. Plant dehydrins--tissue location, structure and function. Cell. Mol. Biol. Lett. 2006, 11, 536–556. [Google Scholar] [CrossRef]
- Herrera-Rodriguez, M.B.; Maldonado, J.M.; Perez-Vicente, R. Light and metabolic regulation of HAS1, HAS1.1 and HAS2, three asparagine synthetase genes in Helianthus annuus. Plant Physiol. Biochem. 2004, 42, 511–518. [Google Scholar] [CrossRef]
- Grauke, L.J.; Wood, B.W.; Harris, M.K. Crop Vulnerability: Carya. HortScience 2016, 51, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Poletto, T.; Muniz, M.F.B.; Poletto, I.; Baggiotto, C. Methods for overcome dormancy of pecan carya illinoinensis (wangenh.) k. koch seeds. Rev. Árvore 2015, 39, 1111–1118. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Jia, G. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [Green Version]
- Xin, Z.; Zhao, Y.; Zheng, Z.L. Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 2005, 139, 1350–1365. [Google Scholar] [CrossRef] [Green Version]
- Lancien, M.; Roberts, M.R. Regulation of Arabidopsis thaliana 14-3-3 gene expression by gamma-aminobutyric acid. Plant Cell Environ. 2006, 29, 1430–1436. [Google Scholar] [CrossRef] [Green Version]
- Avonce, N.; Leyman, B.; Mascorro-Gallardo, J.O.; Van Dijck, P.; Thevelein, J.M.; Iturriaga, G. The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 2004, 136, 3649–3659. [Google Scholar] [CrossRef] [Green Version]
- Guan, X.; Xu, T.; Gao, S.; Qi, M.; Wang, Y.; Liu, X.; Li, T. Temporal and spatial distribution of auxin response factor genes during tomato flower abscission. J. Plant Growth Regul. 2013, 33, 317–327. [Google Scholar] [CrossRef]
- Meir, S.; Philosoph-Hadas, S.; Sundaresan, S.; Selvaraj, K.S.; Burd, S.; Ophir, R.; Kochanek, B.; Reid, M.S.; Jiang, C.Z.; Lers, A. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 2010, 154, 1929–1956. [Google Scholar] [CrossRef]
- Kuang, J.F.; Wu, J.Y.; Zhong, H.Y.; Li, C.Q.; Chen, J.Y.; Lu, W.J.; Li, J.G. Carbohydrate stress affecting fruitlet abscission and expression of genes related to auxin signal transduction pathway in litchi. Int. J. Mol. Sci. 2012, 13, 16084–16103. [Google Scholar] [CrossRef]
- Zuo, X.; Xu, T.; Qi, M.; Lv, S.; Li, J.; Gao, S.; Li, T. Expression patterns of auxin-responsive genes during tomato flower pedicel abscission and potential effects of calcium. Aust. J. Bot. 2012, 60, 68. [Google Scholar] [CrossRef]
- Zhu, H.; Dardick, C.D.; Beers, E.P.; Callanhan, A.M.; Xia, R.; Yuan, R. Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biol. 2011, 11, 138. [Google Scholar] [CrossRef] [Green Version]
- Hou, K.; Wu, W.; Gan, S.S. SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Kant, S.; Bi, Y.M.; Zhu, T.; Rothstein, S.J. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009, 151, 691–701. [Google Scholar] [CrossRef] [Green Version]
