The Role of MRE11 in the IL-6/STAT3 Pathway of Lung Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. shRNA Knockdown Assay
2.3. Cytokine Membrane Array
2.4. Macrophage Recruitment Assay
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
2.6. Quantitative Real-Time PCR
2.7. Western Blot Analysis
2.8. Immunoprecipitation (IP)
2.9. Statistical Analyses
3. Results
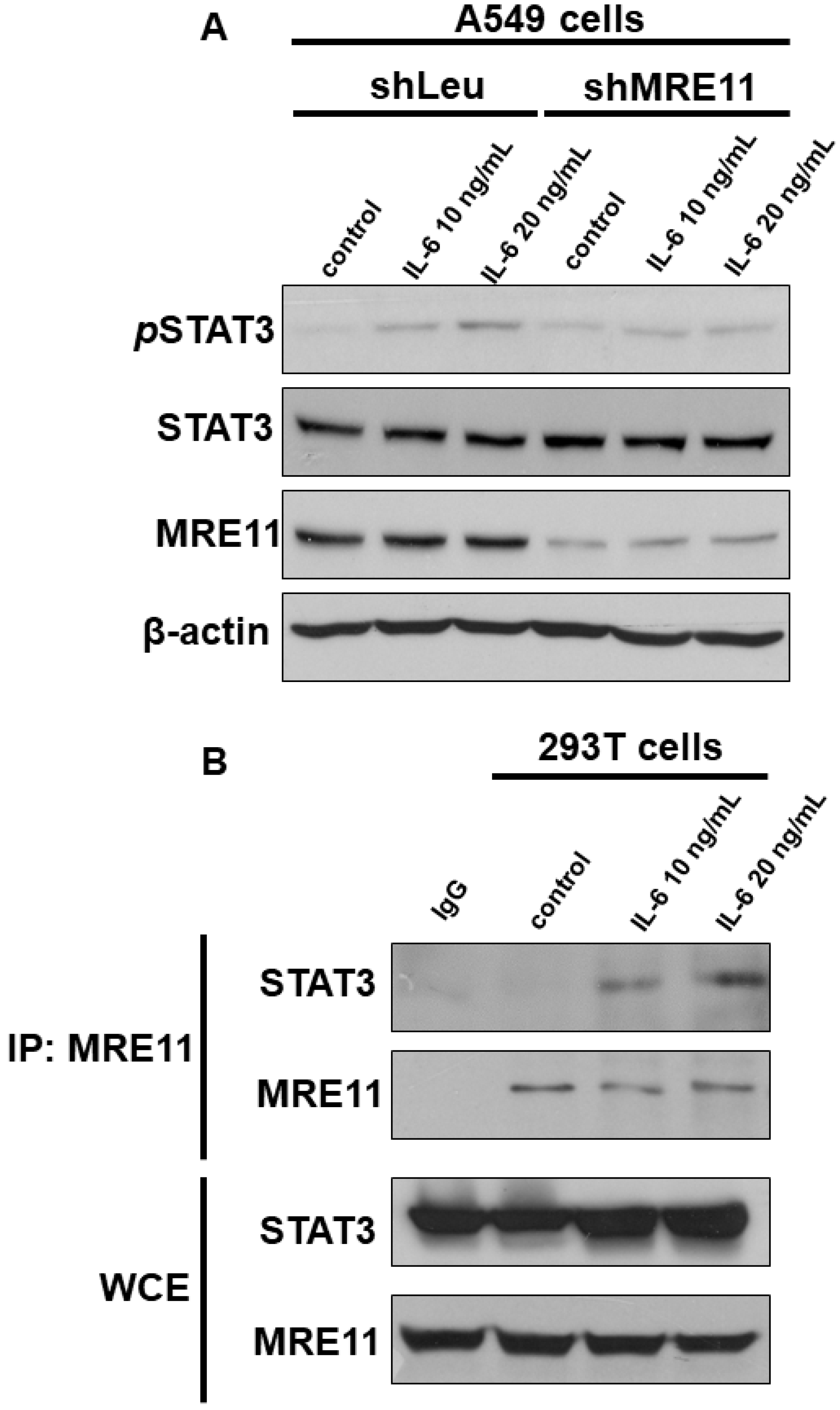
3.1. The Interaction between MRE11 and STAT3
3.2. The Effect of MRE11 on Activation of STAT3 under Treatment with IL-6
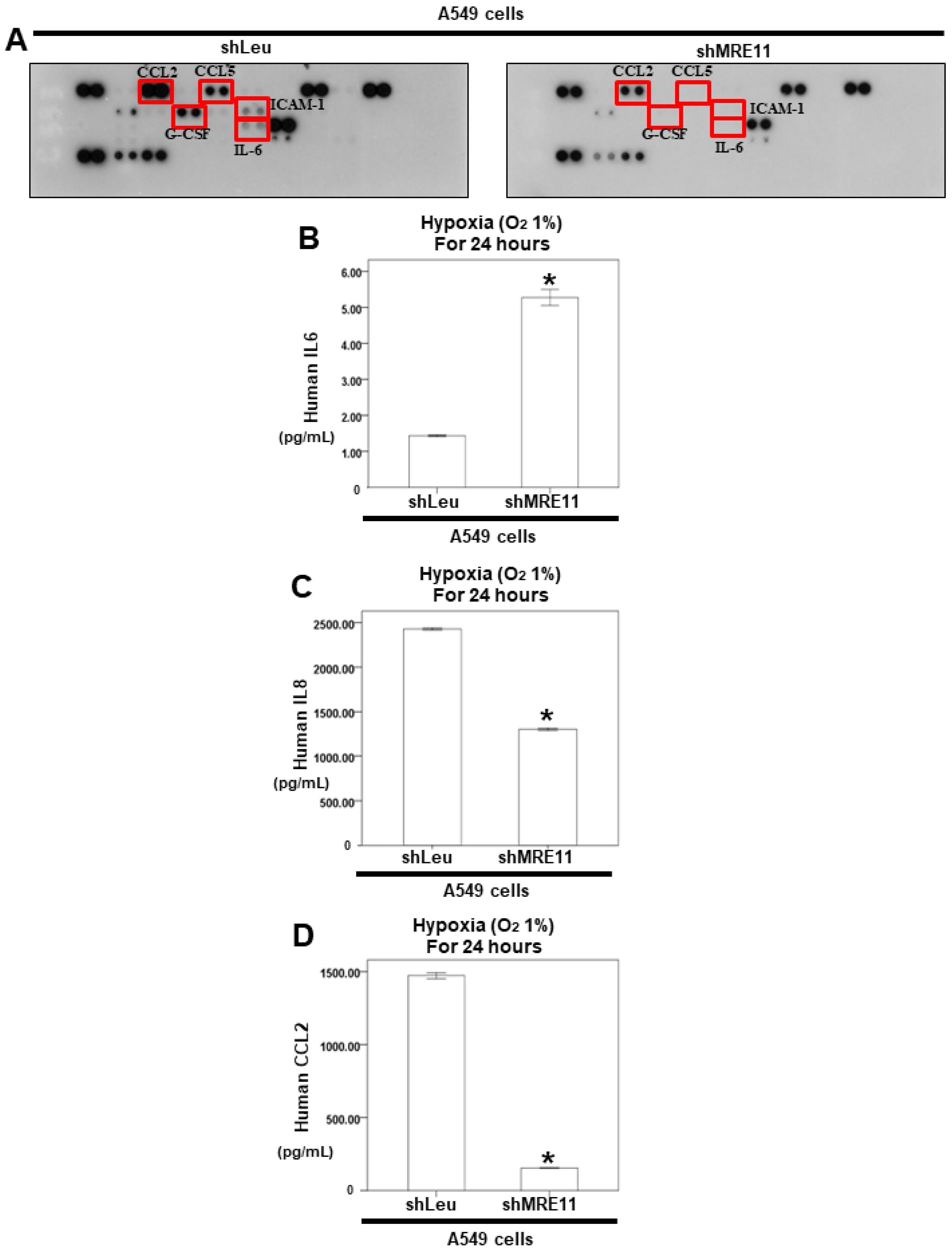
3.3. The Effect of MRE11 on Cytokine Secretion from Lung Cancer Cells
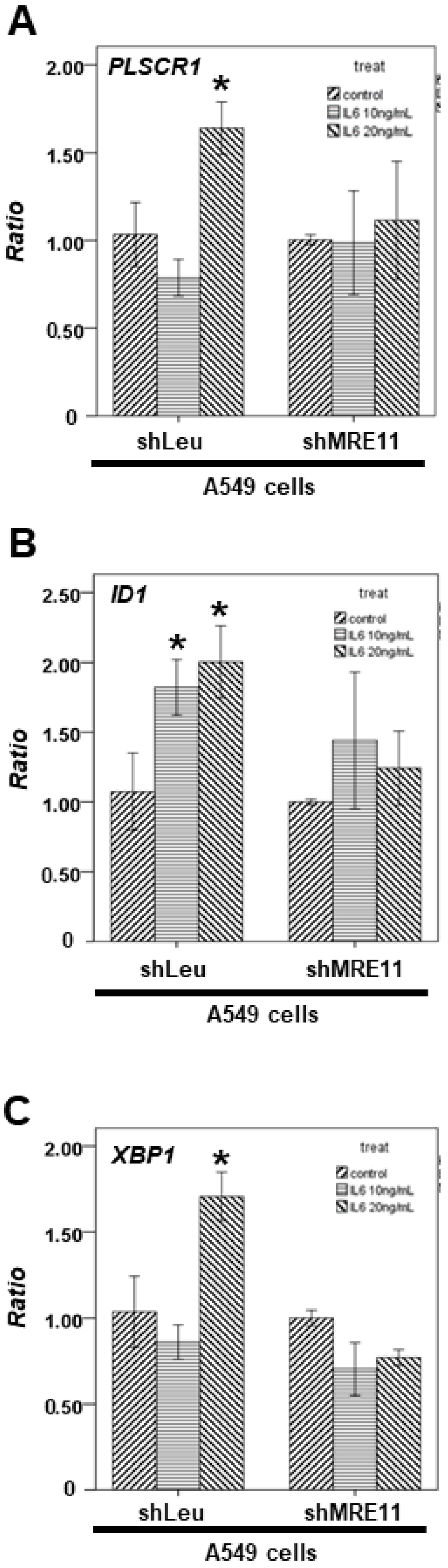
3.4. The Effect of MRE11 on STAT3′s Downstream Genes from Lung Cancer Cells under Activation of Macrophages
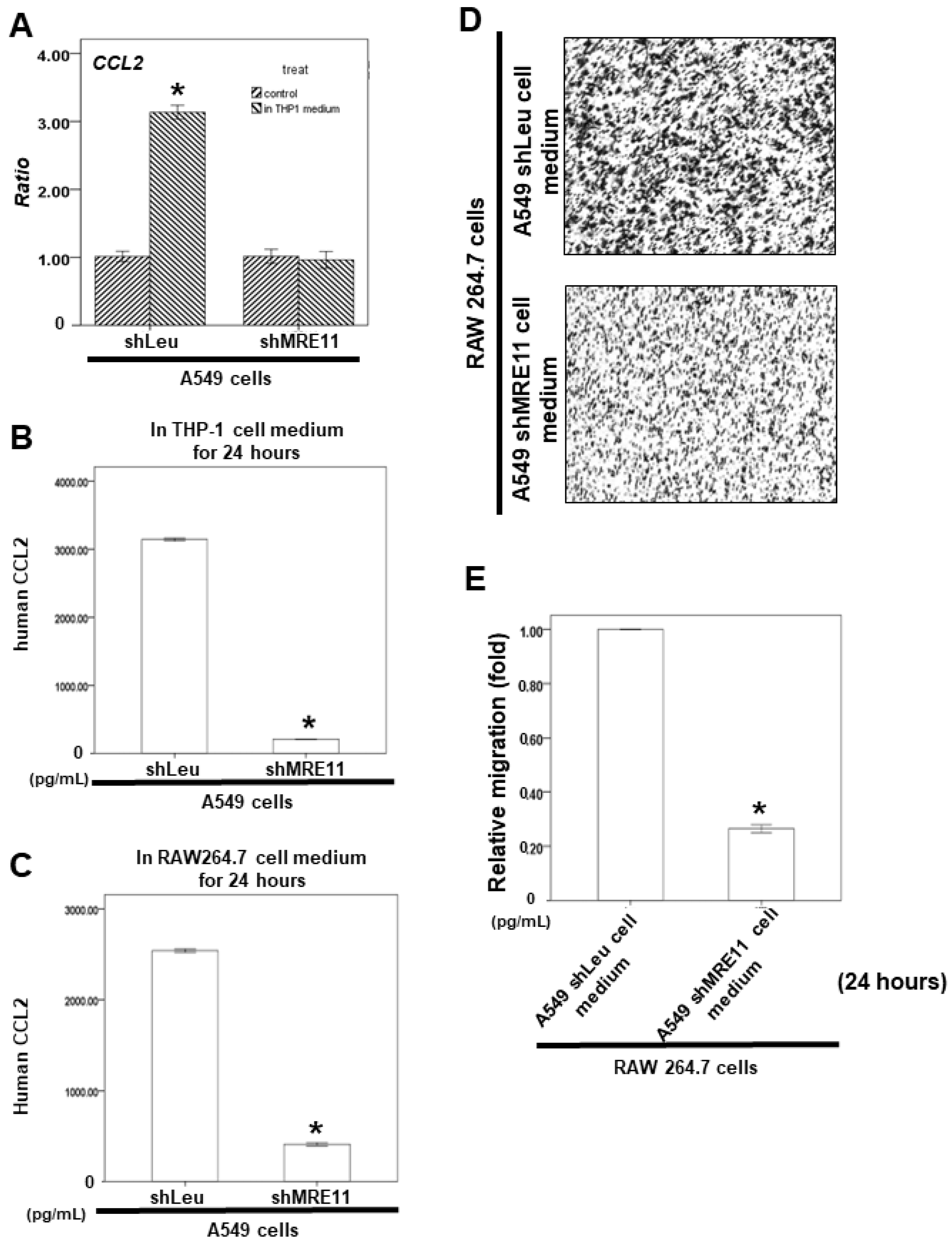
3.5. Effects of MRE11A on RAW 264.7 Cell Recruitment In Vitro
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lavin, M.F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 2007, 26, 7749–7758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Paull, T.T. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007, 26, 7741–7748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beumer, J.H.; Fu, K.Y.; Anyang, B.N.; Siegfried, J.M.; Bakkenist, C.J. Functional analyses of ATM, ATR and Fanconi anemia proteins in lung carcinoma: ATM, ATR and FA in lung carcinoma. BMC Cancer 2015, 15, 649. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ma, L.; Bian, X.; Lv, Y.; Lin, W. FK228 sensitizes radioresistant small cell lung cancer cells to radiation. Clin. Epigenetics 2021, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Ren, D.; Xu, X.; Wang, Y. Molecular cross-talk of IL-6 in tumors and new progress in combined therapy. Thorac. Cancer 2018, 9, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Sun, Y.M. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer (Review). Int. J. Oncol. 2014, 44, 1032–1040. [Google Scholar] [CrossRef] [Green Version]
