Comprehensive Analysis of Glutamate Receptor-like Genes in Rice (Oryza sativa L.): Genome-Wide Identification, Characteristics, Evolution, Chromatin Accessibility, gcHap Diversity, Population Variation and Expression Analysis
Abstract
:1. Introduction
2. Materials and Methods
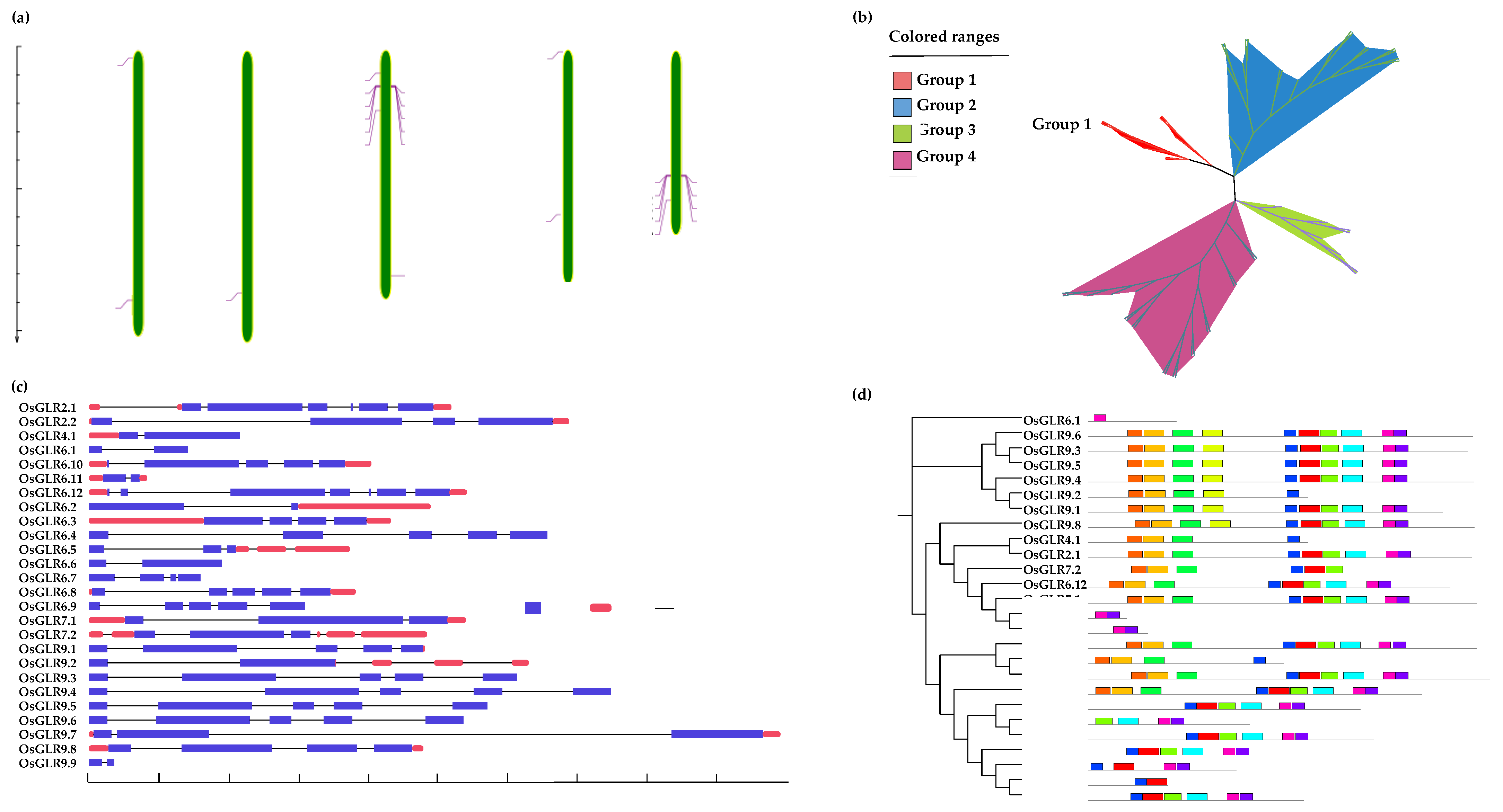
2.1. Identification and Chromosomal Location of OsGLR Genes
2.2. Structural Characteristics and Phylogenetic Analysis of OsGLR Genes
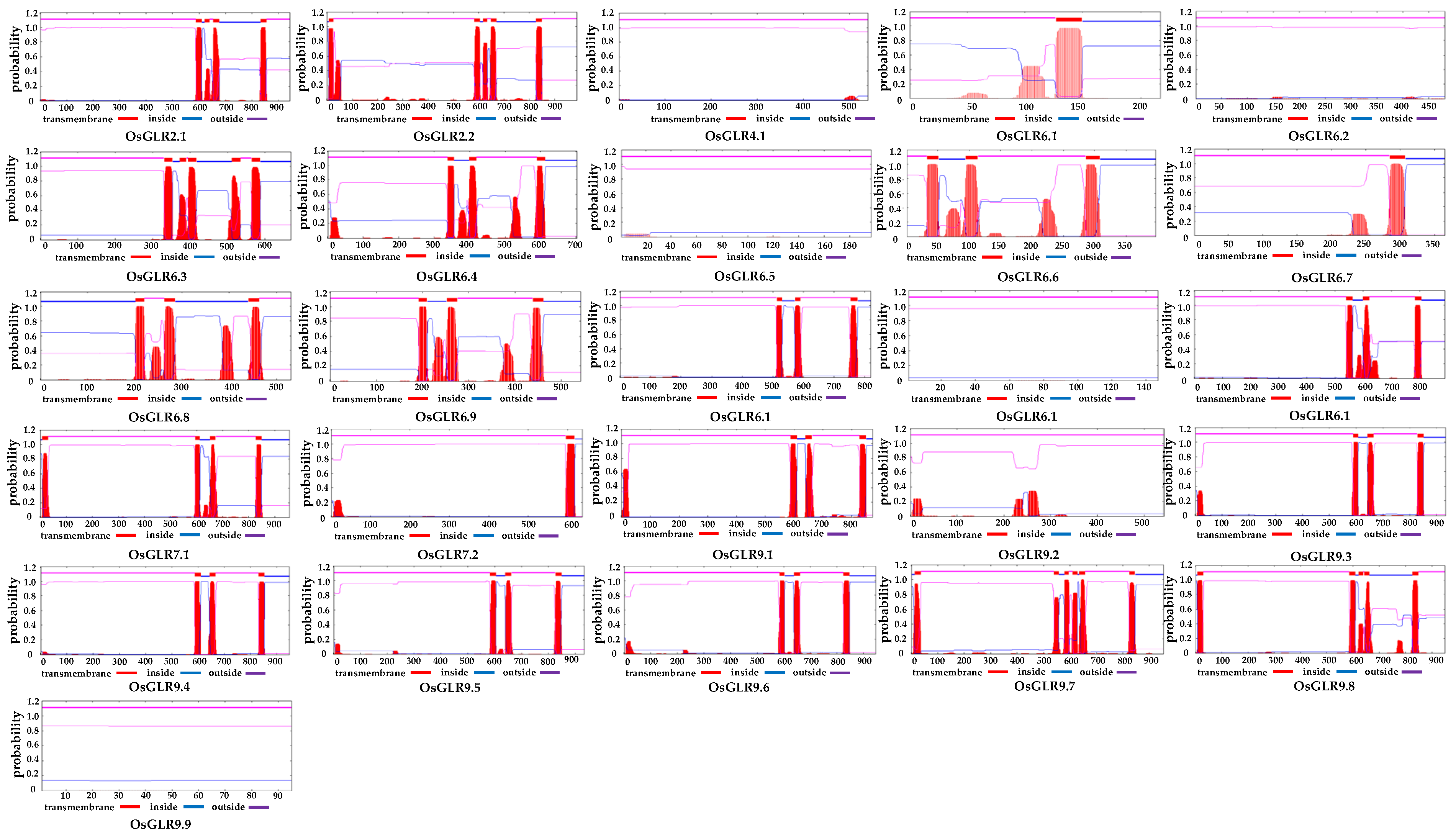
2.3. Prediction of Transmembrane Helices and Signal Peptide of the OsGLR Genes
2.4. Three-Dimensional (3D) Structure and Conserved Motifs of OsGLR Genes
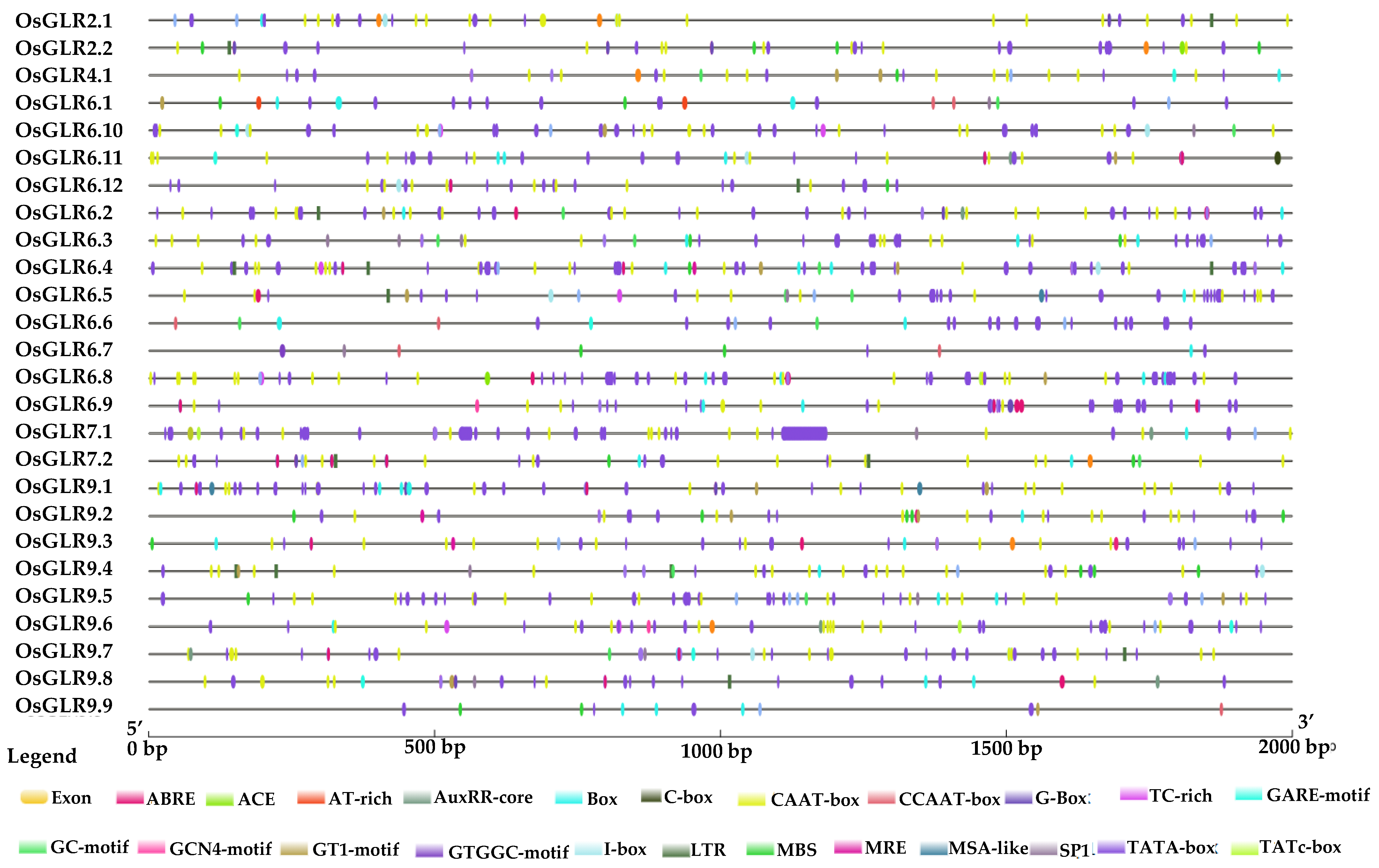
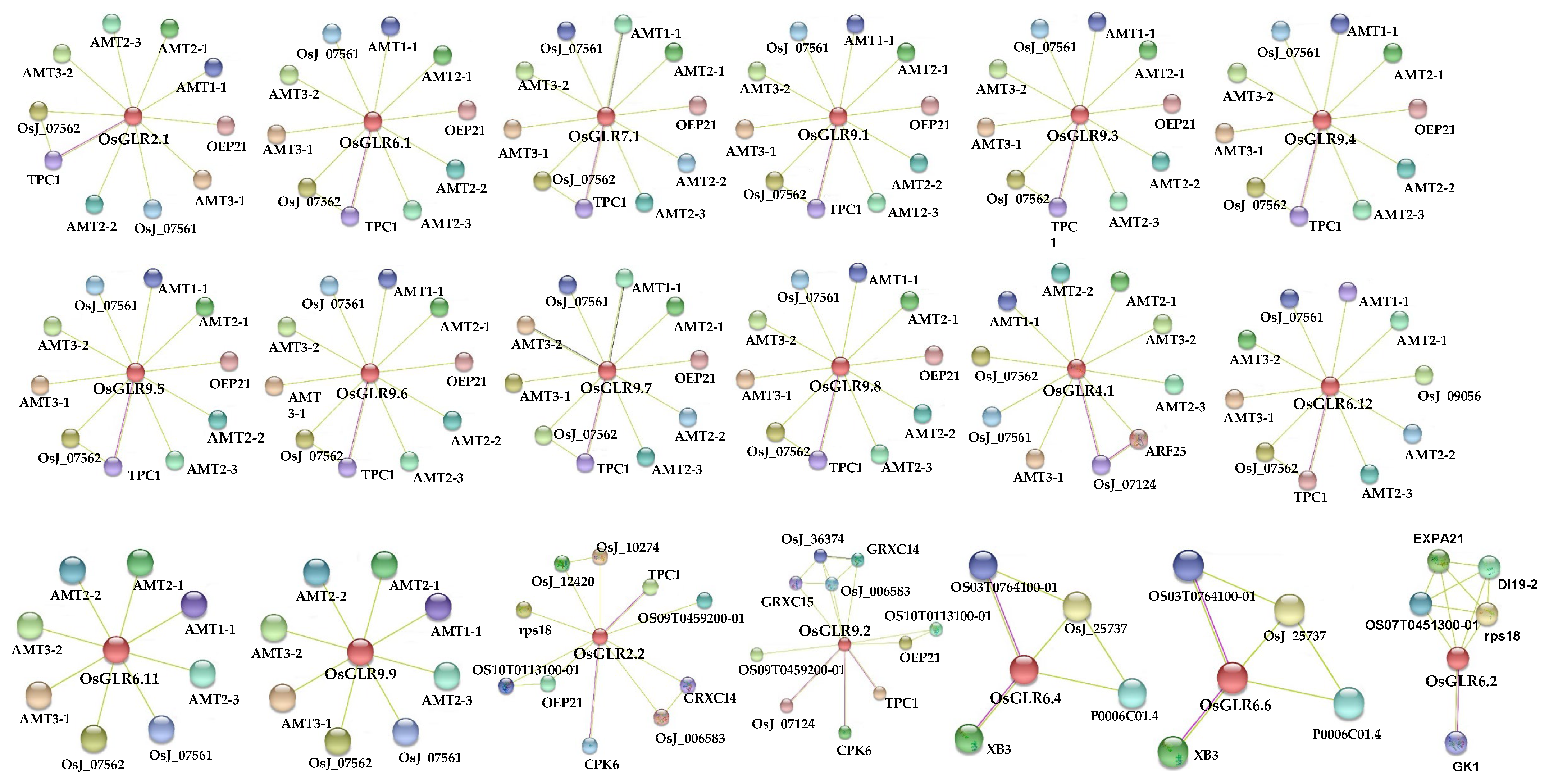
2.5. Cis-Acting Regulatory Elements, Functional Interaction Network and Chromatin Accessibility of OsGLR Genes
2.6. Population Variation, Gene-Coding Sequence Haplotype (gcHap) and KAKS of OsGLR Genes
2.7. Expression Difference of OsGLR Genes under Different Hormone Treatments
2.8. Semi-Quantitative PCR Analysis of OsGLR Genes under Low-Temperature Stress
3. Results
3.1. Genome-Wide Identification and Characterization of the OsGLR Genes in Rice
3.2. Transmembrane (TM) Helices and Signal Peptides of OsGLR Genes
3.3. Three-Dimensional (3D) Structure of OsGLR Genes
3.4. Cis-Acting Regulatory Elements and Functional Interaction Network of the OsGLR Genes
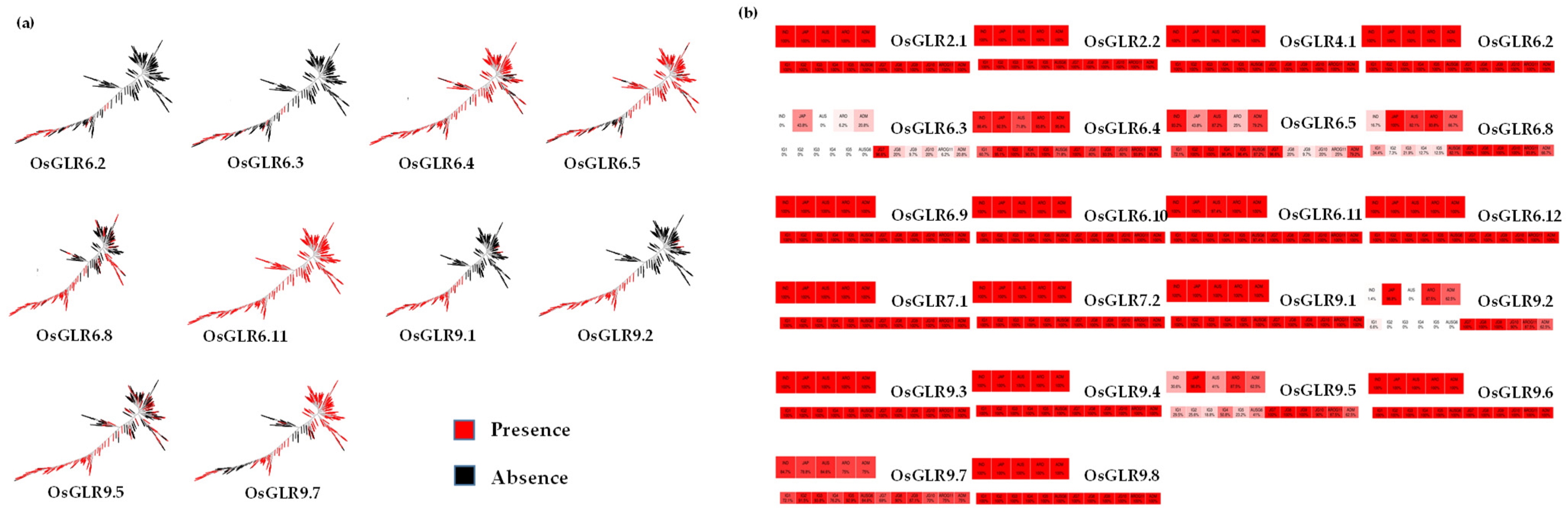
3.5. Chromatin Accessibility of OsGLR Genes
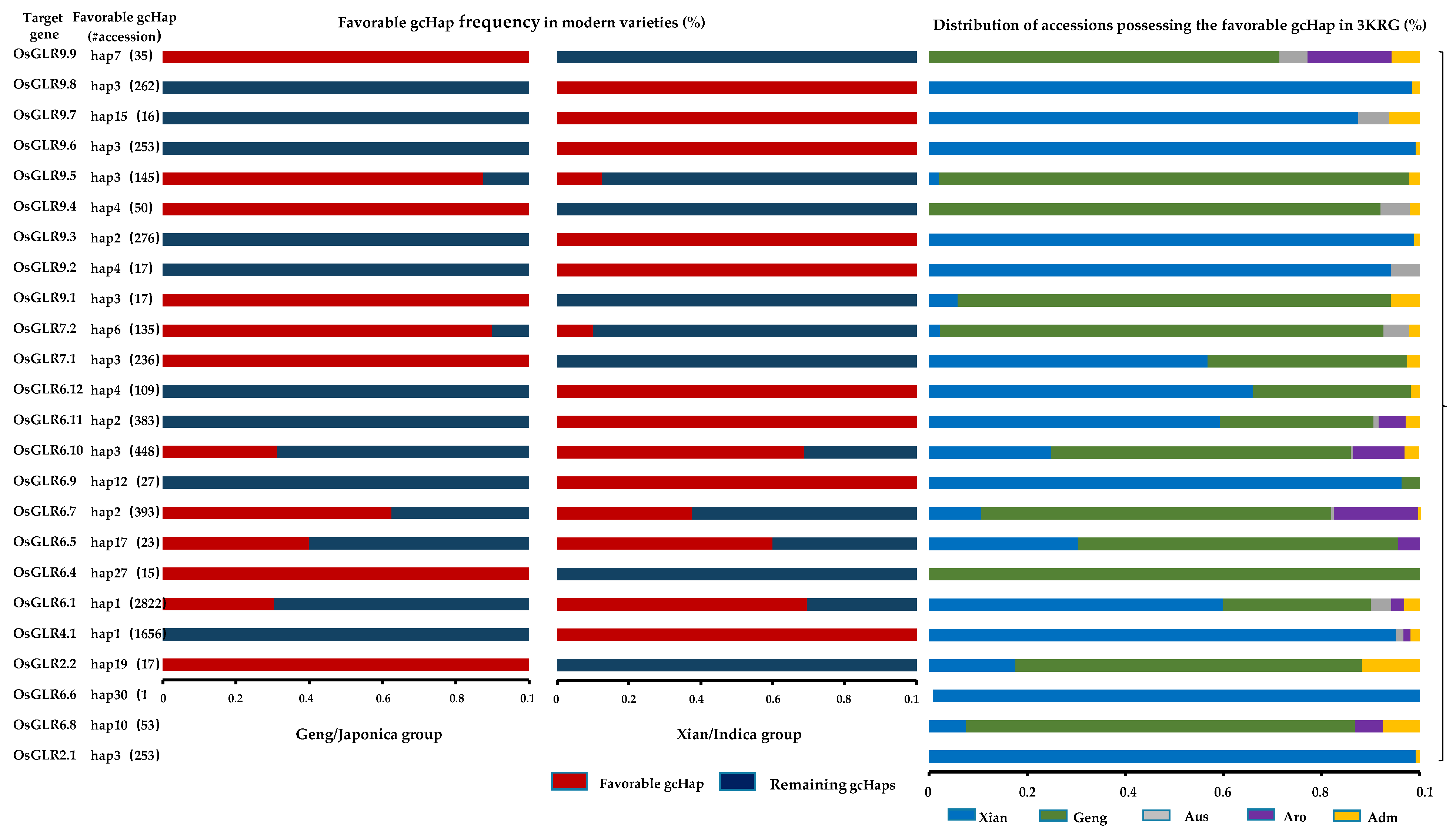
3.6. Evolutionary, Population Variation and Gene-Coding Sequence Haplotype (gcHap) Analysis of the GLR Genes in Rice
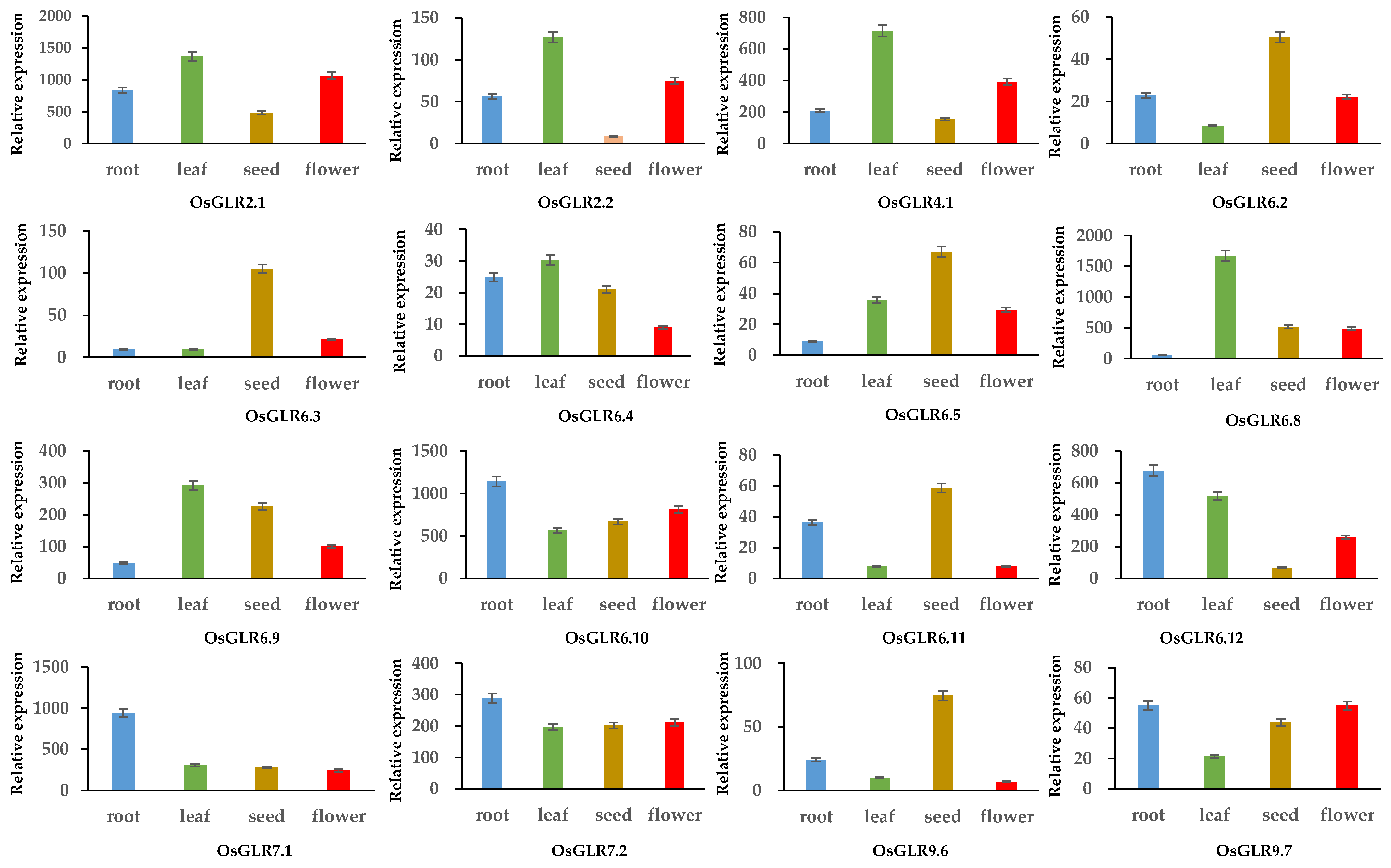
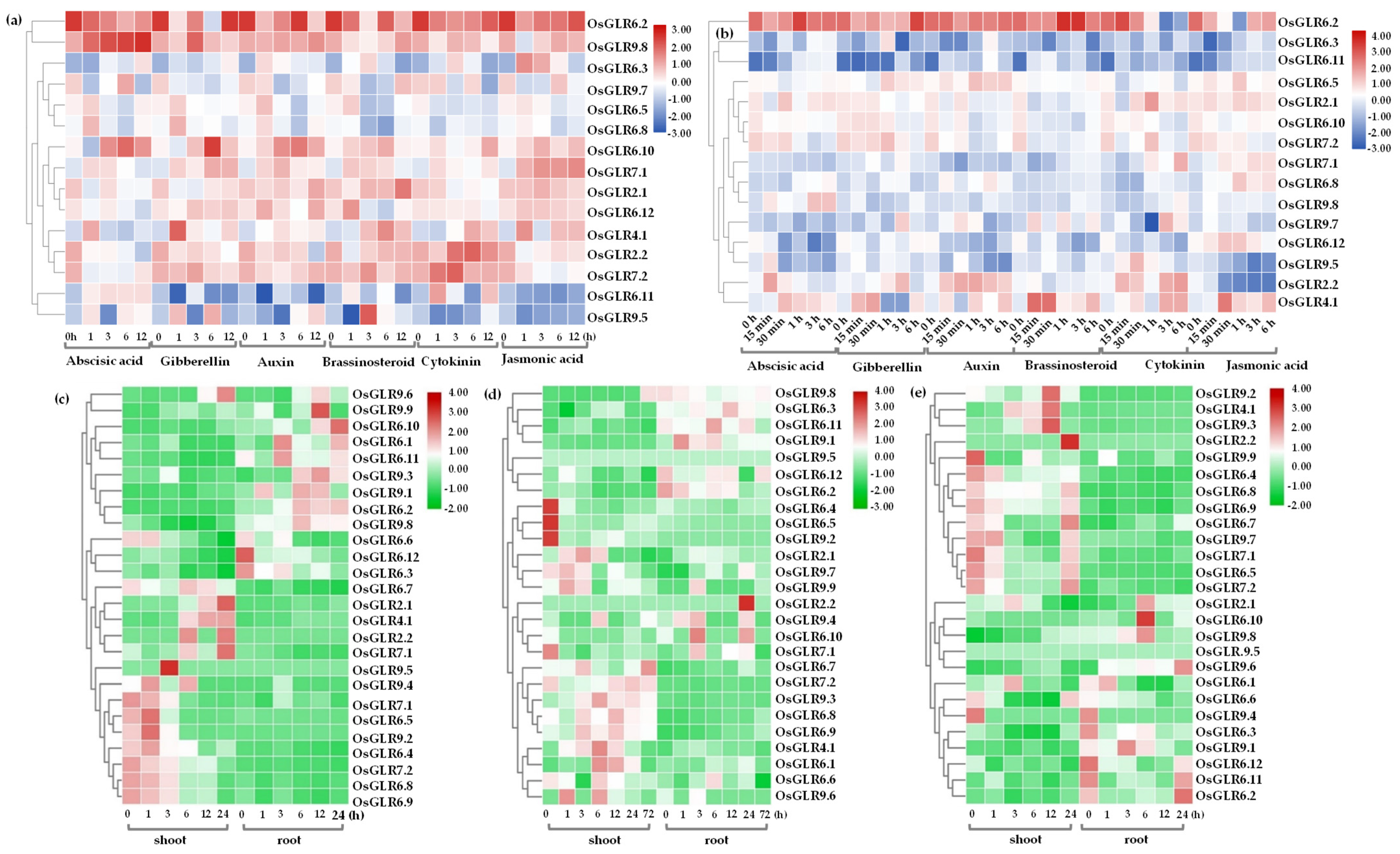
3.7. Expression Analysis of OsGLR Genes under Multiple Abiotic Stress and Hormone Treatments
