DNA Damage and Radiosensitivity in Blood Cells from Subjects Undergoing 45 Days of Isolation and Confinement: An Explorative Study
Abstract
:1. Introduction
2. Results
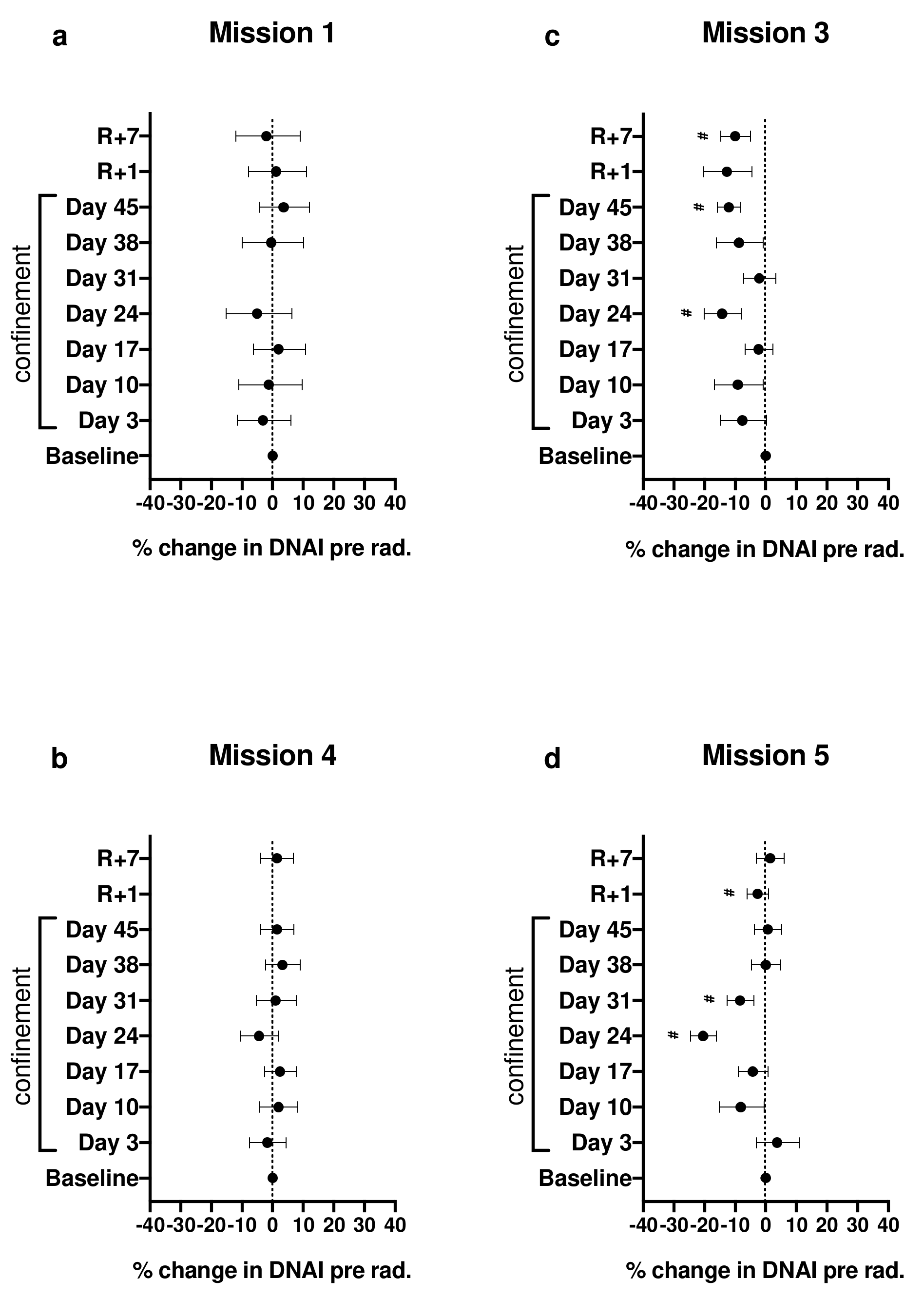
2.1. Effect of Confinement on DNAI before Irradiation
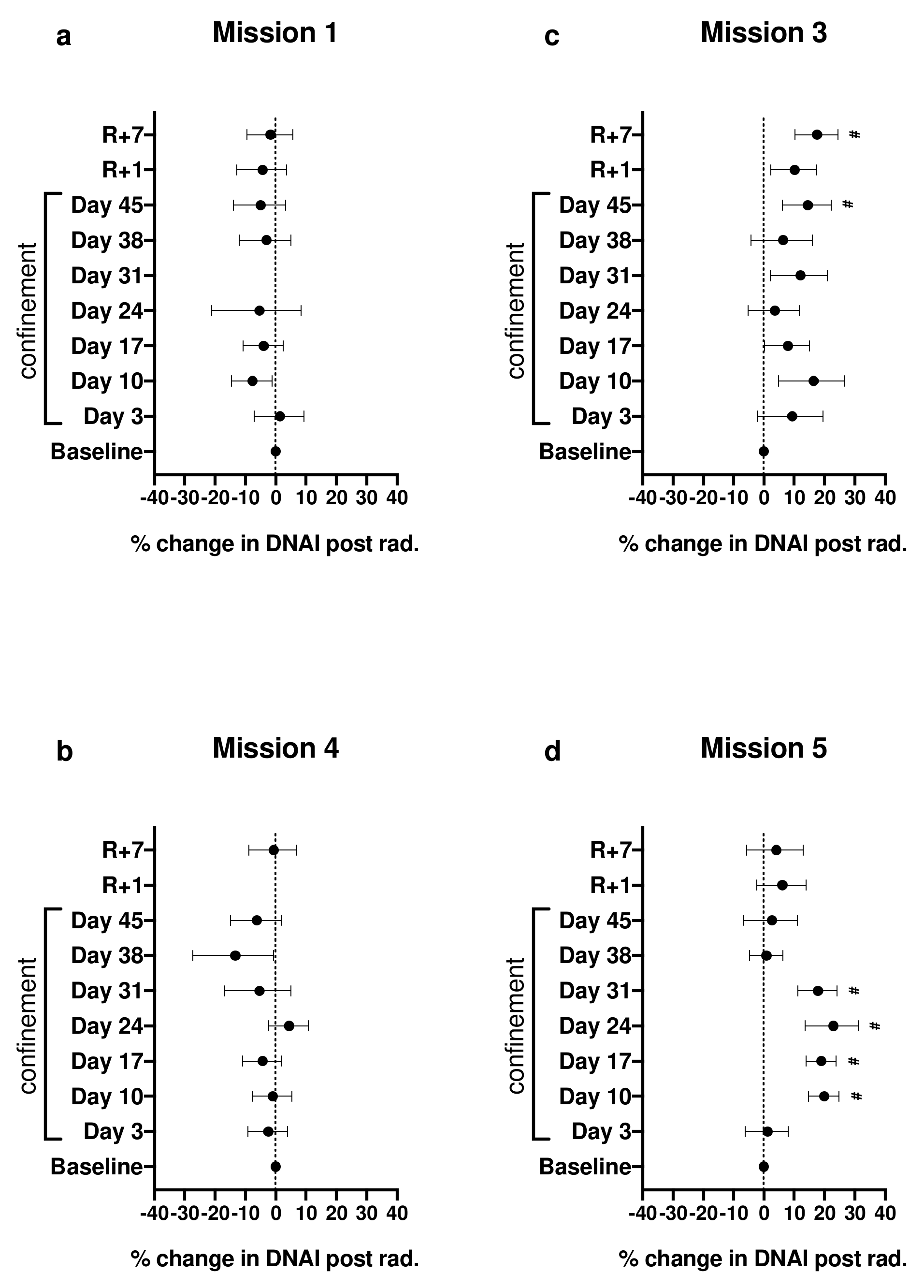
2.2. Effect of Irradiation on DNAI during Confinement
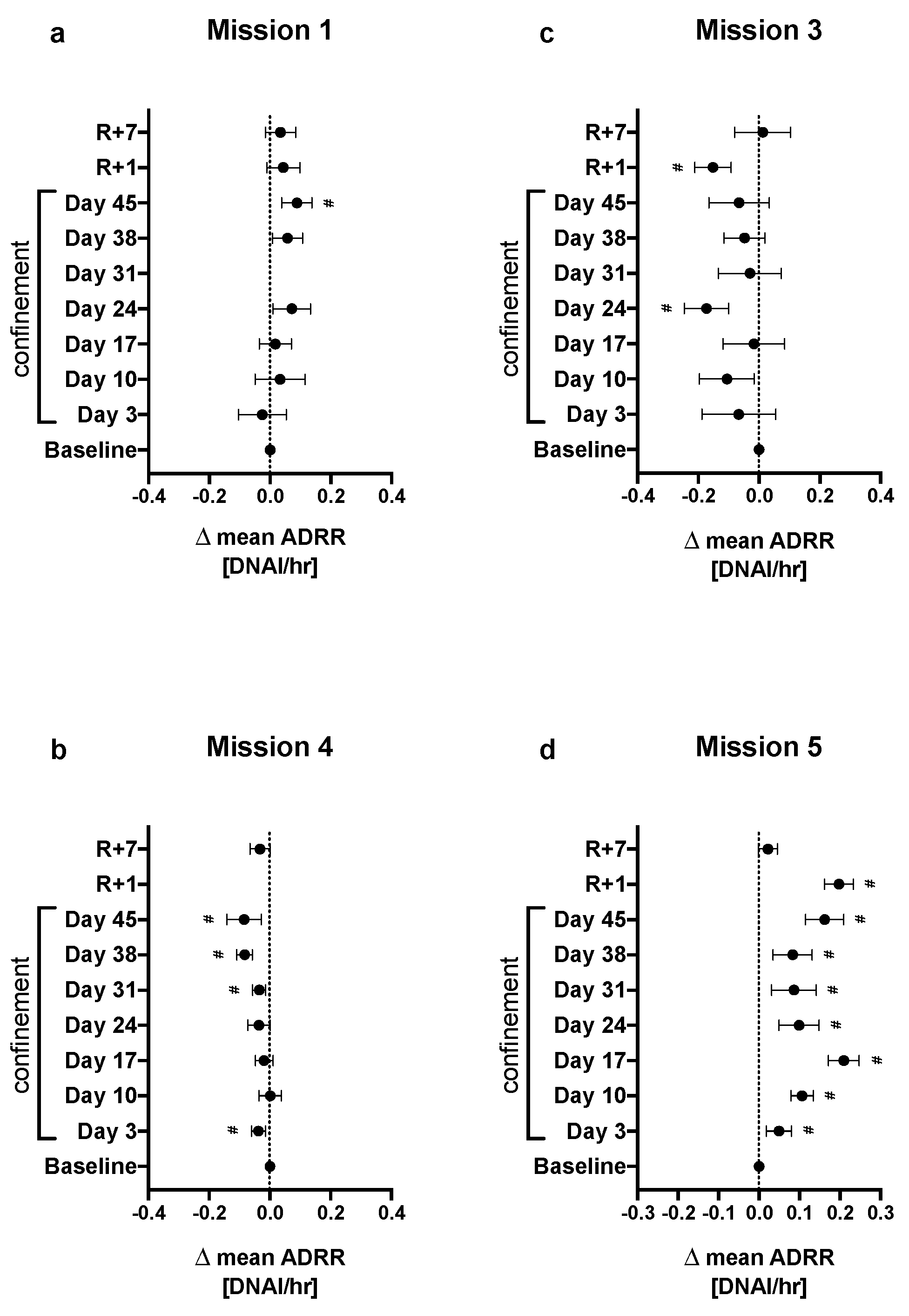
2.3. Effect of Confinement on DNA Repair Rate
2.4. Effect of Age and Sex
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Blood Draw and Sample Processing
4.3. Cell Thawing Procedure
4.4. Induction of DNA Strand Breaks through Irradiation
4.5. Detection of DNA Strand Breaks
4.6. Experimental Design
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
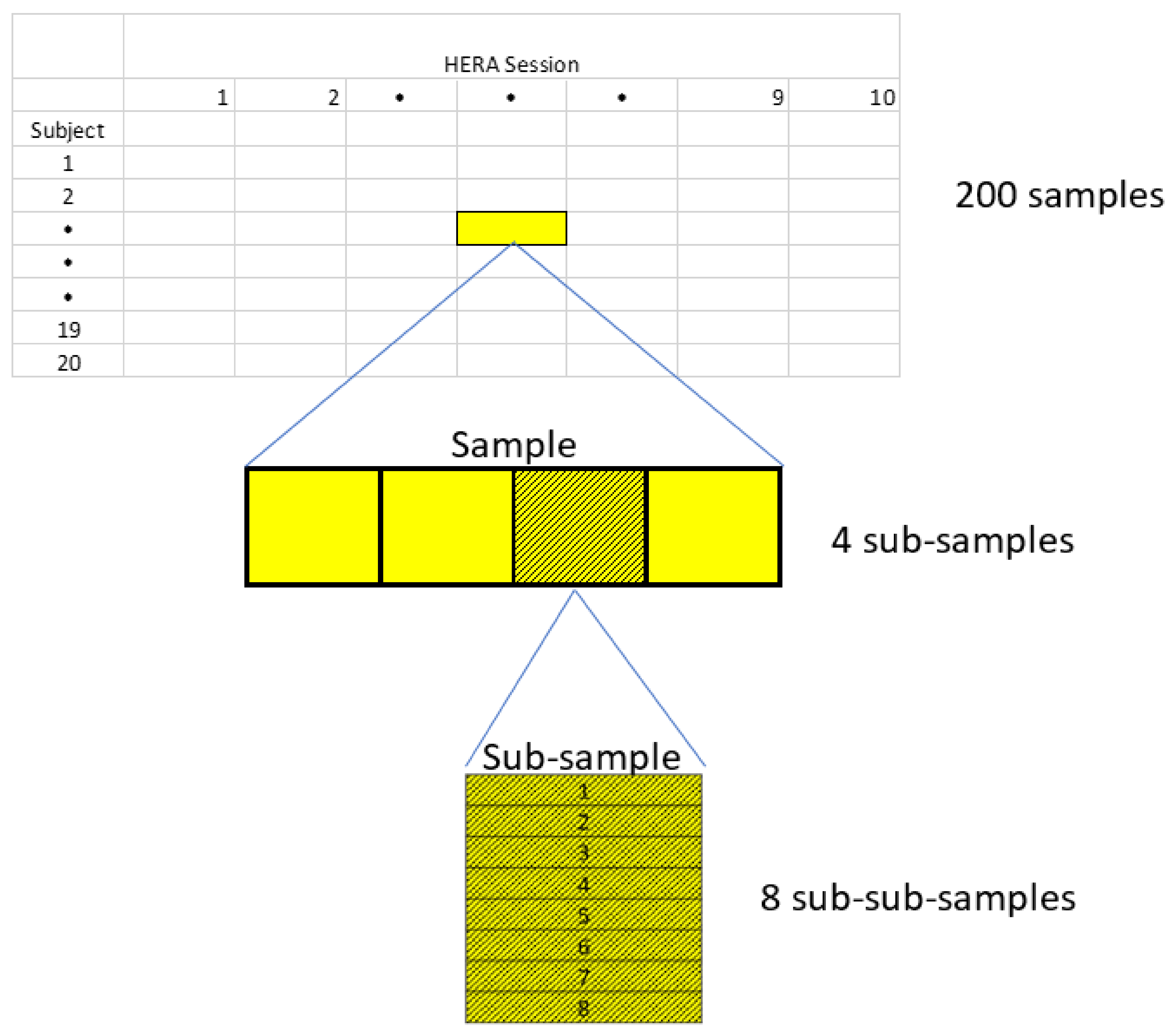
Appendix A.1. Experimental Design (as Originally Planned)
- 20 subjects
- 10 HERA sessions (Table A1)
- Separate sample of lymphocytes for each combination of subject and session = 200 samples total
- 4 subsamples for each sample (“replicates”)
- 100 fluorescence microplates
| HERA Session | Day |
|---|---|
| 1 | pre-confinement |
| 2 | Day 3 |
| 3 | Day 10 |
| 4 | Day 17 |
| 5 | Day 24 |
| 6 | Day 31 |
| 7 | Day 38 |
| 8 | Day 45 |
| 9 | Recovery Day 1 |
| 10 | Recovery Day 7 |
| Sub-Subsample | Exp Condition | X-ray Radiation | Incubation Time (min) |
|---|---|---|---|
| 1 | Viability | none | none |
| 2 | Pre-irradiation | none | none |
| 3 | Time Point 1 | 3.73 Gy | 0 |
| 4 | Time Point 2 | 3.73 Gy | 15 |
| 5 | Time Point 3 | 3.73 Gy | 30 |
| 6 | Time Point 4 | 3.73 Gy | 45 |
| 7 | Time Point 5 | 3.73 Gy | 60 |
| 8 | Time Point 6 | 3.73 Gy | 75 |
| Plate | Subjects | Mission | Session | # of Sub-Samples |
|---|---|---|---|---|
| 1 | 1,2 | 1 | Baseline | 64 * |
| 2 | 3,4 | 1 | Baseline | 64 |
