NADPH–Cytochrome P450 Reductase Mediates the Fatty Acid Desaturation of ω3 and ω6 Desaturases from Mortierella alpina
Abstract
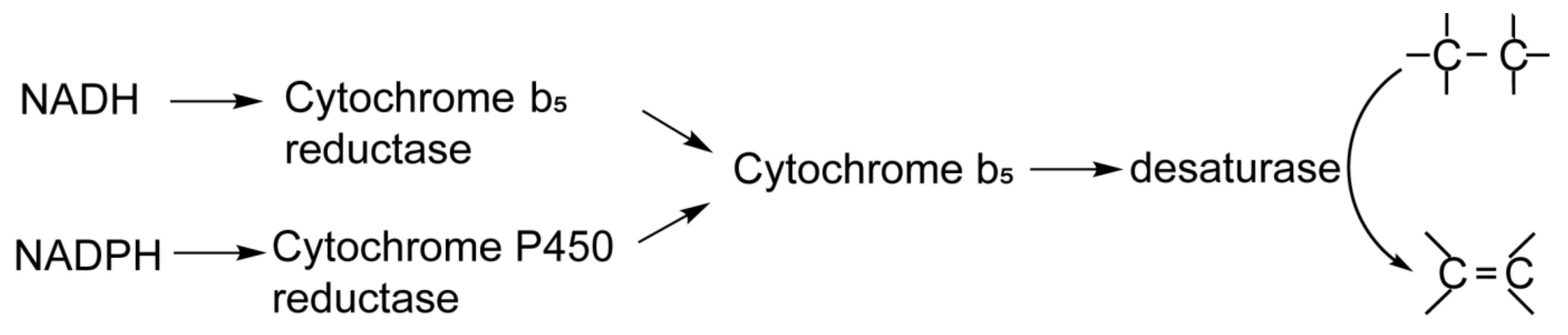
:1. Introduction
2. Materials and Methods
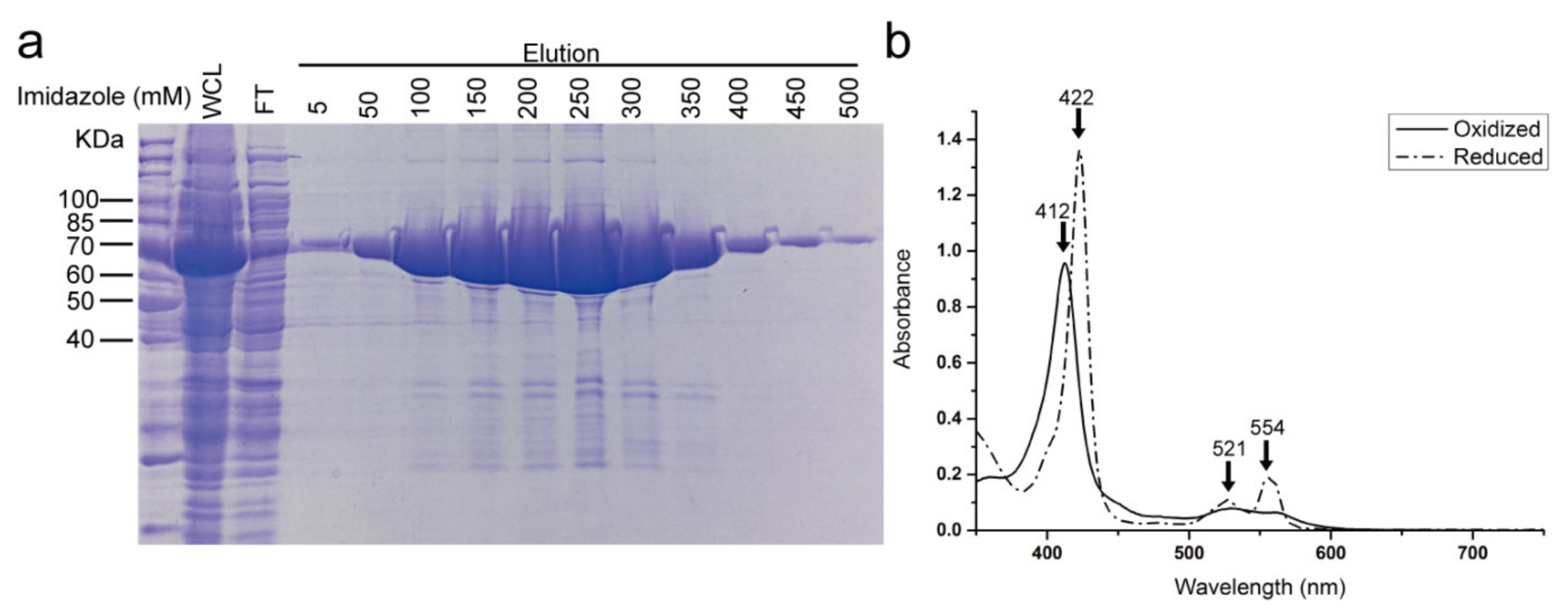
2.1. Expression and Purification of NADPH-CytP450R and Cytb5
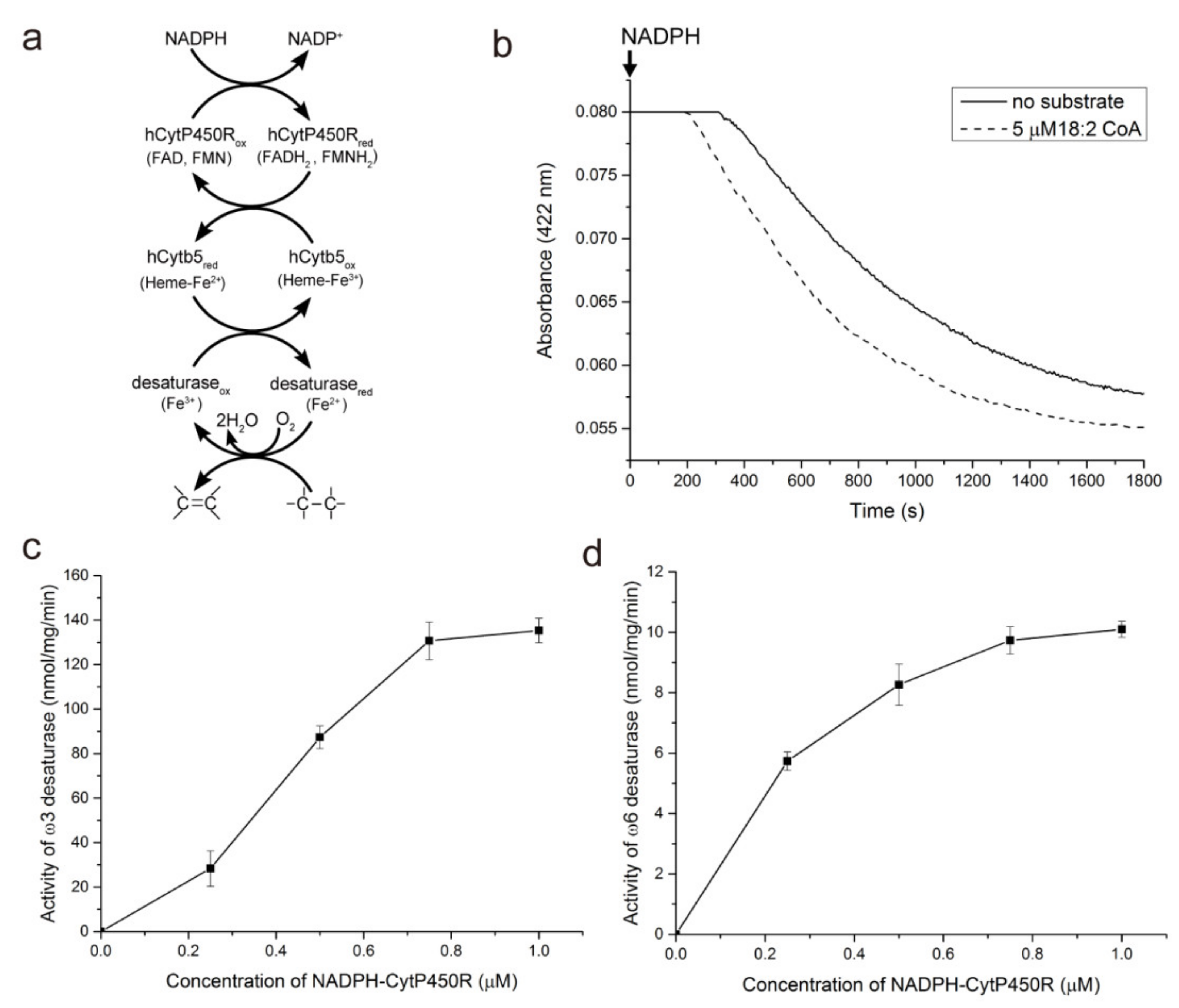
2.2. Assays to Determine the Activity of NADPH-CytP450R and Cytb5 Proteins
2.3. Preparation and Kinetic Analysis of ω3 and ω6 Desaturases in an NADPH-Initiated System
2.4. Nucleotide Sequence Accession Number
3. Results
3.1. Purification of NADPH-CytP450R and the Reduction of Cytb5 by NADPH-CytP450R
3.2. ω3 and ω6 Fatty Acid-CoA Desaturation in the NADPH-CytP450R-Mediated Reaction System
3.3. Kinetic Analysis of ω3 and ω6 Desaturases in NADPH-Dependent Desaturation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bazinet, R.P.; Layé, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Los, D.A.; Murata, N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta 1998, 1394, 3–15. [Google Scholar] [CrossRef]
- Weylandt, K.H.; Chen, Y.Q.; Lim, K.; Su, H.-M.; Cittadini, A.; Calviello, G. ω-3 PUFAs in the prevention and cure of inflammatory, degenerative, and neoplastic diseases 2014. BioMed Res. Int. 2015, 2015, 695875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloomfield, D.; Bloch, K. The formation of δ9-unsaturated fatty acids. J. Biol. Chem. 1960, 235, 337–345. [Google Scholar] [CrossRef]
- Shanklin, J.; Cahoon, E.B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 611–641. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Ω3 fatty acid desaturases from microorganisms: Structure, function, evolution, and biotechnological use. Appl. Microbiol. Biotechnol. 2013, 97, 10255–10262. [Google Scholar] [CrossRef] [Green Version]