3.8. Semi-Quantitative PCR Analysis of OsGLR Genes under Low-Temperature Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Wang, J.Q.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Traynelis, S.F.; Wollmuth, L.P.; McBain, C.J.; Menniti, F.S.; Vance, K.M.; Ogden, K.K.; Hansen, K.B.; Yuan, H.J.; Myers, S.J.; Dingledine, R. Glutamate Receptor Ion Channels: Structure, Regulation, and Function. Pharmacol. Rev. 2010, 62, 405–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, J.C.; Brenner, E.D.; DeSalle, R.; Nitabach, M.N.; Holmes, T.C.; Coruzzi, G.M. Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol. Biol. Evol. 2002, 19, 1066–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubos, C.; Huggins, D.; Grant, G.H.; Knight, M.R.; Campbell, M.M. A role for glycine in the gating of plant NMDA-like receptors. Plant J. 2003, 35, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, S.H.; Song, X.W.; Shen, Y.; Chen, H.M.; Yu, J.; Yi, K.K.; Liu, Y.F.; Karplus, V.J.; Wu, P.; et al. A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell 2006, 18, 340–349. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Yu, Z.M.; Du, G.K.; Zhang, Y.Y.; Taylor, J.L.; Shen, C.J.; Xu, J.; Liu, X.Y.; Wang, Y.F.; Wu, Y.R. Heterologous Expression and Functional Analysis of Rice Glutamate Receptor-Like Family Indicates its Role in Glutamate Triggered Calcium Flux in Rice Roots. Rice 2016, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.H.; Yu, X.Z.; Mo, L.Y.; Lin, Y.J.; Zhang, Q. Involvement of glutamate receptors in regulating calcium influx in rice seedlings under Cr exposure. Ecotoxicology 2019, 28, 650–657. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Mooers, B.H.M. Shortcuts for faster image creation in PyMOL. Protein Sci. 2020, 29, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; Volume 2, pp. 28–36. [Google Scholar]
