Phenotypic Assessment of Probiotic and Bacteriocinogenic Efficacy of Indigenous LAB Strains from Human Breast Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, Processing, and Ethical Statement
2.2. Antimicrobial Spectrum of Bacterial Isolates
2.3. Preliminary Identification of Bacterial Isolates
2.4. Assessment of Probiotic Properties
2.4.1. Tolerance to Lysozyme, Acid, and Bile
2.4.2. Gastrointestinal Digestion
2.4.3. Tolerance to NaCl
2.4.4. Surface Adherence, Auto-Aggregation, and Co-Aggregation Assay
2.4.5. Mucin Adherence
2.4.6. Bile Salt Hydrolase (BSH) Activity
2.5. Evaluation of LAB Isolates for Bacteriocin Production
2.6. Safety Evaluation of Probiotic LAB
2.6.1. Hemolysin Activity
2.6.2. Biofilm Production
2.6.3. Antibiotic Sensitivity Pattern
2.7. Genotypic Identification of Probiotic LAB
2.8. Statistical Analysis
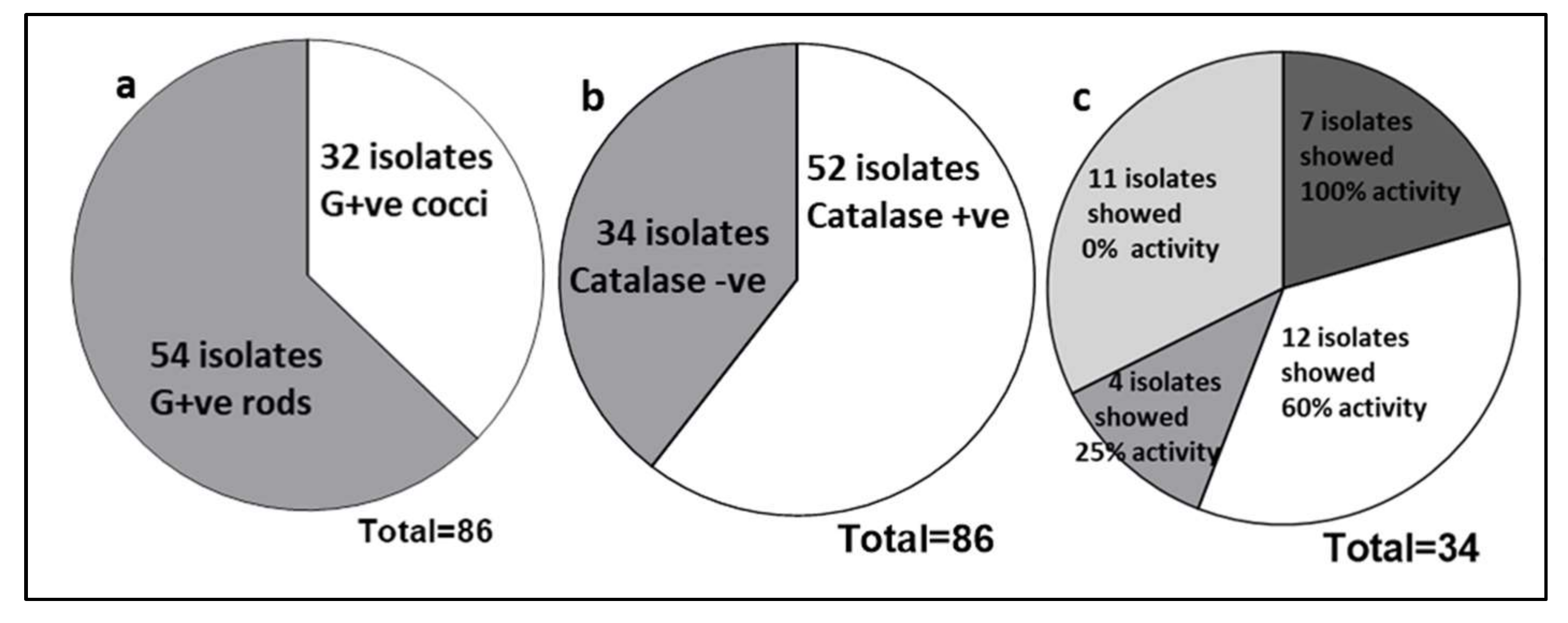
3. Result
3.1. Assessment of Probiotic Potential
Evaluation of Gastrointestinal Survival
3.2. Evaluation of Cell Surface Properties
3.3. Characterization of Antimicrobial Compounds
3.4. Biosafety Assessment of Probiotic LAB
3.5. Genotypic Analysis of Probiotic LAB
4. Discussion
4.1. Gastrointestinal Tolerance
4.2. Bacteriocinogenic Efficacy of LAB Isolates
4.3. Biosafety Assessment of Probiotic LAB
4.4. Identification Probiotic LAB
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interests
References
- Martín, R.; Langa, S.; Reviriego, C.; Jimínez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Lara-Villoslada, F.; Olivares, M.; Sierra, S.; Rodríguez, J.; Boza, J.; Xaus, J. Beneficial effects of probiotic bacteria isolated from breast milk. Br. J. Nutr. 2007, 98, S96–S100. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe 2019, 25, 324–335.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajoka, M.S.R.; Mehwish, H.M.; Siddiq, M.; Haobin, Z.; Zhu, J.; Yan, L.; Shao, D.; Xu, X.; Shi, J. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 2017, 84, 271–280. [Google Scholar] [CrossRef]
- FAO/WHO. Guidelines for the Evaluation of Probiotics in Food; FAO: Paris, France, 2002; pp. 1–11. [Google Scholar]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2012, 6, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Happel, A.-U.; Kullin, B.; Gamieldien, H.; Wentzel, N.; Zauchenberger, C.Z.; Jaspan, H.B.; Dabee, S.; Barnabas, S.L.; Jaumdally, S.Z.; Dietrich, J.; et al. Exploring potential of vaginal Lactobacillus isolates from South African women for enhancing treatment for bacterial vaginosis. PLoS Pathog. 2020, 16, e1008559. [Google Scholar] [CrossRef]
- Bae, J.-M. Prophylactic efficacy of probiotics on travelers’ diarrhea: An adaptive meta-analysis of randomized controlled trials. Epidemiol. Health 2018, 40, e2018043. [Google Scholar] [CrossRef]