| 3 | 5,6 | 2 | Baseline | 64 |
| 4 | 7,8 | 2 | Baseline | 64 |
| 5 | 9,10 | 3 | Baseline | 64 |
| 6 | 11,12 | 3 | Baseline | 64 |
| 7 | 13,14 | 4 | Baseline | 64 |
| 8 | 15,16 | 4 | Baseline | 64 |
| 9 | 17,18 | 5 | Baseline | 64 |
| 10 | 19,20 | 5 | Baseline | 64 |
| 11 | 1,2 | 1 | Day 3 | 64 |
| 12 | 3,4 | 1 | Day 3 | 64 |
| 13 | 5,6 | 2 | Day 3 | 64 |
| 14 | 7,8 | 2 | Day 3 | 64 |
| 15 | 9,10 | 3 | Day 3 | 64 |
| 16 | 11,12 | 3 | Day 3 | 64 |
| 17 | 13,14 | 4 | Day 3 | 64 |
| 18 | 15,16 | 4 | Day 3 | 64 |
| 19 | 17,18 | 5 | Day 3 | 64 |
| 20 | 19,20 | 5 | Day 3 | 64 |
| Plates 21–30: same as above, except session is Day 10 | ||||
| Plates 31–40: same as above, except session is Day 17 | ||||
| Plates 41–50: same as above, except session is Day 24 | ||||
| Plates 51–60: same as above, except session is Day 31 | ||||
| Plates 61–70: same as above, except session is Day 38 | ||||
| Plates 71–80: same as above, except session is Day 45 | ||||
| Plates 81–90: same as above, except session is R + 1 | ||||
| Plates 91–100: same as above, except session is R + 7 | ||||
Appendix A.2. Missing Samples/Data
- The 4 participants (subjects 5–8) in the second mission were required to leave the HERA facility at day 17 due to extreme weather conditions in the Houston area (Hurricane Harvey 2017).
- No data was gathered for day 31 of Mission 1 (Subjects 1–4) because of technical problems with the fluorescence measurements.
- Blood samples were not gathered in the HERA on day R + 1 of Mission 4 (Subjects 13–16).
Appendix A.3. DNAI Mixed-Model Regression Analysis
Appendix A.4. Calculation of Plotted Values Shown in Figures 1 and 2 of the Primary Article
Appendix A.5. Mixed-Model Regression Analysis for Average DNAI Repair Rate (ADRR)
Appendix A.6. Calculation of Plotted Values Shown in Figure 3 of the Primary Article