- Carlson, S.E.; Werkman, S.H.; Peeples, J.M.; Cooke, R.J.; Tolley, E.A. Arachidonic acid status correlates with first year growth in preterm infants. Proc. Natl. Acad. Sci. USA 1993, 90, 1073–1077. [Google Scholar] [CrossRef] [Green Version]
- Connor, K.M.; SanGiovanni, J.P.; Lofqvist, C.; Aderman, C.M.; Chen, J.; Higuchi, A.; Hong, S.; Pravda, E.A.; Majchrzak, S.; Carper, D.; et al. Increased dietary intake of ω-3-polyunsaturated fatty acids reduces pathological retinal angiogenesis. Nat. Med. 2007, 13, 868–873. [Google Scholar] [CrossRef] [Green Version]
- Shimiziu, S.; Kawashima, H.; Shinmen, Y.; Akimoto, K.; Yamada, H. Production of eicosapentaenoic acid by Mortierella fungi. J. Am. Oil Chem. Soc. 1988, 65, 1455–1459. [Google Scholar] [CrossRef]
- Shinmen, Y.; Shimizu, S.; Akimoto, K.; Kawashima, H.; Yamada, H. Production of arachidonic acid by Mortierella fungi. Appl. Microbiol. Biotechnol. 1989, 31, 11–16. [Google Scholar] [CrossRef]
- Wang, M.; Chen, H.; Ailati, A.; Chen, W.; Chilton, F.H.; Todd Lowther, W.; Chen, Y.Q. Substrate specificity and membrane topologies of the iron-containing ω3 and ω6 desaturases from Mortierella alpina. Appl. Microbiol. Biotechnol. 2018, 102, 211–223. [Google Scholar] [CrossRef]
- Enoch, H.G.; Strittmatter, P. Cytochrome b5 reduction by nadph-cytochrome p-450 reductase. J. Biol. Chem. 1979, 254, 8976–8981. [Google Scholar] [CrossRef]
- Porter, T.D. The roles of cytochrome b5 in cytochrome p450 reactions. J. Biochem. Mol. Toxicol. 2002, 16, 311–316. [Google Scholar] [CrossRef]
- Strittmatter, P.; Velick, S.F. The purification and properties of microsomal cytochrome reductase. J. Biol. Chem. 1957, 228, 785–799. [Google Scholar] [CrossRef]