- Zhao, H.; Li, J.C.; Yang, L.; Qin, G.; Xia, C.J.; Xu, X.B.; Su, Y.M.; Liu, Y.M.; Ming, L.C.; Chen, L.L.; et al. An inferred functional impact map of genetic variants in rice. Mol. Plant 2021, 14, 1584–1599. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.C.; Yu, H.; Huang, J.; Wang, W.S.; Faruquee, M.; Zhang, F.; Zhao, X.Q.; Fu, B.Y.; Chen, K.; Zhang, H.L.; et al. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0. Plant Biotechnol. J. 2020, 18, 14–16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wang, C.; Li, M.; Cui, Y.; Shi, Y.; Wu, Z.; Hu, Z.; Wang, W.; Xu, J.; Li, Z. The landscape of gene-CDS-haplotype diversity in rice: Properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol. Plant 2021, 14, 787–804. [Google Scholar] [CrossRef]
- Zeng, W.; Shi, J.; Qiu, C.; Wang, Y.; Rehman, S.; Yu, S.; Huang, S.; He, C.; Wang, W.; Chen, H.; et al. Identification of a genomic region controlling thermotolerance at flowering in maize using a combination of whole genomic re-sequencing and bulked segregant analysis. Theory Appl. Genet. 2020, 133, 2797–2810. [Google Scholar] [CrossRef]
- Manzoor, H.; Kelloniemi, J.; Chiltz, A.; Wendehenne, D.; Pugin, A.; Poinssot, B.; Garcia-Brugger, A. Involvement of the glutamate receptor AtGLR3.3 in plant defense signaling and resistance to Hyaloperonospora arabidopsidis. Plant J. 2013, 76, 466–480. [Google Scholar] [CrossRef]
- Li, H.Z.; Jiang, X.C.; Lv, X.Z.; Ahammed, G.J.; Guo, Z.X.; Qi, Z.Y.; Yu, J.Q.; Zhou, Y.H. Tomato GLR3.3 and GLR3.5 mediate cold acclimation-induced chilling tolerance by regulating apoplastic H2O2 production and redox homeostasis. Plant Cell Environ. 2019, 42, 3326–3339. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Guo, C.; Peng, J.; Li, C.; Wan, F.F.; Zhang, S.M.; Zhou, Y.Y.; Yan, Y.; Qi, L.J.; Sun, K.W.; et al. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytol. 2019, 221, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezhani, S.; Sherameti, I.; Pfannschmidt, T.; Oelmuller, R. A repressor with similarities to prokaryotic and eukaryotic DNA helicases controls the assembly of the CAAT box binding complex at a photosynthesis gene promoter. J. Biol. Chem. 2001, 276, 23785–23789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Xu, W.; Hu, X.S.; Liu, H.J.; Lin, Y.J. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016, 6, 20881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, N.; Guimaraes, J.C.; Gross, T.; Schmidt, A.; Vina-Vilaseca, A.; Nedialkova, D.D.; Aeschimann, F.; Leidel, S.A.; Spang, A.; Zavolan, M. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat. Commun. 2017, 8, 457. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Sun, C.; Hu, Z.; Zheng, T.; Lu, K.; Zhao, Y.; Wang, W.; Shi, J.; Wang, C.; Lu, J.; Zhang, D.; et al. RPAN: Rice pan-genome browser for approximately 3000 rice genomes. Nucleic Acids Res. 2017, 45, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Ju, C.L.; Song, Y.N.; Kong, D.D. Arabidopsis GLR3.5-modulated seed germination involves GA and ROS signaling. Plant Signal. Behav. 2020, 15, 1729537. [Google Scholar] [CrossRef]
- Meyerhoff, O.; Muller, K.; Roelfsema, M.R.; Latz, A.; Lacombe, B.; Hedrich, R.; Dietrich, P.; Becker, D. AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 2005, 222, 418–427. [Google Scholar] [CrossRef]
- Qi, Z.; Stephens, N.R.; Spalding, E.P. Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 2006, 142, 963–971. [Google Scholar] [CrossRef]
- Mousavi, S.A.R.; Chauvin, A.; Pascaud, F.; Kellenberger, S.; Farmer, E.E. GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 2013, 500, 422–426. [Google Scholar] [CrossRef] [Green Version]
- Grenzi, M.; Bonza, M.C.; Alfieri, A.; Costa, A. Structural insights into long-distance signal transduction pathways mediated by plant glutamate receptor-like channels. New Phytol. 2021, 229, 1261–1267. [Google Scholar] [CrossRef]
- Wang, P.H.; Lee, C.E.; Lin, Y.S.; Lee, M.H.; Chen, P.Y.; Chang, H.C.; Chang, I.F. The Glutamate Receptor-Like Protein GLR3.7 Interacts With 14-3-3 omega and Participates in Salt Stress Response in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1169. [Google Scholar] [CrossRef]