References
- Mumtaz, F.; Khan, M.I.; Zubair, M.; Dehpour, A.R. Neurobiology and consequences of social isolation stress in animal model-A comprehensive review. Biomed Pharm. 2018, 105, 1205–1222. [Google Scholar] [CrossRef]
- Takatsu-Coleman, A.L.; Patti, C.L.; Zanin, K.A.; Zager, A.; Carvalho, R.C.; Borcoi, A.R.; Ceccon, L.M.; Berro, L.F.; Tufik, S.; Andersen, M.L.; et al. Short-term social isolation induces depressive-like behaviour and reinstates the retrieval of an aversive task: Mood-congruent memory in male mice? J. Psychiatry Neurosci. 2013, 38, 259–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Famitafreshi, H.; Karimian, M. Assessment of Improvement in Oxidative Stress Indices with Resocialization in Memory Retrieval in Y-Maze in Male Rats. J. Exp. Neurosci. 2018, 12, 1179069518820323. [Google Scholar] [CrossRef] [Green Version]
- Gadek-Michalska, A.; Tadeusz, J.; Bugajski, A.; Bugajski, J. Chronic Isolation Stress Affects Subsequent Crowding Stress-Induced Brain Nitric Oxide Synthase (NOS) Isoforms and Hypothalamic-Pituitary-Adrenal (HPA) Axis Responses. Neurotox. Res. 2019, 36, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Colaianna, M.; Schiavone, S.; Zotti, M.; Tucci, P.; Morgese, M.G.; Backdahl, L.; Holmdahl, R.; Krause, K.H.; Cuomo, V.; Trabace, L. Neuroendocrine profile in a rat model of psychosocial stress: Relation to oxidative stress. Antioxid. Redox Signal. 2013, 18, 1385–1399. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, J.R.; McMahon, E.K.; Boner, W.; Haussmann, M.F. Oxytocin administration prevents cellular aging caused by social isolation. Psychoneuroendocrinology 2019, 103, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Krugel, U.; Fischer, J.; Bauer, K.; Sack, U.; Himmerich, H. The impact of social isolation on immunological parameters in rats. Arch. Toxicol. 2014, 88, 853–855. [Google Scholar] [CrossRef]