- Horosheva, T.V.; Vodyanoy, V.; Sorokulova, I. Efficacy of Bacillus probiotics in prevention of antibiotic-associated diarrhoea: A randomized, double-blind, placebo-controlled clinical trial. JMM Case Rep. 2014, 1. [Google Scholar] [CrossRef] [Green Version]
- Szajewska, H.; Skorka, A. Saccharomyces boulardiifor treating acute gastroenteritis in children: Updated meta-analysis of randomized controlled trials. Aliment. Pharmacol. Ther. 2009, 30, 960–961. [Google Scholar] [CrossRef]
- Al-Sadi, R.; Nighot, P.; Nighot, M.; Haque, M.; Rawat, M.; Ma, T.Y. Lactobacillus acidophilus induces a Strain-specific and Toll-Like Receptor 2–Dependent Enhancement of Intestinal Epithelial Tight Junction Barrier and Protection against Intestinal Inflammation. Am. J. Pathol. 2021, 191, 872–884. [Google Scholar] [CrossRef]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.V.; Sreenivasa, M.Y. Probiotic Properties of Lactic Acid Bacteria Isolated from Neera: A Naturally Fermenting Coconut Palm Nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Abdhul, K.; Ganesh, M.; Shanmughapriya, S.; Vanithamani, S.; Kanagavel, M.; Anbarasu, K.; Natarajaseenivasan, K. Bacteriocinogenic potential of a probiotic strain Bacillus coagulans [BDU3] from Ngari. Int. J. Biol. Macromol. 2015, 79, 800–806. [Google Scholar] [CrossRef]
- Marques, T.M.; Wall, R.; Ross, R.P.; Fitzgerald, G.F.; Ryan, C.A.; Stanton, C. Programming infant gut microbiota: Influence of dietary and environmental factors. Curr. Opin. Biotechnol. 2010, 21, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.; Charbonneau, D.; Sanozky-Dawes, R.; Klaenhammer, T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 2015, 6, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Jamyuang, C.; Phoonlapdacha, P.; Chongviriyaphan, N.; Chanput, W.; Nitisinprasert, S.; Nakphaichit, M. Characterization and probiotic properties of Lactobacilli from human breast milk. 3 Biotech 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Hayat, H.F.; Sarwar, S.; Mehwish, H.M.; Ahmad, F.; Hussain, N.; Shah, S.Z.H.; Khurshid, M.; Siddiqu, M.; Shi, J. Isolation and evaluation of probiotic potential of lactic acid bacteria isolated from poultry intestine. Microbiology 2018, 87, 116–126. [Google Scholar] [CrossRef]
- Poinsot, P.; Penhoat, A.; Mitchell, M.; Sauvinet, V.; Meiller, L.; Louche-Pélissier, C.; Meugnier, E.; Ruiz, M.; Schwarzer, M.; Michalski, M.-C.; et al. Probiotic from human breast milk, Lactobacillus fermentum, promotes growth in animal model of chronic malnutrition. Pediatr. Res. 2020, 88, 374–381. [Google Scholar] [CrossRef]
- NHC-PRC (National Health Commission of the People’s Republic of China). Announcement on Three Strains Including Limosilactobacillus Fermentum CECT5716; NHC-PRC: Beijing, China, 2016. [Google Scholar]
