Hepatoprotective Effect of Silver Nanoparticles at Two Different Particle Sizes: Comparative Study with and without Silymarin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Silver Nanoparticles
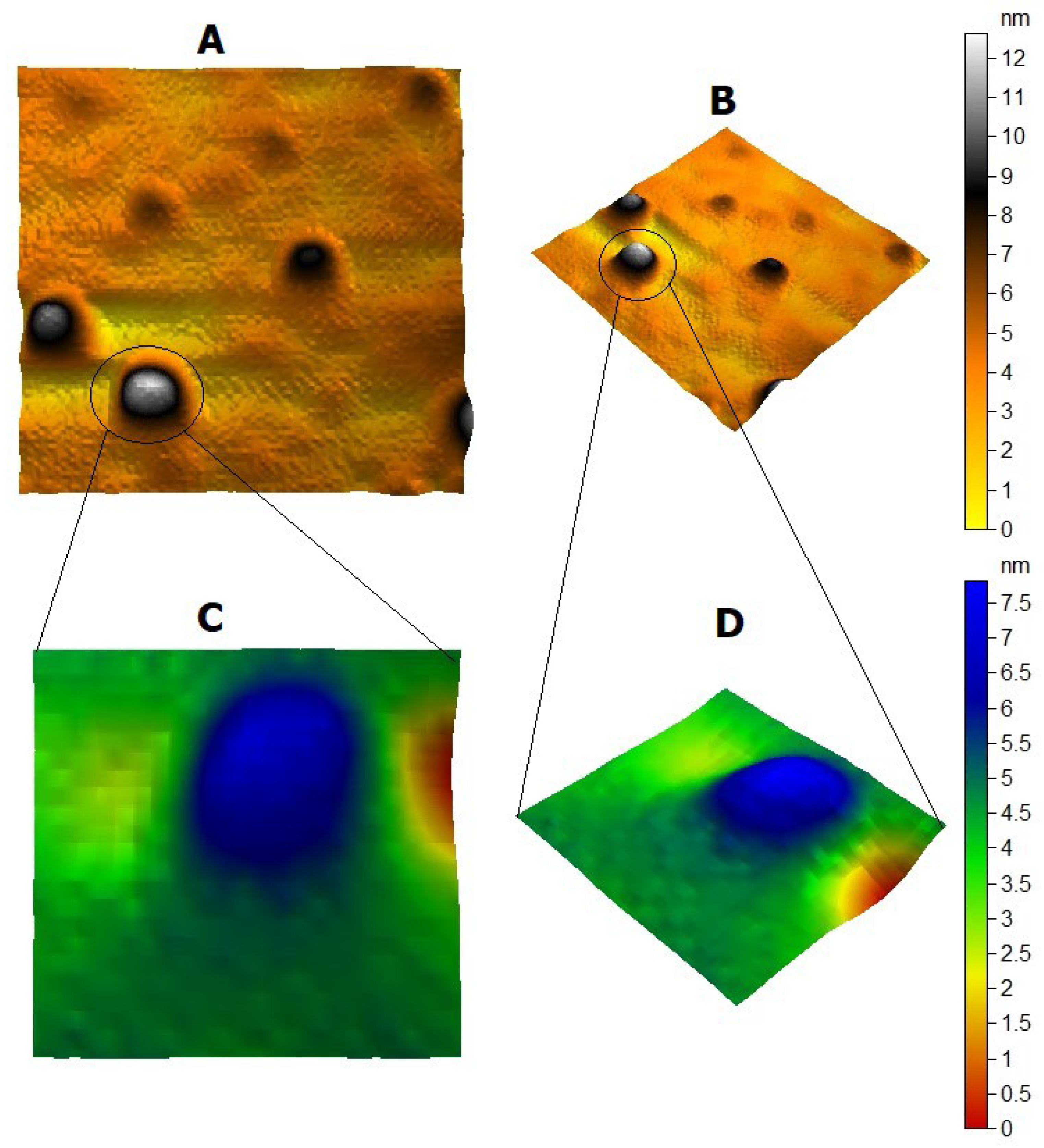
2.2. Silver Nanoparticles’ Characterization