- Aivaliotis, I.L.; Pateras, I.S.; Papaioannou, M.; Glytsou, C.; Kontzoglou, K.; Johnson, E.O.; Zoumpourlis, V. How do cytokines trigger genomic instability? J. Biomed. Biotechnol. 2012, 2012, 536761. [Google Scholar] [CrossRef]
- Akhtar, S.; Najafzadeh, M.; Isreb, M.; Newton, L.; Gopalan, R.C.; Anderson, D. ROS-induced oxidative damage in lymphocytes ex vivo/in vitro from healthy individuals and MGUS patients: Protection by myricetin bulk and nanoforms. Arch. Toxicol. 2020, 94, 1229–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cemeli, E.; Anderson, D. Mechanistic investigation of ROS-induced DNA damage by oestrogenic compounds in lymphocytes and sperm using the comet assay. Int. J. Mol. Sci. 2011, 12, 2783–2796. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, M.S.; Baum, A.; Chambers, W.H.; Jenkins, F.J. Induction of DNA damage, alteration of DNA repair and transcriptional activation by stress hormones. Psychoneuroendocrinology 2007, 32, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, M.; LaRusso, N.F.; Burgart, L.J.; Gores, G.J. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 2000, 60, 184–190. [Google Scholar] [PubMed]
- Moreno-Villanueva, M.; Burkle, A. Stress Hormone-Mediated DNA Damage Response--Implications for Cellular Senescence and Tumour Progression. Curr. Drug Targets 2016, 17, 398–404. [Google Scholar] [CrossRef]
- Rosales, A.L.; Cunningham, J.M.; Bone, A.J.; Green, I.C.; Green, M.H. Repair of cytokine-induced DNA damage in cultured rat islets of Langerhans. Free Radic. Res. 2004, 38, 665–674. [Google Scholar] [CrossRef]
- Thomas, M.; Palombo, P.; Schuhmacher, T.; von Scheven, G.; Bazylianska, V.; Salzwedel, J.; Schafer, N.; Burkle, A.; Moreno-Villanueva, M. Impaired PARP activity in response to the beta-adrenergic receptor agonist isoproterenol. Toxicol. In Vitr. 2018, 50, 29–39. [Google Scholar] [CrossRef]