- Xie, R.; Dong, C.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. Comprehensive analysis of SAUR gene family in citrus and its transcriptional correlation with fruitlet drop from abscission zone A. Funct. Integr. Genom. 2015, 15, 729–740. [Google Scholar] [CrossRef]
- Baena-Gonzalez, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Stitt, M.; Wilke, I.; Feil, R.; Heldt, H.W. Coarse control of sucrose-phosphate synthase in leaves: Alterations of the kinetic properties in response to the rate of photosynthesis and the accumulation of sucrose. Planta 1988, 174, 217–230. [Google Scholar] [CrossRef]
- Signora, L.; Galtier, N.; Skøt, L.; Lucas, H.; Foyer, C.H. Over-expression of sucrose phosphate synthase in Arabidopsis thaliana results in increased foliar sucrose/starch ratios and favours decreased foliar carbohydrate accumulation in plants after prolonged growth with CO2 enrichment. J. Exp. Bot. 1998, 49, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Park, J.Y.; Canam, T.; Kang, K.Y.; Ellis, D.D.; Mansfield, S.D. Over-expression of an arabidopsis family A sucrose phosphate synthase (SPS) gene alters plant growth and fibre development. Transgenic Res. 2008, 17, 181–192. [Google Scholar] [CrossRef]
- Farcuh, M.; Li, B.; Rivero, R.M.; Shlizerman, L.; Sadka, A.; Blumwald, E. Sugar metabolism reprogramming in a non-climacteric bud mutant of a climacteric plum fruit during development on the tree. J. Exp. Bot. 2017, 68, 5813–5828. [Google Scholar] [CrossRef] [Green Version]
- Opassiri, R.; Pomthong, B.; Onkoksoong, T.; Akiyama, T.; Esen, A.; Ketudat Cairns, J.R. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 2006, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Seung, D.; Soyk, S.; Coiro, M.; Maier, B.A.; Eicke, S.; Zeeman, S.C. PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 2015, 13, e1002080. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.M.; Fulton, D.C.; Chia, T.; Thorneycroft, D.; Chapple, A.; Dunstan, H.; Hylton, C.; Zeeman, S.C.; Smith, A.M. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004, 136, 2687–2699. [Google Scholar] [CrossRef] [Green Version]
- Kajiura, H.; Takata, H.; Akiyama, T.; Kakutani, R.; Furuyashiki, T.; Kojima, I.; Harui, T.; Kuriki, T. In vitro synthesis of glycogen: The structure, properties, and physiological function of enzymatically-synthesized glycogen. Biologia 2011, 66, 387–394. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, C.; Gu, Z.; Qiu, Y.; Cheng, L.; Hong, Y.; Li, Z. Retrogradation behavior of corn starch treated with 1,4-α-glucan branching enzyme. Food Chem. 2016, 203, 308–313. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.H.; Huang, L.F.; Li, H.M.; Chen, Y.R.; Yu, S.M. Signal peptide-dependent targeting of a rice α-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 2004, 135, 1367–1377. [Google Scholar] [CrossRef] [Green Version]