- Yadav, R.; Puniya, A.K.; Shukla, P. Probiotic Properties of Lactobacillus plantarum RYPR1 from an Indigenous Fermented Beverage Raabadi. Front. Microbiol. 2016, 7, 1683. [Google Scholar] [CrossRef] [Green Version]
- Jovanović, J.N.; Nikolić, B.; Šeatović, S.; Zavišić, G.; Ćulafić, D.M.; Vuković-Gačić, B.; Knežević-Vukčević, J. Characterization of some potentially probiotic Lactobacillus strains of human origin. Food Sci. Biotechnol. 2015, 24, 1781–1788. [Google Scholar] [CrossRef]
- Kobierecka, P.A.; Wyszyńska, A.; Aleksandrzak-Piekarczyk, T.; Kuczkowski, M.; Tuzimek, A.; Piotrowska, W.; Gorecki, A.; Adamska, I.; Wieliczko, A.; Bardowski, J.; et al. In vitro characteristics ofLactobacillusspp. strains isolated from the chicken digestive tract and their role in the inhibition ofCampylobactercolonization. MicrobiologyOpen 2017, 6, e00512. [Google Scholar] [CrossRef] [Green Version]
- Kos, B.; Šušković, J.; Vuković, S.; Šimpraga, M.; Frece, J.; Matošić, S. Adhesion and aggregation ability of probiotic strainLactobacillus acidophilusM92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Handley, P.S.; Harty, D.W.S.; Wyatt, J.E.; Brown, C.R.; Doran, J.P.; Gibbs, A.C.C. A Comparison of the Adhesion, Coaggregation and Cell-surface Hydrophobicity Properties of Fibrillar and Fimbriate Strains of Streptococcus salivarius. Microbiology 1987, 133, 3207–3217. [Google Scholar] [CrossRef] [Green Version]
- Shehata, M.G.; El Sohaimy, S.A.; El-Sahn, M.A.; Youssef, M.M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016, 61, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Jahromi, M.F.; Ho, Y.W. Probiotic Potential of LactobacillusStrains with Antimicrobial Activity against Some Human Pathogenic Strains. BioMed Res. Int. 2014, 2014, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Coppa, G.V.; Bruni, S.; Morelli, L.; Soldi, S.; Gabrielli, O. The First Prebiotics in Humans. J. Clin. Gastroenterol. 2004, 38, S80–S83. [Google Scholar] [CrossRef]
- Damaceno, Q.S.; Souza, J.P.; Nicoli, J.R.; Paula, R.L.; Assis, G.B.; Figueiredo, H.; Azevedo, V.; Martins, F.S. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob. Proteins 2017, 9, 371–379. [Google Scholar] [CrossRef]
- Rehaiem, A.; Ben Belgacem, Z.; Edalatian, M.R.; Martínez, B.; Rodriguez, A.; Manai, M.; Guerra, N.P. Assessment of potential probiotic properties and multiple bacteriocin encoding-genes of the technological performing strain Enterococcus faecium MMRA. Food Control 2014, 37, 343–350. [Google Scholar] [CrossRef]
- Duraisamy, S.; Balakrishnan, S.; Ranjith, S.; Husain, F.; Sathyan, A.; Peter, A.S.; Prahalathan, C.; Kumarasamy, A. Bacteriocin—A potential antimicrobial peptide towards disrupting and preventing biofilm formation in the clinical and environmental locales. Environ. Sci. Pollut. Res. 2020, 27, 44922–44936. [Google Scholar] [CrossRef] [PubMed]