- Topalovic, D.; Dekanski, D.; Spremo-Potparevic, B.; Djelic, N.; Bajic, V.; Zivkovic, L. Assessment of adrenaline-induced DNA damage in whole blood cells with the comet assay. Arh. Hig. Rada. Toksikol. 2018, 69, 304–308. [Google Scholar] [CrossRef] [Green Version]
- Pagel, J.I.; Chouker, A. Effects of isolation and confinement on humans-implications for manned space explorations. J. Appl. Physiol. 2016, 120, 1449–1457. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.J.; Gavey, S.; NE, R.I.; Kontari, P.; Victor, C. The association between loneliness, social isolation and inflammation: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2020, 112, 519–541. [Google Scholar] [CrossRef]
- Strewe, C.; Muckenthaler, F.; Feuerecker, M.; Yi, B.; Rykova, M.; Kaufmann, I.; Nichiporuk, I.; Vassilieva, G.; Horl, M.; Matzel, S.; et al. Functional changes in neutrophils and psychoneuroendocrine responses during 105 days of confinement. J. Appl. Physiol. 2015, 118, 1122–1127. [Google Scholar] [CrossRef]
- Basner, M.; Dinges, D.F.; Mollicone, D.J.; Savelev, I.; Ecker, A.J.; Di Antonio, A.; Jones, C.W.; Hyder, E.C.; Kan, K.; Morukov, B.V.; et al. Psychological and behavioral changes during confinement in a 520-day simulated interplanetary mission to mars. PLoS ONE 2014, 9, e93298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacubowski, A.; Abeln, V.; Vogt, T.; Yi, B.; Chouker, A.; Fomina, E.; Struder, H.K.; Schneider, S. The impact of long-term confinement and exercise on central and peripheral stress markers. Physiol. Behav. 2015, 152, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.; Matzel, S.; Feuerecker, M.; Horl, M.; Ladinig, C.; Abeln, V.; Chouker, A.; Schneider, S. The impact of chronic stress burden of 520-d isolation and confinement on the physiological response to subsequent acute stress challenge. Behav. Brain Res. 2015, 281, 111–115. [Google Scholar] [CrossRef]