- Iyanagi, T.; Mason, H.S. Properties of hepatic reduced nicotinamide adenine dinucleotide phosphate-cytochrome c reductase. Biochemistry 1973, 12, 2297–2308. [Google Scholar] [CrossRef]
- Enoch, H.G.; Catala, A.; Strittmatter, P. Mechanism of rat liver microsomal stearyl-coa desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J. Biol. Chem. 1976, 251, 5095–5103. [Google Scholar] [CrossRef]
- Okayasu, T.; Nagao, M.; Ishibashi, T.; Imai, Y. Purification and partial characterization of linoleoyl-coa desaturase from rat liver microsomes. Arch. Biochem. Biophys. 1981, 206, 21–28. [Google Scholar] [CrossRef]
- Mitchell, A.G.; Martin, C.E. A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae δ-9 fatty acid desaturase (∗). J. Biol. Chem. 1995, 270, 29766–29772. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Gu, Z.; Zhang, H.; Wang, M.; Chen, W.; Lowther, W.T.; Chen, Y.Q. Expression and purification of integral membrane fatty acid desaturases. PLoS ONE 2013, 8, e58139. [Google Scholar] [CrossRef]
- Xia, C.; Panda, S.P.; Marohnic, C.C.; Martásek, P.; Masters, B.S.; Kim, J.-J.P. Structural basis for human nadph-cytochrome p450 oxidoreductase deficiency. Proc. Natl. Acad. Sci. USA 2011, 108, 13486–13491. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Chen, W.; Feng, Y.; Ren, Y.; Gu, Z.; Chen, H.; Wang, H.; Thomas, M.J.; Zhang, B.; Berquin, I.M.; et al. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS ONE 2011, 6, e28319. [Google Scholar] [CrossRef] [Green Version]
- Napier, A.J.; Hey, J.S.; Lacey, J.D.; Shewry, R.P. Identification of a caenorhabditis elegans δ6-fatty-acid-desaturase by heterologous expression in Saccharomyces cerevisiae. Biochem. J. 1998, 330, 611–614. [Google Scholar] [CrossRef]