- Corzo, G.; Gilliland, S.E. Bile salt hydrolase activity of three strains of Lactobacillus acidophi-lus. J. Dairy Sci. 1999, 82, 472–480. [Google Scholar] [CrossRef]
- Sumeri, I.; Arike, L.; Stekolštšikova, J.; Uusna, R.; Adamberg, S.; Adamberg, K.; Paalme, T. Effect of stress pretreatment on survival of probiotic bacteria in gastrointestinal tract simulator. Appl. Microbiol. Biotechnol. 2010, 86, 1925–1931. [Google Scholar] [CrossRef]
- Berrada, N.; Lemeland, J.-F.; Laroche, G.; Thouvenot, P.; Piaia, M. Bifidobacterium from Fermented Milks: Survival during Gastric Transit. J. Dairy Sci. 1991, 74, 409–413. [Google Scholar] [CrossRef]
- Samedi, L.; Charles, A.L. Isolation and characterization of potential probiotic Lactobacilli from leaves of food plants for possible additives in pellet feeding. Ann. Agric. Sci. 2019, 64, 55–62. [Google Scholar] [CrossRef]
- Yateem, A.; Balba, M.; Al-Surraya, T.; Al-Mutairi, B.; Al-Daher, R. Isolation of Lactic Acid Bacteria with Probiotic Potential from Camel Milk. Int. J. Dairy Sci. 2008, 3, 194–199. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Zhang, X.; Li, S.; Li, C.; Li, D.; Yang, Z. Evaluation of probiotic properties of Lactobacillus plantarum strains isolated from Chinese sauerkraut. World J. Microbiol. Biotechnol. 2012, 29, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, I.; Saeed, M.; Khan, W.A.; Khaliq, A.; Chughtai, M.F.J.; Iqbal, R.; Tehseen, S.; Naz, S.; Liaqat, A.; Mehmood, T.; et al. In Vitro Probiotic Potential and Safety Evaluation (Hemolytic, Cytotoxic Activity) of Bifidobacterium Strains Isolated from Raw Camel Milk. Microorganisms 2020, 8, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.-Q.; Shah, N.P. Invited review: Characterization of new probiotics from dairy and nondairy products—Insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J. Dairy Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef] [PubMed]
- Begley, M.; Hill, C.; Gahan, C.G.M. Bile Salt Hydrolase Activity in Probiotics. Appl. Environ. Microbiol. 2006, 72, 1729–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.I.A.; McCartney, A.L.; Gibson, G.R. An In Vitro Study of the Probiotic Potential of a Bile-Salt-Hydrolyzing Lactobacillus fermentum Strain, and Determination of Its Cholesterol-Lowering Properties. Appl. Environ. Microbiol. 2003, 69, 4743–4752. [Google Scholar] [CrossRef] [Green Version]