- Bonastre, E.; Verdura, S.; Zondervan, I.; Facchinetti, F.; Lantuejoul, S.; Chiara, M.D.; Rodrigo, J.P.; Carretero, J.; Condom, E.; Vidal, A.; et al. PARD3 Inactivation in Lung Squamous Cell Carcinomas Impairs STAT3 and Promotes Malignant Invasion. Cancer Res. 2015, 75, 1287–1297. [Google Scholar] [CrossRef] [Green Version]
- Devarajan, E.; Huang, S. STAT3 as a central regulator of tumor metastases. Curr. Mol. Med. 2009, 9, 626–633. [Google Scholar] [CrossRef]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Gao, S.P.; Mark, K.G.; Leslie, K.; Pao, W.; Motoi, N.; Gerald, W.L.; Travis, W.D.; Bornmann, W.; Veach, D.; Clarkson, B.; et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J. Clin. Investig. 2007, 117, 3846–3856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarkson, R.W.; Boland, M.P.; Kritikou, E.A.; Lee, J.M.; Freeman, T.C.; Tiffen, P.G.; Watson, C.J. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol. Endocrinol. 2006, 20, 675–685. [Google Scholar] [CrossRef]
- Valente, A.J.; Graves, D.T.; Vialle-Valentin, C.E.; Delgado, R.; Schwartz, C.J. Purification of a monocyte chemotactic factor secreted by nonhuman primate vascular cells in culture. Biochemistry 1988, 27, 4162–4168. [Google Scholar] [CrossRef]
- Yoshimura, T.; Robinson, E.A.; Tanaka, S.; Appella, E.; Kuratsu, J.; Leonard, E.J. Purification and amino acid analysis of two human glioma-derived monocyte chemoattractants. J. Exp. Med. 1989, 169, 1449–1459. [Google Scholar] [CrossRef] [Green Version]