- Sakuradani, E.; Kobayashi, M.; Ashikari, T.; Shimizu, S. Identification of δ12-fatty acid desaturase from arachidonic acid-producing mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. Eur. J. Biochem. 1999, 261, 812–820. [Google Scholar] [CrossRef]
- Sakuradani, E.; Abe, T.; Iguchi, K.; Shimizu, S. A novel fungal ω3-desaturase with wide substrate specificity from arachidonic acid-producing Mortierella alpina 1s-4. Appl. Microbiol. Biotechnol. 2005, 66, 648–654. [Google Scholar] [CrossRef]
- Strittmatter, P.; Spatz, L.; Corcoran, D.; Rogers, M.J.; Setlow, B.; Redline, R. Purification and properties of rat liver microsomal stearyl coenzyme a desaturase. Proc. Natl. Acad. Sci. USA 1974, 71, 4565–4569. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Hao, G.; Wang, L.; Wang, H.; Gu, Z.; Liu, L.; Zhang, H.; Chen, W.; Chen, Y.Q. Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci. Rep. 2015, 5, 11247. [Google Scholar] [CrossRef]
- Wasylenko, T.M.; Ahn, W.S.; Stephanopoulos, G. The oxidative pentose phosphate pathway is the primary source of nadph for lipid overproduction from glucose in yarrowia lipolytica. Metab. Eng. 2015, 30, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Chen, H.; Du, K.; Huang, X.; Song, Y.; Gu, Z.; Wang, L.; Zhang, H.; Chen, W.; Chen, Y.Q. Increased fatty acid unsaturation and production of arachidonic acid by homologous over-expression of the mitochondrial malic enzyme in Mortierella alpina. Biotechnol. Lett. 2014, 36, 1827–1834. [Google Scholar] [CrossRef]
- Hao, G.; Chen, H.; Wang, L.; Gu, Z.; Song, Y.; Zhang, H.; Chen, W.; Chen Yong, Q.; Cullen, D. Role of malic enzyme during fatty acid synthesis in the oleaginous fungus Mortierella alpina. Appl. Environ. Microbiol. 2014, 80, 2672–2678. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen Yong, Q.; Cullen, D. Metabolic engineering of Mortierella alpina for enhanced arachidonic acid production through the nadph-supplying strategy. Appl. Environ. Microbiol. 2016, 82, 3280–3288. [Google Scholar] [CrossRef] [Green Version]