- Yi, B.; Rykova, M.; Feuerecker, M.; Jager, B.; Ladinig, C.; Basner, M.; Horl, M.; Matzel, S.; Kaufmann, I.; Strewe, C.; et al. 520-d Isolation and confinement simulating a flight to Mars reveals heightened immune responses and alterations of leukocyte phenotype. Brain Behav. Immun. 2014, 40, 203–210. [Google Scholar] [CrossRef]
- Shearer, W.T.; Lee, B.N.; Cron, S.G.; Rosenblatt, H.M.; Smith, E.O.; Lugg, D.J.; Nickolls, P.M.; Sharp, R.M.; Rollings, K.; Reuben, J.M. Suppression of human anti-inflammatory plasma cytokines IL-10 and IL-1RA with elevation of proinflammatory cytokine IFN-gamma during the isolation of the Antarctic winter. J. Allergy Clin. Immunol. 2002, 109, 854–857. [Google Scholar] [CrossRef]
- Smith, S.M.; Davis-Street, J.E.; Fesperman, J.V.; Smith, M.D.; Rice, B.L.; Zwart, S.R. Nutritional status changes in humans during a 14-day saturation dive: The NASA Extreme Environment Mission Operations V project. J. Nutr. 2004, 134, 1765–1771. [Google Scholar] [CrossRef] [Green Version]
- Zwart, S.R.; Jessup, J.M.; Ji, J.; Smith, S.M. Saturation diving alters folate status and biomarkers of DNA damage and repair. PLoS ONE 2012, 7, e31058. [Google Scholar] [CrossRef] [PubMed]
- Zwart, S.R.; Kala, G.; Smith, S.M. Body iron stores and oxidative damage in humans increased during and after a 10- to 12-day undersea dive. J. Nutr. 2009, 139, 90–95. [Google Scholar] [CrossRef]
- Hallingberg, B.; Turley, R.; Segrott, J.; Wight, D.; Craig, P.; Moore, L.; Murphy, S.; Robling, M.; Simpson, S.A.; Moore, G. Exploratory studies to decide whether and how to proceed with full-scale evaluations of public health interventions: A systematic review of guidance. Pilot Feasibility Stud. 2018, 4, 104. [Google Scholar] [CrossRef]