- Schmall, A.; Al-Tamari, H.M.; Herold, S.; Kampschulte, M.; Weigert, A.; Wietelmann, A.; Vipotnik, N.; Grimminger, F.; Seeger, W.; Pullamsetti, S.S.; et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am. J. Respir. Crit. Care Med. 2015, 191, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, J.; Chen, S.; Lu, M.; Luo, X.; Yao, S.; Liu, S.; Qin, Y.; Chen, H. Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung Cancer 2011, 74, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Shimamura, S.; Kuriyama, S.; Maeda, D.; Goto, A.; Aiba, N. SKAP2 Promotes Podosome Formation to Facilitate Tumor-Associated Macrophage Infiltration and Metastatic Progression. Cancer Res. 2016, 76, 358–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komohara, Y.; Horlad, H.; Ohnishi, K.; Fujiwara, Y.; Bai, B.; Nakagawa, T.; Suzu, S.; Nakamura, H.; Kuratsu, J.; Takeya, M. Importance of direct macrophage-tumor cell interaction on progression of human glioma. Cancer Sci. 2012, 103, 2165–2172. [Google Scholar] [CrossRef]
- Li, X.; Tai, H.H. Activation of thromboxane A2 receptor (TP) increases the expression of monocyte chemoattractant protein -1 (MCP-1)/chemokine (C-C motif) ligand 2 (CCL2) and recruits macrophages to promote invasion of lung cancer cells. PLoS ONE 2013, 8, e54073. [Google Scholar] [CrossRef]
- Huo, N.; Cong, R.; Sun, Z.J.; Li, W.C.; Zhu, X.; Xue, C.Y.; Chen, Z.; Ma, L.Y.; Chu, Z.; Han, Y.C.; et al. STAT3/LINC00671 axis regulates papillary thyroid tumor growth and metastasis via LDHA-mediated glycolysis. Cell Death Dis. 2021, 12, 799. [Google Scholar] [CrossRef]
- Cheng, H.; Hao, Y.; Gao, Y.; He, Y.; Luo, C.; Sun, W.; Yuan, M.; Wu, X. PLCepsilon promotes urinary bladder cancer cells proliferation through STAT3/LDHA pathwaymediated glycolysis. Oncol. Rep. 2019, 41, 2844–2854. [Google Scholar] [CrossRef] [Green Version]