- Chary, S.N.; Hicks, G.R.; Choi, Y.G.; Carter, D.; Raikhel, N.V. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 2008, 146, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Mo, Z.; Feng, G.; Su, W.; Liu, Z.; Peng, F. Transcriptomic analysis provides insights into grafting union development in pecan (Carya illinoinensis). Genes 2018, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Osuna, D.; Usadel, B.; Morcuende, R.; Gibon, Y.; Blasing, O.E.; Hohne, M.; Gunter, M.; Kamlage, B.; Trethewey, R.; Scheible, W.R.; et al. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 2007, 49, 463–491. [Google Scholar] [CrossRef]
- Paul, M.; Pellny, T.; Goddijn, O. Enhancing photosynthesis with sugar signals. Trends Plant Sci. 2001, 6, 197–200. [Google Scholar] [CrossRef]
- Dai, N.; Petreikov, M.; Portnoy, V.; Katzir, N.; Pharr, D.M.; Schaffer, A.A. Cloning and expression analysis of a UDP-galactose/glucose pyrophosphorylase from melon fruit provides evidence for the major metabolic pathway of galactose metabolism in raffinose oligosaccharide metabolizing plants. Plant Physiol. 2006, 142, 294–304. [Google Scholar] [CrossRef] [Green Version]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [Green Version]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Liu, X.; Wang, T.; Bartholomew, E.; Black, K.; Dong, M.; Zhang, Y.; Yang, S.; Cai, Y.; Xue, S.; Weng, Y.; et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.). Hortic. Res. 2018, 5, 31. [Google Scholar] [CrossRef]
- Jia, D.; Jiang, Z.; Fu, H.; Chen, L.; Liao, G.; He, Y.; Huang, C.; Xu, X. Genome-wide identification and comprehensive analysis of NAC family genes involved in fruit development in kiwifruit (Actinidia). BMC Plant Biol. 2021, 21, 44. [Google Scholar] [CrossRef]
| Group Name | Persisting Fruitlets (P1) | Abscised Fruitlets (A1) |
|---|---|---|
| No. of clean bases (×108) | 68.24 ± 1.00 | 56.40 ± 0.69 |
| No. of total reads (×106) | 45.30 ± 0.68 | 37.48 ± 0.47 |
| No. of mapped reads (×106) | 42.57 ± 0.65 | 35.32 ± 0.40 |
| Mapped percentage (%) | 93.98 ± 0.0.3 | 94.25 ± 0.16 |
| No. of unique mapped reads (×106) | 41.45 ± 0.62 | 34.39 ± 0.47 |
| No. of sequenced reference genes | 24,307 ± 73 | 21,723 ± 360 |
| Percentage of sequenced reference genes (%) | 78.23 ± 0.24 | 69.91 ± 1.16 |
| No. of sequenced novel genes | 1874.00 ± 14.73 | 1701.33 ± 22.68 |
| Percentage of sequenced novel genes (%) | 89.80 ± 0.71 | 81.52 ± 1.09 |
| Sequenced total genes | 26,181.00 ± 88.10 | 23,424.33 ± 382.98 |
| Percentage of sequenced total genes (%) | 78.96 ± 0.27 | 70.64 ± 1.15 |