- Campos, A.C.; Fogaca, M.V.; Aguiar, D.C.; Guimaraes, F.S. Animal models of anxiety disorders and stress. Braz. J. Psychiatry 2013, 35 (Suppl. 2), S101–S111. [Google Scholar] [CrossRef] [Green Version]
- Nagel, Z.D.; Chaim, I.A.; Samson, L.D. Inter-individual variation in DNA repair capacity: A need for multi-pathway functional assays to promote translational DNA repair research. DNA Repair 2014, 19, 199–213. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.R.; Fogarty, J.A.; Richard, E.E. Human health and performance risk management—An approach for exploration missions. Acta Astronaut. 2008, 63, 98. [Google Scholar] [CrossRef]
- Geuna, S.; Brunelli, F.; Perino, M.A. Stressors, stress, and stress consequences during long-duration manned space missions: A descriptive model. Acta Astronaut. 1996, 36, 347–356. [Google Scholar] [CrossRef]
- Morphew, M.E. Psychological and human factors in long duration spaceflight. McGill J. Med. 2001, 6, 74–80. [Google Scholar] [CrossRef]
- Vanhove, A.J.; Herian, M.N.; Harms, P.D.; Luthans, F. Resilience and Growth in Long-Duration Isolated, Confined and Extreme (ICE) Missions; NASA/TM-2015-218566; National Aeronautics and Space Administration: Washington, DC, USA, 2015.
- NASA. Human Research Program Human Research Analog (HERA) Facility and Capabilities Information; NASA: Washington, DC, USA, 2019.
- Douglas, G.L.; Crucian, B.E.; Lorenzi, H.; Smith, S.M.; Stowe, R.P.; Young, M.H.; Zwart, S.R. The Integrated Impact of Diet on Human Immune Response, the Gut Microbiota, and Nutritional Status during Adaptation to Spaceflight. Available online: https://humanresearchroadmap.nasa.gov/tasks/task.aspx?i=2068 (accessed on 27 November 2021).