- Adachi, Y.; Aoki, C.; Yoshio-Hoshino, N.; Takayama, K.; Curiel, D.T.; Nishimoto, N. Interleukin-6 induces both cell growth and VEGF production in malignant mesotheliomas. Int. J. Cancer 2006, 119, 1303–1311. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Xiao, J.; Dai, Z.; Chen, Q. Membrane progesterone receptor alpha (mPRalpha) enhances hypoxia-induced vascular endothelial growth factor secretion and angiogenesis in lung adenocarcinoma through STAT3 signaling. J. Transl. Med. 2022, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Bae, S.C.; Chuang, L.S. The RUNX family: Developmental regulators in cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, J.; Cai, C.; Xie, X. N-myc Downstream-Regulated Gene 2 (NDRG2) Promotes Bone Morphogenetic Protein 2 (BMP2)-Induced Osteoblastic Differentiation and Calcification by Janus Kinase 3 (JAK3)/Signal Transducer and Activator of Transcription 3 (STAT3) Signaling Pathway. Med. Sci. Monit. 2020, 26, e918541. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Shi, J.; Chen, S.; Xu, L.; Li, F.; Dong, N. IL-21 promotes osteoblastic differentiation of human valvular interstitial cells through the JAK3/STAT3 pathway. Int. J. Med. Sci. 2020, 17, 3065–3072. [Google Scholar] [CrossRef]
- Lee, H.W.; Choi, H.J.; Ha, S.J.; Lee, K.T.; Kwon, Y.G. Recruitment of monocytes/macrophages in different tumor microenvironments. Biochim. Biophys. Acta 2013, 1835, 170–179. [Google Scholar] [CrossRef]
- Murdoch, C.; Muthana, M.; Coffelt, S.B.; Lewis, C.E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer 2008, 8, 618–631. [Google Scholar] [CrossRef]
- Xu, X.; Padilla, M.T.; Li, B.; Wells, A.; Kato, K.; Tellez, C.; Belinsky, S.A.; Kim, K.C.; Lin, Y. MUC1 in macrophage: Contributions to cigarette smoke-induced lung cancer. Cancer Res. 2014, 74, 460–470. [Google Scholar] [CrossRef] [Green Version]