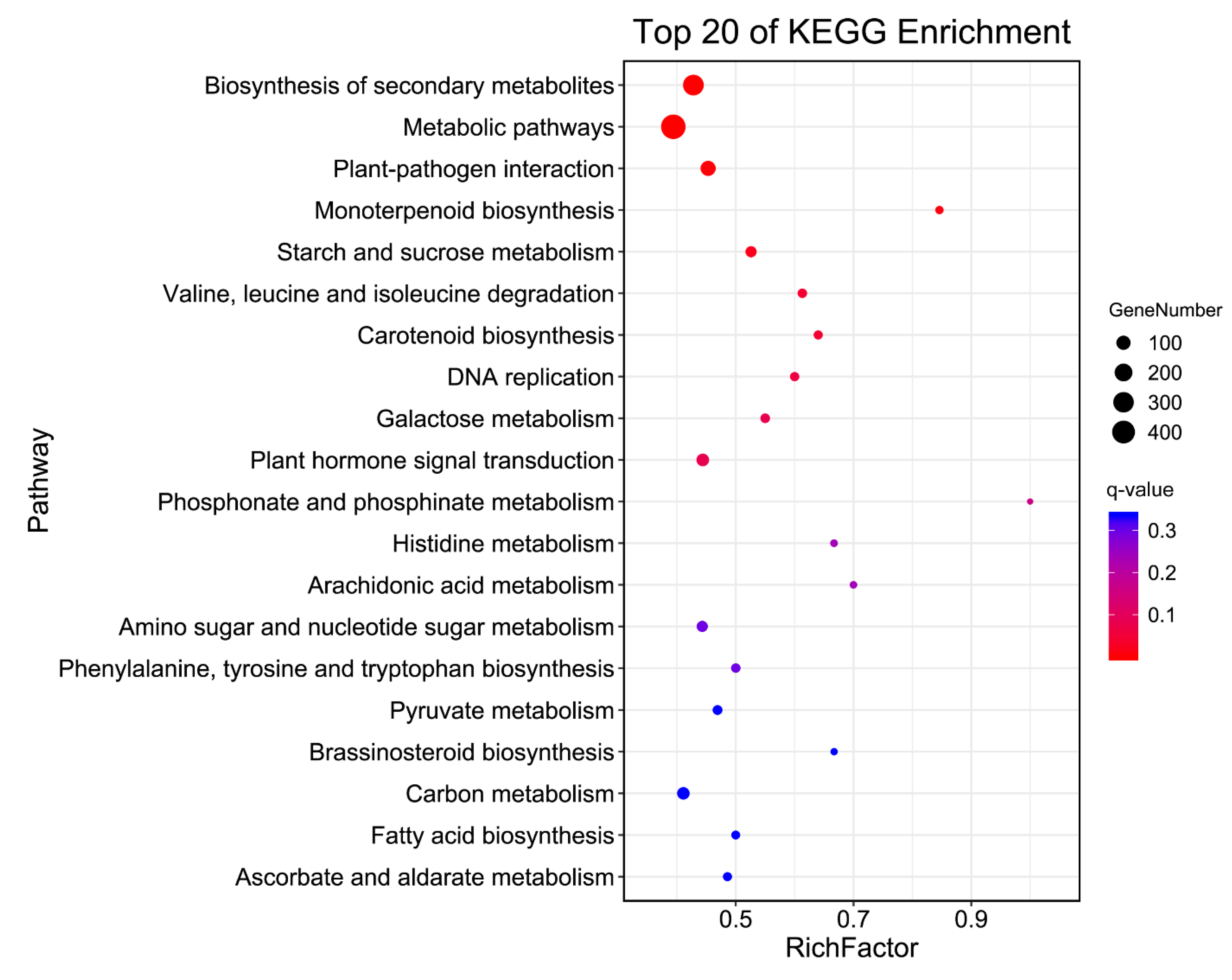
| Pathway | DEGs | p-Value | q-Value | Pathway ID |
|---|---|---|---|---|
| Biosynthesis of secondary metabolites | 306 | 8.64 × 10−8 | 1.11 × 10−5 | ko01110 |
| Metabolic pathways | 492 | 1.50 × 10−6 | 9.69 × 10−5 | ko01100 |
| Plant–pathogen interaction | 120 | 9.58 × 10−5 | 0.004 | ko04626 |
| Monoterpenoid biosynthesis | 11 | 2.94 × 10−4 | 0.009 | ko00902 |
| Starch and sucrose metabolism | 41 | 6.91 × 10−4 | 0.018 | ko00500 |
| Valine, leucine, and isoleucine degradation | 19 | 1.99 × 10−3 | 0.043 | ko00280 |
| Carotenoid biosynthesis | 16 | 2.40 × 10−3 | 0.044 | ko00906 |
| DNA replication | 18 | 3.64 × 10−3 | 0.059 | ko03030 |
| Galactose metabolism | 22 | 5.96 × 10−3 | 0.083 | ko00052 |
| Plant hormone signal transduction | 67 | 6.40 × 10−3 | 0.083 | ko04075 |
| Gene ID | Description | Symbol | Name | log2(A/P) | p-Value |
|---|---|---|---|---|---|
| Abscisic acid | |||||
| CIL1308S0009 | Abscisic acid receptor PYL9 | PYR/PYL | PYL9 | −1.938 | 4.18 × 10−15 |
| CIL1506S0033 | Protein phosphatase 2C 53 | PP2C | HAB1 | −2.868 | 6.3 × 10−9 |
| CIL1317S0086 | Protein phosphatase 2C 16 | PP2C | HAB1 | −1.024 | 0.029075 |
| CIL1562S0028 | Protein phosphatase 2C 75 | PP2C | AHG1 | −2.910 | 2.89 × 10−8 |
| CIL0302S0016 | Serine/threonine-protein kinase SAPK7 | SnRK2 | SRK2H | −2.478 | 6.01 × 10−8 |
| CIL0409S0006 | Serine/threonine-protein kinase SAPK2 | SnRK2 | SAPK2 | −7.128 | 2.1 × 10−9 |
| CIL0045S0005 | ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ABF | DPBF3 | 2.287 | 1.66 × 10−59 |
| CIL1371S0045 | ABSCISIC ACID-INSENSITIVE 5-like protein 6 | ABF | ABF2 | −2.671 | 1.13 × 10−6 |
| CIL1387S0048 | ABSCISIC ACID-INSENSITIVE 5-like protein 2 | ABF | DPBF3 | −1.484 | 0.000172 |
| Auxin | |||||
| CIL1565S0004 | Auxin transporter-like protein 3 | AUX1 | LAX3 | 1.772 | 1.09 × 10−7 |
| CIL1464S0004 | TRANSPORT INHIBITOR RESPONSE protein | TIR | TIR1 | −2.584 | 3.62 × 10−6 |
| CIL0202S0015 | Auxin-responsive protein IAA16-like | AUX/IAA | IAA16 | −16.109 | 3.02 × 10−23 |
| CIL0203S0027 | Auxin-responsive protein IAA9-like | AUX/IAA | IAA9 | −3.587 | 1.04 × 10−39 |
| CIL0344S0014 | Auxin-responsive protein IAA18-like | AUX/IAA | IAA26 | −2.671 | 2.16 × 10−11 |
| CIL0732S0001 | Auxin-responsive protein IAA9-like | AUX/IAA | IAA9 | −10.617 | 0.000112 |
| CIL1268S0077 | Auxin-responsive protein IAA27-like | AUX/IAA | IAA8 | −3.986 | 6.35 × 10−5 |
| CIL1294S0084 | Auxin-responsive protein IAA20-like | AUX/IAA | IAA20 | −9.765 | 0.001964 |
| CIL1320S0049 | Auxin-responsive protein IAA27-like | AUX/IAA | IAA27 | −4.217 | 2.06 × 10−11 |
| CIL1358S0014 | Auxin-responsive protein IAA29-like | AUX/IAA | IAA11 | −10.951 | 8.53 × 10−7 |
| CIL1490S0013 | Auxin response factor 18 | ARF | ARF9 | −8.323 | 1.83 × 10−54 |