- Singh, S.K.; Chien, C.T.; Chang, I.F. The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J. Exp. Bot. 2016, 67, 1853–1869. [Google Scholar] [CrossRef] [Green Version]
- Vincill, E.D.; Clarin, A.E.; Molenda, J.N.; Spalding, E.P. Interacting glutamate receptor-like proteins in Phloem regulate lateral root initiation in Arabidopsis. Plant Cell 2013, 25, 1304–1313. [Google Scholar] [CrossRef] [Green Version]
- Wudick, M.M.; Portes, M.T.; Michard, E.; Rosas-Santiago, P.; Lizzio, M.A.; Nunes, C.O.; Campos, C.; Damineli, D.S.C.; Carvalho, J.C.; Lima, P.T.; et al. CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca2+ homeostasis. Science 2018, 360, 533–536. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R.; Mori, I.C.; Kamizono, N.; Shichiri, Y.; Shimatani, T.; Miyata, F.; Honda, K.; Iwai, S. Glutamate functions in stomatal closure in Arabidopsis and fava bean. J. Plant Res. 2016, 129, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Kong, D.; Hu, H.C.; Okuma, E.; Lee, Y.; Lee, H.S.; Munemasa, S.; Cho, D.; Ju, C.; Pedoeim, L.; Rodriguez, B.; et al. L-Met Activates Arabidopsis GLR Ca2+ Channels Upstream of ROS Production and Regulates Stomatal Movement. Cell Rep. 2016, 17, 2553–2561. [Google Scholar] [CrossRef]
- Lv, A.M.; Fan, N.N.; Xie, J.P.; Yuan, S.L.; An, Y.; Zhou, P. Expression of CdDHN4, a Novel YSK2-Type Dehydrin Gene from Bermudagrass, Responses to Drought Stress through the ABA-Dependent Signal Pathway. Front. Plant Sci. 2017, 8, 748. [Google Scholar] [CrossRef]
- Zheng, Y.; Luo, L.; Wei, J.; Chen, Q.; Yang, Y.; Hu, X.; Kong, X. The glutamate receptors AtGLR1.2 and AtGLR1.3 increase cold tolerance by regulating jasmonate signaling in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 506, 895–900. [Google Scholar] [CrossRef]