- Knarreborg, A.; Engberg, R.M.; Jensen, S.K.; Jensen, B.B. Quantitative Determination of Bile Salt Hydrolase Activity in Bacteria Isolated from the Small Intestine of Chickens. Appl. Environ. Microbiol. 2002, 68, 6425–6428. [Google Scholar] [CrossRef] [Green Version]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Jeong, H.S.; Lee, H.Y.; Ahn, J. Assessment of cell surface properties and adhesion potential of selected probiotic strains. Lett. Appl. Microbiol. 2009, 49, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Mojgani, N.; Hussaini, F.; Vaseji, N. Characterization of Indigenous Lactobacillus Strains for Probiotic Properties. Jundishapur J. Microbiol. 2015, 8, e17523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.M.A.; Pires, M.C.V.; Leão, T.L.; Hernández, Z.P.; Rodriguez, M.L.; Martins, A.K.S.; Miranda, L.S.; Martins, F.; Nicoli, J.R. Selection of Lactobacillus strains as potential probiotics for vaginitis treatment. Microbiology 2016, 162, 1195–1207. [Google Scholar] [CrossRef]
- Gil, N.F.; Martinez, R.C.; Gomes, B.C.; Nomizo, A.; De Martinis, E.C.P. Vaginal lactobacilli as potential probiotics against Candida spp. Braz. J. Microbiol. 2010, 41, 6–14. [Google Scholar] [CrossRef] [Green Version]
- Al Kassaa, I.; Hamze, M.; Hober, D.; Chihib, N.-E.; Drider, D. Identification of Vaginal Lactobacilli with Potential Probiotic Properties Isolated from Women in North Lebanon. Microb. Ecol. 2014, 67, 722–734. [Google Scholar] [CrossRef]
- Del Re, B.; Sgorbati, B.; Miglioli, M.; Palenzona, D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000, 31, 438–442. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Prete, R.; Battista, N.; Corsetti, A. Adhesion Properties of Food-Associated Lactobacillus plantarum Strains on Human Intestinal Epithelial Cells and Modulation of IL-8 Release. Front. Microbiol. 2018, 9, 2392. [Google Scholar] [CrossRef]
- Tallon, R.; Arias, S.; Bressollier, P.; Urdaci, M. Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J. Appl. Microbiol. 2007, 102, 442–451. [Google Scholar] [CrossRef]
- Jensen, H.; Roos, S.; Jonsson, H.; Rud, I.; Grimmer, S.; Van Pijkeren, J.-P.; Britton, R.A.; Axelsson, L. Role of Lactobacillus reuteri cell and mucus-binding protein A (CmbA) in adhesion to intestinal epithelial cells and mucus in vitro. Microbiology 2014, 160, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Valeriano, V.D.; Parungao-Balolong, M.M.; Kang, D.-K. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J. Appl. Microbiol. 2014, 117, 485–497. [Google Scholar] [CrossRef]
- Banerjee, S.P.; Dora, K.C.; Chowdhury, S. Detection, partial purification and characterization of bacteriocin produced by Lactobacillus brevis FPTLB3 isolated from freshwater fish. J. Food Sci. Technol. 2011, 50, 17–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, A.; Akkoç, N.; Akçelik, M.; Kumari, A.; Akkoç, N.; Akçelik, M. Purification and partial characterization of bacteriocin produced by Lactococcus lactis ssp. lactis LL171. World J. Microbiol. Biotechnol. 2011, 28, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, D.; Liu, S.; Zhang, L. Garviecin LG34, a novel bacteriocin produced by Lactococcus garvieae isolated from traditional Chinese fermented cucumber. Food Control. 2015, 50, 896–900. [Google Scholar] [CrossRef]