| CIL1564S0002 | Auxin response factor 9 | ARF | ARF9 | −3.530 | 1.08 × 10−38 |
| CIL1354S0026 | Auxin response factor 19 | ARF | ARF7 | −2.515 | 3.06 × 10−13 |
| CIL1313S0055 | Probable indole-3-acetic acid-amido synthetase GH3.6 | GH3 | GH3.6 | 2.440 | 5.86 × 10−18 |
| CIL1405S0075 | Indole-3-acetic acid-amido synthetase GH3.6 | GH3 | GH3.6 | −4.055 | 6.98 × 10−7 |
| CIL1456S0019 | Probable indole-3-acetic acid-amido synthetase GH3.1 | GH3 | GH3.1 | −2.721 | 1.89 × 10−6 |
| CIL1295S0016 | Auxin-responsive protein SAUR36 | SAUR | SAUR36 | 3.508 | 3.6 × 10−26 |
| CIL0367S0005 | Auxin-induced protein X15 | SAUR | SAUR50 | −3.078 | 0.003923 |
| MSTRG.7503 | Auxin-responsive protein SAUR32 | SAUR | SAUR32 | 6.343 | 4.96 × 10−90 |
| CIL1294S0056 | Indole-3-acetic acid-induced protein ARG7 | SAUR | SAUR36 | 10.129 | 0.000393 |
| Cytokinin | |||||
| CIL1595S0016 | Histidine kinase 2 | CRE1 | AHK2 | −4.205 | 3.31 × 10−19 |
| CIL1384S0015 | Histidine kinase 2 | CRE1 | AHK2 | −1.934 | 0.004026 |
| CIL0037S0021 | Histidine-containing phosphotransfer protein 1 | AHP | AHP1 | −12.078 | 3.11 × 10−9 |
| CIL1268S0039 | Histidine-containing phosphotransfer protein 4 | AHP | PHP5 | −6.299 | 2.73 × 10−5 |
| CIL1268S0040 | Histidine-containing phosphotransfer protein 4 | AHP | AHP4 | 10.326 | 1.02 × 10−6 |
| CIL1369S0025 | Histidine-containing phosphotransfer protein 1 | AHP | AHP1 | −1.748 | 3.06 × 10−9 |
| MSTRG.14062 | Histidine-containing phosphotransfer protein 1 | AHP | AHP1 | 1.101 | 0.001041 |
| CIL1575S0022 | Two-component response regulator ARR12 | B-ARR | RR23 | −2.035 | 5.52 × 10−13 |
| CIL0004S0012 | Two-component response regulator ARR8 | A-ARR | ARR8 | −11.423 | 0.000187 |
| CIL0338S0017 | Two-component response regulator ARR9 | A-ARR | ARR9 | −1.943 | 0.030964 |
| CIL1308S0026 | Two-component response regulator ARR5 | A-ARR | ARR4 | −4.320 | 2.19 × 10−12 |
| CIL1596S0045 | Two-component response regulator ARR5 | A-ARR | ARR15 | −5.520 | 3.19 × 10−12 |
| Ethylene | |||||
| CIL1354S0046 | Ethylene-responsive transcription factor 1B | EBF1/2 | ERF1B | 2.436 | 0.000283 |
| CIL1358S0043 | EIN3-binding F-box protein 1 | EBF1/2 | EBF2 | 2.280 | 7.01 × 10−36 |
| CIL1493S0026 | Ethylene receptor 2 | ETR | ETR2 | 4.072 | 1.15 × 10−73 |
| Gibberellin | |||||
| CIL1324S0067 | DELLA protein SLN1 | DELLA | GAI1 | −3.061 | 1.6 × 10−16 |
| CIL1294S0080 | Transcription factor PIF3 | TF | PIL15 | −5.458 | 8.24 × 10−14 |
| CIL1495S0010 | Transcription factor PIF1 | TF | PIF1 | 2.298 | 5.46 × 10−8 |
| Brassinosteroid | |||||
| CIL1492S0038 | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | BAK1 | BAK1 | 1.609 | 1.12 × 10−22 |
| CIL1312S0044 | Brassinosteroid LRR receptor kinase | BRI1 | CURL3 | −8.773 | 1.46 × 10−58 |