- Hernández-Coronado, M.; Dias Araujo, P.C.; Ip, P.L.; Nunes, C.O.; Rahni, R.; Wudick, M.M.; Lizzio, M.A.; Feijó, J.A.; Birnbaum, K.D. Plant glutamate receptors mediate a bet-hedging strategy between regeneration and defense. Dev. Cell 2022, 57, 451–465.e456. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Ramírez, C.; Michard, E.; Simon, A.A.; Damineli, D.S.C.; Hernández-Coronado, M.; Becker, J.D.; Feijó, J.A. Glutamate Receptor-Like channels are essential for chemotaxis and reproduction in mosses. Nature 2017, 549, 91–95. [Google Scholar] [CrossRef] [PubMed]


| Gene id | Gene Name | The Upstream Primer (5’-3’) | Downstream Primers (5’-3’) |
|---|---|---|---|
| Os09t0431100 | OsGLR9.7 | GATGGTTGCTGATGGGGCATTC | CTTCAGGAACACCCACGTCC |
| Os09t0429000 | OsGLR9.3 | ATAACAGCTTCTGGGTATGCGAT | ATGGCTGCATCATATACCCCTAG |
| Os06t0680500 | OsGLR6.12 | CTAAACACAATCGACGAGTACGC | AATCTCTCTGGAATGCGAATCCC |
| Os04t0585200 | OsGLR4.1 | ACCGTCAATTTGTATCAGTTGATAG | ATAAGCTCCGAATAGCTTGGATTC |
| Os06t0188700 | OsGLR6.4 | TCGTGGTGGACATGACGAGC | CTTGATAAGGTCTTCGGCGGC |
| Os07t0103100 | OsGLR7.1 | ATCATCCAAGGTCTGCAGGTGAT | AGGAAGTAAGGGTATTGGGAGGA |
| Os02t0787600 | OsGLR2.2 | TGTTCGACGAGGTCATGAAGATT | CATCTGCCTTCTGTGACGACA |
| Os09t0428300 | OsGLR9.1 | AGAAAGGCAGAGGAATTCCATGT | CCAGCAAATCAAGAACTGCAGAT |
| Os03t0234200 | OsUBQ | AAGAAGCTGAAGCATCCAGC | CCAGGACAAGATGATCTGCC |
| Subfamily | RAP ID | Gene Name | No. of Amino Acids | Molecular Weight (Da) | Isoelectric Point (PI) | Aliphatic Index | Start (bp) | End (bp) | Location |
|---|---|---|---|---|---|---|---|---|---|
| I | Os02g0787600 | OsGLR2.2 | 988 | 107,252.1 | 6.4 | 83.94 | 33,469,138 | 33,475,994 | P(9/14),Ch(3/14) |
| I | Os06g0155000 | OsGLR6.1 | 217 | 22,390.23 | 6.29 | 76.54 | 2,836,446 | 2,837,851 | Ch(11/14),N(3/14) |
| I | Os06g0188400 | OsGLR6.2 | 481 | 52,305.65 | 8.34 | 91.73 | 4,459,007 | 4,463,882 | Cy(7/14),Ch(3/14) |
| I | Os09g0431100 | OsGLR9.7 | 955 | 100,789.9 | 6.18 | 86.15 | 15,752,949 | 15,762,817 | P(7/14),V(3/14) |
| II | Os09g0428300 | OsGLR9.1 | 872 | 96,824.56 | 7.99 | 96.9 | 15,564,675 | 15,569,470 | P(12/14) |