| Desaturase | Substrate | Specific Activity (nmol min−1 mg−1) | Km (µM) | kcat (min−1) | kcat/Km (µM−1 min−1) |
|---|---|---|---|---|---|
| ω6 | 18:1 ω9 | 10.0 ± 0.5 a | 5.4 ± 0.8 | 0.5 ± 0.02 | 0.09 |
| ω6 | 16:1 ω7 | 3.5 ± 0.2 | 3.9 ± 0.9 | 0.2 ± 0.01 | 0.04 |
| ω3 | 18:2 ω6 | 138.9 ± 7.8 | 16.0 ± 2.2 | 6.6 ± 0.6 | 0.41 |
| ω3 | 18:3 ω6 | 213.4 ± 9.0 | 87.8 ± 9.9 | 10.1 ± 0.4 | 0.12 |
| ω3 | 20:4 ω6 | 28.8 ± 1.3 | 157.0 ± 23.8 | 1.4 ± 0.1 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Li, J.; Cong, W.; Zhang, J. NADPH–Cytochrome P450 Reductase Mediates the Fatty Acid Desaturation of ω3 and ω6 Desaturases from Mortierella alpina. Curr. Issues Mol. Biol. 2022, 44, 1828-1837. https://doi.org/10.3390/cimb44050125
Wang M, Li J, Cong W, Zhang J. NADPH–Cytochrome P450 Reductase Mediates the Fatty Acid Desaturation of ω3 and ω6 Desaturases from Mortierella alpina. Current Issues in Molecular Biology. 2022; 44(5):1828-1837. https://doi.org/10.3390/cimb44050125
Chicago/Turabian StyleWang, Mingxuan, Jing Li, Wenjie Cong, and Jianguo Zhang. 2022. "NADPH–Cytochrome P450 Reductase Mediates the Fatty Acid Desaturation of ω3 and ω6 Desaturases from Mortierella alpina" Current Issues in Molecular Biology 44, no. 5: 1828-1837. https://doi.org/10.3390/cimb44050125
APA StyleWang, M., Li, J., Cong, W., & Zhang, J. (2022). NADPH–Cytochrome P450 Reductase Mediates the Fatty Acid Desaturation of ω3 and ω6 Desaturases from Mortierella alpina. Current Issues in Molecular Biology, 44(5), 1828-1837. https://doi.org/10.3390/cimb44050125