- Burysek, L.; Syrovets, T.; Simmet, T. The serine protease plasmin triggers expression of MCP-1 and CD40 in human primary monocytes via activation of p38 MAPK and janus kinase (JAK)/STAT signaling pathways. J. Biol. Chem. 2002, 277, 33509–33517. [Google Scholar] [CrossRef]
- Lin, T.H.; Izumi, K.; Lee, S.O.; Lin, W.J.; Yeh, S.; Chang, C. Anti-androgen receptor ASC-J9 versus anti-androgens MDV3100 (Enzalutamide) or Casodex (Bicalutamide) leads to opposite effects on prostate cancer metastasis via differential modulation of macrophage infiltration and STAT3-CCL2 signaling. Cell Death Dis. 2013, 4, e764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumi, K.; Fang, L.Y.; Mizokami, A.; Namiki, M.; Li, L.; Lin, W.J.; Chang, C. Targeting the androgen receptor with siRNA promotes prostate cancer metastasis through enhanced macrophage recruitment via CCL2/CCR2-induced STAT3 activation. EMBO Mol. Med. 2013, 5, 1383–1401. [Google Scholar] [CrossRef]
- Tamm, M.; Bihl, M.; Eickelberg, O.; Stulz, P.; Perruchoud, A.P.; Roth, M. Hypoxia-induced interleukin-6 and interleukin-8 production is mediated by platelet-activating factor and platelet-derived growth factor in primary human lung cells. Am. J. Respir. Cell Mol. Biol. 1998, 19, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Mojsilovic-Petrovic, J.; Callaghan, D.; Cui, H.; Dean, C.; Stanimirovic, D.B.; Zhang, W. Hypoxia-inducible factor-1 (HIF-1) is involved in the regulation of hypoxia-stimulated expression of monocyte chemoattractant protein-1 (MCP-1/CCL2) and MCP-5 (Ccl12) in astrocytes. J. Neuroinflammation 2007, 4, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neta, R.; Perlstein, R.; Vogel, S.N.; Ledney, G.D.; Abrams, J. Role of interleukin 6 (IL-6) in protection from lethal irradiation and in endocrine responses to IL-1 and tumor necrosis factor. J. Exp. Med. 1992, 175, 689–694. [Google Scholar] [CrossRef] [Green Version]