| II | Os09g0428600 | OsGLR9.2 | 541 | 60,230.77 | 6.15 | 88.84 | 15,574,058 | 15,580,334 | E(5/14), Ch(3/14), V(3/14) |
| II | Os09g0429000 | OsGLR9.3 | 933 | 104,346.78 | 5.96 | 92.18 | 15,588,269 | 15,594,378 | P(13/14) |
| II | Os09g0429200 | OsGLR9.4 | 948 | 105,520.29 | 6.33 | 90.88 | 15,599,149 | 15,606,593 | P(8/14),ER(3/14) |
| II | Os09g0429400 | OsGLR9.5 | 934 | 104,377.7 | 5.97 | 91.75 | 15,610,715 | 15,616,401 | P(11/14) |
| II | Os09g0429500 | OsGLR9.6 | 946 | 105,792.31 | 5.94 | 92.32 | 15,620,335 | 15,625,678 | P(13/14) |
| II | Os09g0431200 | OsGLR9.8 | 950 | 106,149.72 | 5.41 | 90.39 | 15,765,945 | 15,770,717 | P(13/14) |
| III | Os06g0188600 | OsGLR6.3 | 670 | 74,295.45 | 5.87 | 80.9 | 4,472,352 | 4,476,661 | P(8/14) |
| III | Os06g0188700 | OsGLR6.4 | 702 | 76,300.5 | 9.52 | 81.15 | 4,481,240 | 4,487,783 | P(10/14) |
| III | Os06g0188800 | OsGLR6.5 | 197 | 21,457.38 | 8.45 | 84.67 | 4,489,588 | 4,493,909 | Ch(10/14) |
| III | Os06g0189100 | OsGLR6.6 | 397 | 43,568.45 | 6.08 | 82.92 | 4,500,766 | 4,502,665 | P(6/14),V(4/14),ER(3/14) |
| III | Os06g0190000 | OsGLR6.7 | 365 | 39,821.64 | 4.88 | 79.01 | 4,556,487 | 4,558,077 | E(4/14),V(3/14) |
| III | Os06g0190500 | OsGLR6.8 | 531 | 59,380.02 | 9.1 | 89.76 | 4,571,098 | 4,574,904 | P(8/14),M(3/14) |
| III | Os06g0190700 | OsGLR6.9 | 542 | 59,762.91 | 8.73 | 82.2 | 4,587,987 | 4,591,068 | P(8/14) |
| III | Os06g0190800 | OsGLR6.10 | 820 | 91,791.68 | 8.41 | 92.21 | 4,593,916 | 4,597,945 | P(9/14), V(3/14) |
| IV | Os02g0117500 | OsGLR2.1 | 944 | 105,303.79 | 5.67 | 94.29 | 918,797 | 923,968 | P(13/14) |
| IV | Os04g0585200 | OsGLR4.1 | 540 | 59,717.23 | 6.36 | 95.67 | 29,565,754 | 29,570,387 | P(10/14) |
| IV | Os06g0246600 | OsGLR6.11 | 147 | 15,686.37 | 4.05 | 90.27 | 7,607,108 | 7,607,939 | Ch(4/14),E(5/14),M(3/14) |
| IV | Os06g0680500 | OsGLR6.12 | 890 | 98,881.67 | 5.34 | 90.39 | 28,345,366 | 28,350,758 | P(11/14) |