| MSTRG.23225 | Serine/threonine-protein kinase | BSK | BSK1 | −1.699 | 0.003421 |
| CIL1383S0002 | BES1/BZR1 homolog protein 2 | BZR1/2 | BEH2 | −1.441 | 0.013346 |
| CIL0182S0015 | HMA domain-containing protein | BZR1/2 | – | 5.157 | 0.000157 |
| CIL1577S0019 | Xyloglucan endotransglucosylase/hydrolase protein 22 | TCH4 | XTH22 | −3.772 | 2.16 × 10−5 |
| CIL1371S0059 | Cyclin-D3-1 | CYCD3 | CYCD3-3 | −11.257 | 4 × 10−9 |
| JA | |||||
| CIL1295S0055 | Protein TIFY 10A | JAZ | TIFY10B | 2.236 | 3.57 × 10−13 |
| CIL1338S0009 | Protein TIFY 6B | JAZ | TIFY6B | −4.081 | 4.52 × 10−18 |
| CIL1390S0024 | Protein TIFY 6B | JAZ | TIFY6B | −5.604 | 1.59 × 10−14 |
| CIL1565S0018 | Protein TIFY 6B | JAZ | TIFY6B | −3.637 | 1.25 × 10−12 |
| MSTRG.9204 | Protein TIFY 10A | JAZ | – | −3.812 | 0.016461 |
| CIL1399S0034 | Transcription factor MYC2 | MYC2 | BHLH14 | −3.273 | 9.07 × 10−10 |
| MSTRG.9232 | Transcription factor MYC4 | MYC2 | MYC4 | 6.314 | 2 × 10−22 |
| SA | |||||
| CIL1326S0021 | Transcription factor TGA7-like | TGA | TGA7 | 1.407 | 1.05 × 10−15 |
| CIL0037S0002 | Pathogenesis-related protein 1 | PR-1 | PRB1 | 8.827 | 1.24 × 10−60 |
| CIL0037S0003 | Pathogenesis-related protein 1 | PR-1 | PRB1 | 3.015 | 1.31 × 10−16 |
| CIL0037S0006 | Pathogenesis-related protein 1 | PR-1 | At2g14610 | 8.831 | 1.4 × 10−139 |
| CIL0037S0007 | Basic form of pathogenesis-related protein 1 | PR-1 | PRMS | 10.196 | 6.07 × 10−5 |
| CIL0232S0001 | Pathogenesis-related protein 1 | PR-1 | PRB1 | −4.242 | 9.5 × 10−24 |
| Category | Total | Upregulated | Downregulated |
|---|---|---|---|
| MYB | 40 | 10 | 30 |
| NAC | 24 | 13 | 11 |
| bHLH | 24 | 4 | 20 |
| WRKY | 24 | 17 | 7 |
| bZIP | 4 | 1 | 3 |
| C2 | 2 | 0 | 2 |
| C2H2 | 1 | 0 | 1 |
| MADS | 3 | 0 | 3 |
| ERF | 25 | 11 | 14 |
| AUX/IAA | 25 | 4 | 21 |
| zinc finger | 69 | 15 | 54 |
| B3 | 4 | 1 | 4 |
| Gene ID | Description | EC No. | Symbol | log2(fc) | p-Value |
|---|---|---|---|---|---|
| CIL1395S0001 | Alpha-glucosidase | 3.2.1.20 | – | 1.594 | 0.012 |
| MSTRG.2545 | Alpha-glucosidase | 3.2.1.20 | Os06g0675700 | 1.429 | 2.71 × 10−8 |
| CIL1595S0049 | Glucose-1-phosphate adenylyltransferase | 2.7.7.27 | glgC | −2.746 | 2.81 × 10−16 |
| CIL0203S0004 | Glucose-1-phosphate adenylyltransferase | 2.7.7.27 | glgC | −3.281 | 1.24 × 10−10 |
| MSTRG.12720 | Alpha-amylase-like isoform X1 | 3.2.1.1 | AMY1.1 | 3.719 | 1.74 × 10−76 |
| MSTRG.2528 | Alpha-amylase-like isoform X1 | 3.2.1.1 | AMY1.1 | 1.358 | 6.01 × 10−13 |
| CIL1383S0034 | Glucan endo-1,3-beta-glucosidase 1 | 3.2.1.39 | At1g11820 | −3.735 | 1.49 × 10−32 |
| CIL1482S0014 | Glucan endo-1,3-beta-glucosidase 4-like | 3.2.1.39 | At3g13560 | −3.409 | 2.95 × 10−15 |