- Kanwal, H.; Di Cerbo, A.; Zulfiqar, F.; Sabia, C.; Nawaz, A.; Siddiqui, F.M.; Aqeel, M.; Ghazanfar, S. Probiotic Characterization and Population Diversity Analysis of Gut-Associated Pediococcus acidilactici for Its Potential Use in the Dairy Industry. Appl. Sci. 2021, 11, 9586. [Google Scholar] [CrossRef]
- Peres, C.M.; Alves, M.; Hernandez-Mendoza, A.; Moreira, L.; Silva, S.; Bronze, M.R.; Vilas-Boas, L.; Peres, C.; Malcata, F.X. Novel isolates of lactobacilli from fermented Portuguese olive as potential probiotics. LWT 2014, 59, 234–246. [Google Scholar] [CrossRef] [Green Version]
- Casarotti, S.; Carneiro, B.; Todorov, S.; Nero, L.A.; Rahal, P.; Penna, A.L.B. In vitro assessment of safety and probiotic potential characteristics of Lactobacillus strains isolated from water buffalo mozzarella cheese. Ann. Microbiol. 2017, 67, 289–301. [Google Scholar] [CrossRef] [Green Version]
- Arboleya, S.; Ruas-Madiedo, P.; Margolles, A.; Solís, G.; Salminen, S.; de los Reyes-Gavilán, C.G.; Gueimonde, M. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 2011, 149, 28–36. [Google Scholar] [CrossRef] [Green Version]








| Tests | Breast Milk Bacterial Isolates | ||||||
|---|---|---|---|---|---|---|---|
| BDUMBT08 | BDUMBT09 | BDUMBT10 | BDUMBT11 | BDUMBT12 | BDUMBT13 | M2403 | |
| Morphological | |||||||
| Gram’s staining | +ve rods | +ve rods | +ve rods | +ve rods | +ve rods | +ve rods | +ve rods |
| Endospore production | – | – | – | – | – | – | – |
| Motility | – | – | – | – | – | – | – |
| Physiological | |||||||
| Catalase production | – | – | – | – | – | – | – |
| Oxidase production | – | – | – | – | – | – | + |
| Indole production | – | – | – | – | – | – | – |
| Acid production | – | – | – | – | – | – | – |
| Acetoin production | – | – | – | – | – | – | – |
| Citrate hydrolysis | – | – | – | – | – | – | – |
| Urease production | – | – | – | – | – | – | – |
| Starch hydrolysis | – | – | – | – | – | – | + |
| Carbohydrate fermentation | |||||||
| Lactose | + | + | + | + | + | + | + |
| Glucose | + | + | + | + | + | + | + |
| Mannitol | + | – | + | + | + | + | – |
| Fructose | + | + | + | + | + | + | + |
| Sorbitol | + | + | + | + | + | + | + |
| Maltose | + | + | + | + | + | + | + |
| Galactose | + | + | + | + | + | + | + |
| Mannose | + | + | – | + | + | + | + |
| Arabinose | – | – | – | – | – | – | – |
| Growth at different temperature | |||||||
| 4 | – | – | – | – | – | – | – |