- Fenech, M. Principles of Nutrigenetics and Nutrigenomics Fundamentals of Individualized Nutrition. In Chapter 4—The Role of Nutrition in DNA Replication, DNA Damage Prevention and DNA Repair; Raffaele, C., Martinez, A., Kohlmeier, M., Eds.; Elsevier Inc. Academic Press: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Fenech, M.F. Dietary reference values of individual micronutrients and nutriomes for genome damage prevention: Current status and a road map to the future. Am. J. Clin. Nutr. 2010, 91, 1438S–1454S. [Google Scholar] [CrossRef] [Green Version]
- Caple, F.; Williams, E.A.; Spiers, A.; Tyson, J.; Burtle, B.; Daly, A.K.; Mathers, J.C.; Hesketh, J.E. Inter-individual variation in DNA damage and base excision repair in young, healthy non-smokers: Effects of dietary supplementation and genotype. Br. J. Nutr. 2010, 103, 1585–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohrenweiser, H.W.; Xi, T.; Vazquez-Matias, J.; Jones, I.M. Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiol. Biomark. Prev. 2002, 11, 1054–1064. [Google Scholar]
- Shen, M.R.; Jones, I.M.; Mohrenweiser, H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998, 58, 604–608. [Google Scholar] [PubMed]
- Wilson, D.M., 3rd; Kim, D.; Berquist, B.R.; Sigurdson, A.J. Variation in base excision repair capacity. Mutat. Res. 2011, 711, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Santa-Gonzalez, G.A.; Gomez-Molina, A.; Arcos-Burgos, M.; Meyer, J.N.; Camargo, M. Distinctive adaptive response to repeated exposure to hydrogen peroxide associated with upregulation of DNA repair genes and cell cycle arrest. Redox Biol. 2016, 9, 124–133. [Google Scholar] [CrossRef] [Green Version]
- Clementi, E.; Inglin, L.; Beebe, E.; Gsell, C.; Garajova, Z.; Markkanen, E. Persistent DNA damage triggers activation of the integrated stress response to promote cell survival under nutrient restriction. BMC Biol. 2020, 18, 36. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Villanueva, M.; Eltze, T.; Dressler, D.; Bernhardt, J.; Hirsch, C.; Wick, P.; von Scheven, G.; Lex, K.; Burkle, A. The automated FADU-assay, a potential high-throughput in vitro method for early screening of DNA breakage. ALTEX 2011, 28, 295–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Villanueva, M.; Pfeiffer, R.; Sindlinger, T.; Leake, A.; Muller, M.; Kirkwood, T.B.; Burkle, A. A modified and automated version of the ’Fluorimetric Detection of Alkaline DNA Unwinding’ method to quantify formation and repair of DNA strand breaks. BMC Biotechnol. 2009, 9, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newson, R.B. Frequentist q-values for multiple-test procedures. Stata J. 2010, 10, 568–584. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feiveson, A.H.; Krieger, S.S.; von Scheven, G.; Crucian, B.E.; Bürkle, A.; Stahn, A.C.; Wu, H.; Moreno-Villanueva, M. DNA Damage and Radiosensitivity in Blood Cells from Subjects Undergoing 45 Days of Isolation and Confinement: An Explorative Study. Curr. Issues Mol. Biol. 2022, 44, 654-669. https://doi.org/10.3390/cimb44020046
Feiveson AH, Krieger SS, von Scheven G, Crucian BE, Bürkle A, Stahn AC, Wu H, Moreno-Villanueva M. DNA Damage and Radiosensitivity in Blood Cells from Subjects Undergoing 45 Days of Isolation and Confinement: An Explorative Study. Current Issues in Molecular Biology. 2022; 44(2):654-669. https://doi.org/10.3390/cimb44020046
Chicago/Turabian StyleFeiveson, Alan H., Stephanie S. Krieger, Gudrun von Scheven, Brian E. Crucian, Alexander Bürkle, Alexander C. Stahn, Honglu Wu, and María Moreno-Villanueva. 2022. "DNA Damage and Radiosensitivity in Blood Cells from Subjects Undergoing 45 Days of Isolation and Confinement: An Explorative Study" Current Issues in Molecular Biology 44, no. 2: 654-669. https://doi.org/10.3390/cimb44020046
APA StyleFeiveson, A. H., Krieger, S. S., von Scheven, G., Crucian, B. E., Bürkle, A., Stahn, A. C., Wu, H., & Moreno-Villanueva, M. (2022). DNA Damage and Radiosensitivity in Blood Cells from Subjects Undergoing 45 Days of Isolation and Confinement: An Explorative Study. Current Issues in Molecular Biology, 44(2), 654-669. https://doi.org/10.3390/cimb44020046