| CIL1332S0070 | Glucan endo-1,3-beta-glucosidase 5-like | 3.2.1.39 | At4g31140 | −1.632 | 4.40 × 10−4 |
| CIL1347S0008 | Glucan endo-1,3-beta-glucosidase 6 | 3.2.1.39 | At5g58090 | −6.006 | 5.41 × 10−32 |
| CIL1359S0017 | Hexokinase-3-like [Juglans regia] | 2.7.1.1 | At1g50460 | −1.211 | 3.43 × 10−3 |
| MSTRG.14644 | Hexokinase-3-like isoform X3 | 2.7.1.1 | At1g50460 | −2.125 | 2.03 × 10−7 |
| CIL1459S0007 | Hexokinase-3-like isoform X2 | 2.7.1.1 | At1g50460 | −2.411 | 5.93 × 10−7 |
| CIL0282S0003 | Hexokinase-1-like isoform X2 | 2.7.1.1 | HXK1 | 1.958 | 1.53 × 10−12 |
| CIL1518S0008 | Hexokinase-2 | 2.7.1.1 | HXK2 | −2.598 | 0.005 |
| CIL1568S0010 | Probable fructokinase-7 | 2.7.1.4 | At5g51830 | 2.623 | 2.65 × 10−36 |
| CIL0508S0004 | Beta-glucosidase 12-like isoform X3 | 3.2.1.21 | BGLU12 | −3.745 | 0.003 |
| CIL1537S0001 | Beta-glucosidase 12-like | 3.2.1.21 | BGLU12 | −8.471 | 5.16 × 10−51 |
| CIL0493S0002 | Beta-glucosidase 12-like | 3.2.1.21 | BGLU13 | 11.919 | 2.12 × 10−11 |
| MSTRG.6474 | Beta-glucosidase 12-like | 3.2.1.21 | BGLU13 | 10.737 | 1.03 × 10−7 |
| CIL0508S0002 | Beta-glucosidase 13-like isoform X2 | 3.2.1.21 | BGLU13 | −2.922 | 1.26 × 10−4 |
| MSTRG.7161 | Beta-glucosidase 12-like | 3.2.1.21 | BGLU24 | −9.010 | 1.78 × 10−54 |
| CIL1405S0071 | Beta glucosidase 41 isoform 2 | 3.2.1.21 | BGLU25 | −3.444 | 1.12 × 10−11 |
| CIL0391S0004 | Beta-glucosidase 42 isoform X1 | 3.2.1.21 | BGLU42 | −4.571 | 2.02 × 10−52 |
| CIL1320S0039 | Beta-glucosidase 47-like isoform X1 | 3.2.1.21 | BGLU47 | 4.020 | 9.97 × 10−18 |
| CIL1407S0038 | Endoglucanase 8-like | 3.2.1.4 | CEL1 | −6.822 | 8.06 × 10−27 |
| MSTRG.23283 | Beta-glucosidase | 3.2.1.21 | RE1 | 2.092 | 1.75 × 10−3 |
| CIL1317S0076 | Beta-fructofuranosidase | 3.2.1.26 | CWINV1 | 4.017 | 8.53 × 10−14 |
| CIL1506S0011 | Beta-fructofuranosidase | 3.2.1.26 | CWINV3 | −5.878 | 1.01 × 10−4 |
| CIL1264S0043 | Acid beta-fructofuranosidase-like | 3.2.1.26 | INV*DC4 | −3.426 | 1.78 × 10−25 |
| CIL0525S0001 | Nudix hydrolase 14, chloroplastic | 3.6.1.21 | NUDT14 | −1.662 | 8.32 × 10−4 |
| CIL1568S0006 | Phosphoglucomutase, chloroplastic | 5.4.2.2 | PGMP | −1.205 | 0.011 |
| CIL0360S0002 | Sucrose-phosphate synthase 1 | 2.4.1.14 | SPS1 | −1.765 | 8.57 × 10−3 |
| CIL1271S0008 | Sucrose-phosphate synthase 1 | 2.4.1.14 | SPS1 | −7.197 | 1.16 × 10−29 |
| CIL1417S0045 | 1,4-alpha-glucan-branching enzyme 3 | 2.4.1.18 | GBE3, glgB, SBE3 | −4.466 | 5.87 × 10−16 |
| CIL1531S0004 | 1,4-alpha-glucan-branching enzyme 1 | 2.4.1.18 | GBE3, glgB, SBEI | −2.321 | 4.31 × 10−18 |
| CIL0218S0017 | Granule-bound starch synthase 1 | 2.4.1.242 | WAXY, GBSS1 | −3.748 | 8.21 × 10−14 |
| CIL0389S0009 | Granule-bound starch synthase 1 | 2.4.1.242 | WAXY, GBSS1 | −12.135 | 6.74 × 10−12 |
| CIL0176S0049 | Granule-bound starch synthase 2 | 2.4.1.21 | SS2 | −5.083 | 1.69 × 10−20 |