- Centurione, L.; Aiello, F.B. DNA Repair and Cytokines: TGF-beta, IL-6, and Thrombopoietin as Different Biomarkers of Radioresistance. Front. Oncol. 2016, 6, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhang, F.; Tsai, Y.; Yang, X.; Yang, L.; Duan, S.; Wang, X.; Keng, P.; Lee, S.O. IL-6 signaling promotes DNA repair and prevents apoptosis in CD133+ stem-like cells of lung cancer after radiation. Radiat. Oncol. 2015, 10, 227. [Google Scholar] [CrossRef] [Green Version]
- Kondo, T.; Kobayashi, J.; Saitoh, T.; Maruyama, K.; Ishii, K.J.; Barber, G.N.; Komatsu, K.; Akira, S.; Kawai, T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA 2013, 110, 2969–2974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Niimi, A.; Yasuhara, T.; Permata, T.B.M.; Hagiwara, Y.; Isono, M.; Nuryadi, E.; Sekine, R.; Oike, T.; Kakoti, S.; et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 2017, 8, 1751. [Google Scholar] [CrossRef]
- Yuan, S.S.; Hou, M.F.; Hsieh, Y.C.; Huang, C.Y.; Lee, Y.C.; Chen, Y.J.; Lo, S. Role of MRE11 in cell proliferation, tumor invasion, and DNA repair in breast cancer. J. Natl. Cancer Inst. 2012, 104, 1485–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsushima, K.; Larsen, C.G.; DuBois, G.C.; Oppenheim, J.J. Purification and characterization of a novel monocyte chemotactic and activating factor produced by a human myelomonocytic cell line. J. Exp. Med. 1989, 169, 1485–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Lin, J.; Xu, A.; Lou, J.; Qian, C.; Li, X.; Wang, Y.; Yu, W.; Tao, H. CCL2: An Important Mediator Between Tumor Cells and Host Cells in Tumor Microenvironment. Front. Oncol. 2021, 11, 722916. [Google Scholar] [CrossRef] [PubMed]