| 25 | ++ | ++ | ++ | ++ | ++ | ||
| 37 | +++ | +++ | +++ | +++ | +++ | ||
| 45 | – | – | – | – | – | – | + |
| Bacterial Isolates | Tolerance to Lysozyme | Tolerance to Acid | Tolerance to Bile | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At pH 2 | At pH 3 | At pH 4 | Control | At 0.5% | At 1% | Control | |||||||||
| Log CFU | S (%) | Control | Log CFU | S (%) | Log CFU | S (%) | Log CFU | S (%) | Log CFU | Log CFU | S (%) | Log CFU | S (%) | Log CFU | |
| BDUMBT08 | 6.7 ± 0.16 | 70 | 9.6 ± 0.7 | 5.7 ± 0.13 | 63 | 7.1 ± 0.13 | 78 | 8.5 ± 0.13 | 93 | 9.1 ± 0.03 | 6.8 ± 0.18 | 71 | 5.2 ± 0.16 | 54 | 9.6 ± 0.7 |
| BDUMBT09 | 7.7 ± 0.18 | 81 | 9.5 ± 1.5 | 5.9 ± 0.18 | 64 | 7.4 ± 0.23 | 80 | 8.6 ± 0.08 | 94 | 9.2 ± 0.08 | 7.0 ± 0.13 | 71 | 5.0 ± 0.13 | 51 | 9.9 ± 1.5 |
| BDUMBT10 | 7.4 ± 0.13 | 78 | 9.5 ± 1.9 | 5.2 ± 0.19 | 58 | 6.9 ± 0.35 | 77 | 8.2 ± 0.11 | 91 | 9.0 ± 0.16 | 7.1 ± 0.17 | 75 | 5.6 ± 1.0 | 59 | 9.5 ± 1.9 |
| BDUMBT11 | 6.8 ± 0.34 | 72 | 9.5 ± 0.7 | 5.6 ± 0.09 | 59 | 7.2 ± 0.31 | 76 | 8.4 ± 0.16 | 88 | 9.5 ± 0.16 | 6.9 ± 0.23 | 71 | 4.8 ± 0.5 | 50 | 9.7 ± 0.7 |
| BDUMBT12 | 7.6 ± 0.38 | 80 | 9.5 ± 0.4 | 5.1 ± 0.16 | 55 | 7.4 ± 0.13 | 80 | 8.3 ± 0.18 | 89 | 9.3 ± 0.18 | 6.3 ± 0.21 | 67 | 4.4 ± 0.8 | 47 | 9.4 ± 0.4 |
| BDUMBT13 | 7.4 ± 0.21 | 79 | 9.4 ± 0.13 | 5.0 ± 0.13 | 53 | 7.2 ± 0.41 | 76 | 7.8 ± 0.13 | 82 | 9.5 ± 0.13 | 6.5 ± 0.13 | 68 | 4.9 ± 1.3 | 52 | 9.5 ± 0.31 |
| M2403 | 7.0 ± 0.27 | 74 | 9.5 ± 0.18 | 4.5 ± 0.18 | 49 | 6.1 ± 0.17 | 66 | 7.3 ± 0.16 | 79 | 9.2 ± 0.0 | 6.7 ± 0.2 | 67 | 4.7 ± 0.14 | 50 | 9.5 ± 1.9 |
| Bacterial Isolates | Tolerance to AGF | Tolerance to AIF | Control | ||
|---|---|---|---|---|---|
| Log CFU | S (%) | Log CFU | S (%) | ||
| BDUMBT08 | 5.0 ± 0.06 | 52 | 7.2 ± 0.13 | 75 | 9.6 ± 0.7 |
| BDUMBT09 | 5.4 ± 0.18 | 55 | 7.0 ± 0.02 | 71 | 9.9 ± 1.5 |
| BDUMBT10 | 5.2 ± 0.12 | 55 | 6.0 ± 0.58 | 60 | 9.5 ± 1.9 |
| BDUMBT11 | 6.0 ± 0.15 | 51 | 7.1 ± 0.09 | 73 | 9.7 ± 0.7 |
| BDUMBT12 | 4.7 ± 0.08 | 50 | 5.9 ± 0.08 | 63 | 9.4 ± 0.4 |
| BDUMBT13 | 5.0 ± 0.07 | 53 | 6.9 ± 0.15 | 67 | 9.5 ± 0.13 |
| M2403 | 4.8 ± 0.04 | 51 | 6.3 ± 0.15 | 66 | 9.5 ± 1.9 |
| Breast Milk Isolates | Hemolysis Activity | Biofilm Production | Zone of Inhibition (mm) and Score * as Resistant and Sensitive | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CEP | Car | VAN | CIP | NOR | RIF | C | ERY | GEN | STREP | TRI | SX | |||
| BDUMBT08 | – | – | 23 S | 24 S | 20 S | 21 S | 18 S | 20 S | 21 S | 23 S | 25 S | 21 S | 19 S | 24 S | 23 S |
| BDUMBT09 | – | – | 25 S | 22 S | 22 S | 26 S | 20 S | 17 S | 19 S | 7R | 0 R | 18 S | 8R | 20 S | 23 S |
| BDUMBT10 | – | – | 26 S | 26 S | 18 S | 18 S | 21 S | 19 S | 19 S | 10 S | 7 R | 20 S | 9 R | 20 S | 20 S |