| CIL1531S0021 | Trehalose-phosphate phosphatase | 3.1.3.12 | TPP | 1.896 | 3.70 × 10−5 |
| CIL1310S0034 | Trehalose 6-phosphate synthase | 2.4.1.15 | TPS | 1.535 | 1.66 × 10−12 |
| CIL0021S0018 | Alpha-galactosidase 1-like | 3.2.1.22 | AGAL1 | −2.014 | 3.88 × 10−12 |
| CIL1429S0021 | Aldose 1-epimerase | 5.1.3.3 | Galm | 1.077 | 4.95 × 10−48 |
| CIL0309S0003 | Inositol 3-alpha-galactosyltransferase 1 | 2.4.1.123 | GOLS1 | 1.063 | 1.44 × 10−11 |
| MSTRG.21476 | Inositol 3-alpha-galactosyltransferase 1 | 2.4.1.123 | GOLS1 | −13.936 | 3.70 × 10−15 |
| MSTRG.20754 | Inositol 3-alpha-galactosyltransferase 2 | 2.4.1.123 | GOLS2 | −2.722 | 0.013 |
| MSTRG.20755 | Inositol 3-alpha-galactosyltransferase 2 | 2.4.1.123 | GOLS2 | −2.837 | 0.021 |
| CIL0344S0021 | 6-phosphofructokinase 1 | 2.7.1.1 | PFK3 | −4.090 | 2.05 × 10−8 |
| CIL1568S0006 | Phosphoglucomutase | 5.4.2.2 | PGMP | −1.205 | 0.011 |
| CIL1358S0005 | Raffinose synthase | 2.4.1.82 | RFS6 | 3.211 | 2.01 × 10−33 |
| CIL0272S0009 | UDP-glucose 4-epimerase GEPI48-like | 5.1.3.2 | UGE5 | 1.349 | 1.70 × 10−11 |
| CIL1564S0017 | UDP-glucose 4-epimerase GEPI48 | 5.1.3.2 | UGE5 | 3.159 | 8.72 × 10−43 |
| CIL1297S0013 | UDP-sugar pyrophosphorylase | 2.7.7.64 | USP | −3.752 | 4.18 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, T.; Zhang, F.; Liu, Y.; Wang, G. Comparative Analysis of the Transcriptomes of Persisting and Abscised Fruitlets: Insights into Plant Hormone and Carbohydrate Metabolism Regulated Self-Thinning of Pecan Fruitlets during the Early Stage. Curr. Issues Mol. Biol. 2022, 44, 176-193. https://doi.org/10.3390/cimb44010013
Zhang J, Wang T, Zhang F, Liu Y, Wang G. Comparative Analysis of the Transcriptomes of Persisting and Abscised Fruitlets: Insights into Plant Hormone and Carbohydrate Metabolism Regulated Self-Thinning of Pecan Fruitlets during the Early Stage. Current Issues in Molecular Biology. 2022; 44(1):176-193. https://doi.org/10.3390/cimb44010013
Chicago/Turabian StyleZhang, Jiyu, Tao Wang, Fan Zhang, Yongzhi Liu, and Gang Wang. 2022. "Comparative Analysis of the Transcriptomes of Persisting and Abscised Fruitlets: Insights into Plant Hormone and Carbohydrate Metabolism Regulated Self-Thinning of Pecan Fruitlets during the Early Stage" Current Issues in Molecular Biology 44, no. 1: 176-193. https://doi.org/10.3390/cimb44010013
APA StyleZhang, J., Wang, T., Zhang, F., Liu, Y., & Wang, G. (2022). Comparative Analysis of the Transcriptomes of Persisting and Abscised Fruitlets: Insights into Plant Hormone and Carbohydrate Metabolism Regulated Self-Thinning of Pecan Fruitlets during the Early Stage. Current Issues in Molecular Biology, 44(1), 176-193. https://doi.org/10.3390/cimb44010013