| IV | Os07g0103100 | OsGLR7.1 | 956 | 104,440.94 | 6.22 | 92.01 | 191,053 | 196,429 | P(10/14),ER(3/14) |
| IV | Os07g0522600 | OsGLR7.2 | 637 | 69,593.23 | 5.35 | 93.05 | 20,221,077 | 20,225,924 | P(5/14),ER(4/14),V(3/14) |
| IV | Os09g0485700 | OsGLR9.9 | 95 | 10,497.73 | 4.54 | 83.16 | 18,740,826 | 18,741,187 | P(10/14),Cy(3/14) |
| Seq_1 | Seq_2 | Ka | Ks | Ka_Ks | Mya |
|---|---|---|---|---|---|
| OsGLR6.8 | OsGLR6.9 | 0.17 | 0.32 | 0.53 | 24.53 |
| OsGLR9.5 | OsGLR9.3 | 0.06 | 0.27 | 0.22 | 20.93 |
| OsGLR9.5 | OsGLR9.6 | 0.07 | 0.28 | 0.25 | 21.61 |
| OsGLR9.6 | OsGLR9.3 | 0.06 | 0.27 | 0.22 | 21.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Zeng, W.; Xu, M.; Li, H.; Zhang, F.; Chen, Z.; Anicet, G.; Huang, S.; Huang, Y.; Wang, X.; et al. Comprehensive Analysis of Glutamate Receptor-like Genes in Rice (Oryza sativa L.): Genome-Wide Identification, Characteristics, Evolution, Chromatin Accessibility, gcHap Diversity, Population Variation and Expression Analysis. Curr. Issues Mol. Biol. 2022, 44, 6404-6427. https://doi.org/10.3390/cimb44120437
Shi Y, Zeng W, Xu M, Li H, Zhang F, Chen Z, Anicet G, Huang S, Huang Y, Wang X, et al. Comprehensive Analysis of Glutamate Receptor-like Genes in Rice (Oryza sativa L.): Genome-Wide Identification, Characteristics, Evolution, Chromatin Accessibility, gcHap Diversity, Population Variation and Expression Analysis. Current Issues in Molecular Biology. 2022; 44(12):6404-6427. https://doi.org/10.3390/cimb44120437
Chicago/Turabian StyleShi, Yingyao, Wei Zeng, Minhui Xu, Hua Li, Fanlin Zhang, Zulong Chen, Gatera Anicet, Shiji Huang, Yuheng Huang, Xiyu Wang, and et al. 2022. "Comprehensive Analysis of Glutamate Receptor-like Genes in Rice (Oryza sativa L.): Genome-Wide Identification, Characteristics, Evolution, Chromatin Accessibility, gcHap Diversity, Population Variation and Expression Analysis" Current Issues in Molecular Biology 44, no. 12: 6404-6427. https://doi.org/10.3390/cimb44120437