| BDUMBT11 | – | – | 24 S | 25 S | 15 S | 26 S | 21 S | 21 S | 23 S | 20 S | 17 S | 18 S | 18 S | 22 S | 26 S |
| BDUMBT12 | – | – | 22 S | 19 S | 15 S | 21 S | 18 S | 23 S | 21 S | 20 S | 0 R | 18 S | 0 R | 17 S | 23 S |
| BDUMBT13 | – | – | 24 S | 18 S | 17 S | 27 S | 20 S | 17 S | 17 S | 23 S | 0 R | 21 S | 0 R | 17 S | 20 S |
| M2403 | – | – | 22 S | 19 S | 19 S | 21 S | 18 S | 20 S | 17 S | 23 S | 0 R | 20 S | 0 R | 20 S | 18 S |
| Breast Milk Bacterial Isolates | NCBI Accession No. of the Isolates | The Isolates Highly Matched with Species from Genbank | % of Query Coverage | E Value | % of Identity |
|---|---|---|---|---|---|
| BDUMBT08 | MT673657 | Levilactobacillus brevis strain BSO464 | 100 | 0 | 97 |
| BDUMBT09 | MT774596 | Lactobacillus gastricus strain 32-154 | 100 | 0 | 97 |
| BDUMBT10 | MT775430 | Lactobacillus paracasei strain Lp02 | 100 | 0 | 97 |
| BDUMBT11 | MW785062 | Levilactobacillus brevis ATCC14869 | 100 | 0 | 98 |
| BDUMBT12 | MW785063 | Lactobacillus casei subsp. casei ATCC393 | 99 | 0 | 99 |
| BDUMBT13 | MW785178 | Lactobacillus casei strain BCRC10697 | 99 | 0 | 98 |
| M2403 | MK371781 | Brevibacillus brevis strain HK544 | 99 | 0 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duraisamy, S.; Husain, F.; Balakrishnan, S.; Sathyan, A.; Subramani, P.; Chidambaram, P.; Arokiyaraj, S.; Al-Qahtani, W.H.; Rajabathar, J.; Kumarasamy, A. Phenotypic Assessment of Probiotic and Bacteriocinogenic Efficacy of Indigenous LAB Strains from Human Breast Milk. Curr. Issues Mol. Biol. 2022, 44, 731-749. https://doi.org/10.3390/cimb44020051
Duraisamy S, Husain F, Balakrishnan S, Sathyan A, Subramani P, Chidambaram P, Arokiyaraj S, Al-Qahtani WH, Rajabathar J, Kumarasamy A. Phenotypic Assessment of Probiotic and Bacteriocinogenic Efficacy of Indigenous LAB Strains from Human Breast Milk. Current Issues in Molecular Biology. 2022; 44(2):731-749. https://doi.org/10.3390/cimb44020051
Chicago/Turabian StyleDuraisamy, Senbagam, Fazal Husain, Senthilkumar Balakrishnan, Aswathy Sathyan, Prabhu Subramani, Prahalathan Chidambaram, Selvaraj Arokiyaraj, Wahidah H. Al-Qahtani, Jothiramalingam Rajabathar, and Anbarasu Kumarasamy. 2022. "Phenotypic Assessment of Probiotic and Bacteriocinogenic Efficacy of Indigenous LAB Strains from Human Breast Milk" Current Issues in Molecular Biology 44, no. 2: 731-749. https://doi.org/10.3390/cimb44020051
APA StyleDuraisamy, S., Husain, F., Balakrishnan, S., Sathyan, A., Subramani, P., Chidambaram, P., Arokiyaraj, S., Al-Qahtani, W. H., Rajabathar, J., & Kumarasamy, A. (2022). Phenotypic Assessment of Probiotic and Bacteriocinogenic Efficacy of Indigenous LAB Strains from Human Breast Milk. Current Issues in Molecular Biology, 44(2), 731-749. https://doi.org/10.3390/cimb44020051