- Martin, O.A.; Redon, C.E.; Nakamura, A.J.; Dickey, J.S.; Georgakilas, A.G.; Bonner, W.M. Systemic DNA damage related to cancer. Cancer Res. 2011, 71, 3437–3441. [Google Scholar] [CrossRef] [Green Version]
- Burgess, J.T.; Rose, M.; Boucher, D.; Plowman, J.; Molloy, C.; Fisher, M.; O’Leary, C.; Richard, D.J.; O’Byrne, K.J.; Bolderson, E. The Therapeutic Potential of DNA Damage Repair Pathways and Genomic Stability in Lung Cancer. Front. Oncol. 2020, 10, 1256. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Y.D.; Zhan, Y.T.; Zhu, Y.H.; Li, Y.; Xie, D.; Guan, X.Y. High levels of CCL2 or CCL4 in the tumor microenvironment predict unfavorable survival in lung adenocarcinoma. Thorac. Cancer 2018, 9, 775–784. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-Y.; Shu, L.-H.; Liu, H.-T.; Cheng, Y.-C.; Wu, Y.-H.; Wu, Y.-H. The Role of MRE11 in the IL-6/STAT3 Pathway of Lung Cancer Cells. Curr. Issues Mol. Biol. 2022, 44, 6132-6144. https://doi.org/10.3390/cimb44120418
Wu C-Y, Shu L-H, Liu H-T, Cheng Y-C, Wu Y-H, Wu Y-H. The Role of MRE11 in the IL-6/STAT3 Pathway of Lung Cancer Cells. Current Issues in Molecular Biology. 2022; 44(12):6132-6144. https://doi.org/10.3390/cimb44120418
Chicago/Turabian StyleWu, Ching-Yuan, Li-Hsin Shu, Hung-Te Liu, Yu-Ching Cheng, Yu-Huei Wu, and Yu-Heng Wu. 2022. "The Role of MRE11 in the IL-6/STAT3 Pathway of Lung Cancer Cells" Current Issues in Molecular Biology 44, no. 12: 6132-6144. https://doi.org/10.3390/cimb44120418
APA StyleWu, C.-Y., Shu, L.-H., Liu, H.-T., Cheng, Y.-C., Wu, Y.-H., & Wu, Y.-H. (2022). The Role of MRE11 in the IL-6/STAT3 Pathway of Lung Cancer Cells. Current Issues in Molecular Biology, 44(12), 6132-6144. https://doi.org/10.3390/cimb44120418