2.2.1. Identification Class
2.2.2. Index Class
2.2.3. Microscopic Class
2.3. Animals
2.4. Determination of IL-6, NO, and TNF-α Biomarkers
2.5. Determination of COX2 Enzyme by Quantitative Real-Time PCR
2.5.1. RNA Extraction
2.5.2. cDNA Synthesis
2.5.3. Real-Time qPCR Using SYBR Green I
2.5.4. Preparation of the Reaction Master Mix for Q-PCR
2.5.5. Calculation of Relative Quantification (RQ) (Relative Expression)
2.6. Statistical Analysis
3. Result
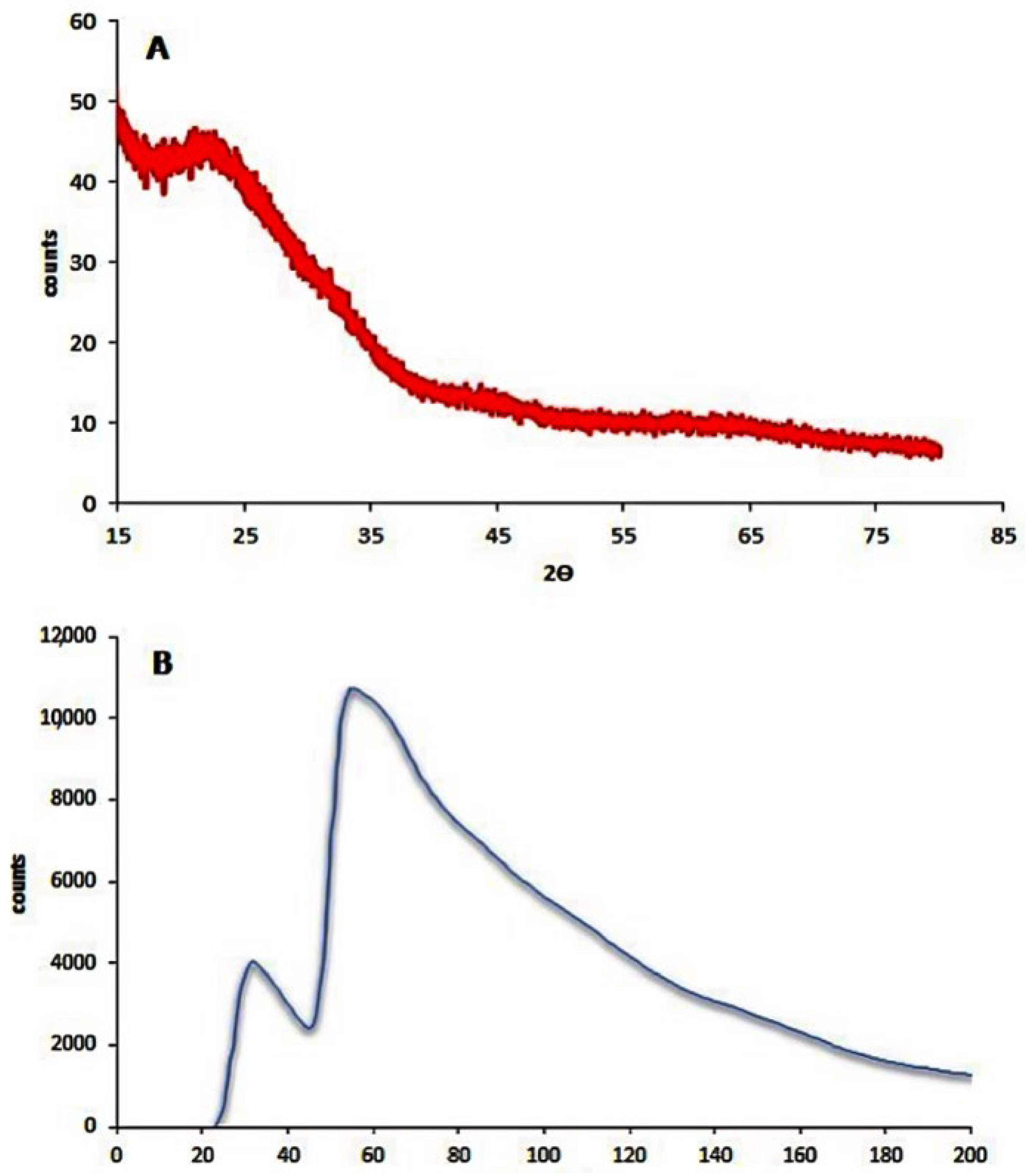
3.1. Silver Nanoparticles’ Characterization
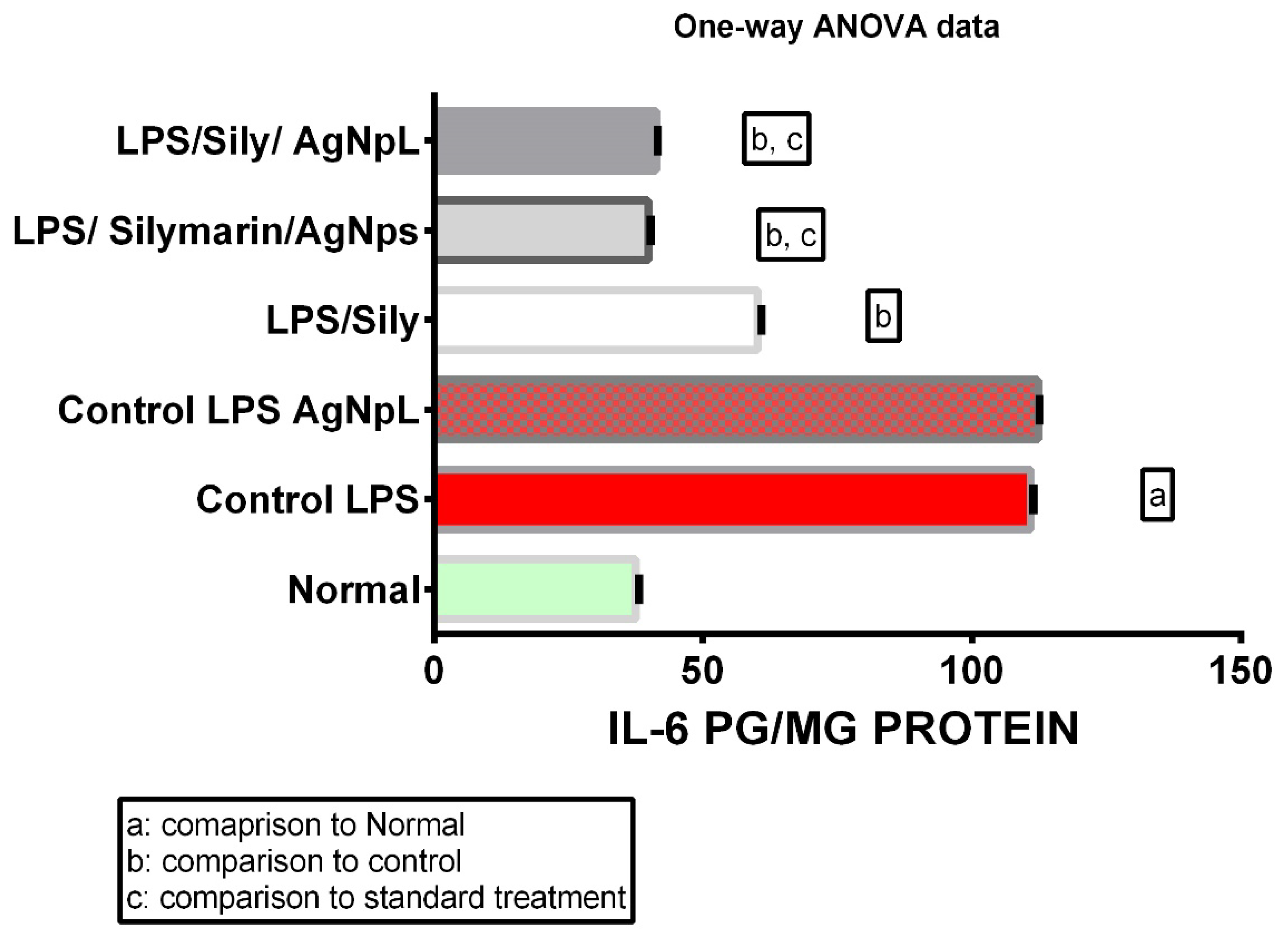
3.2. IL-6
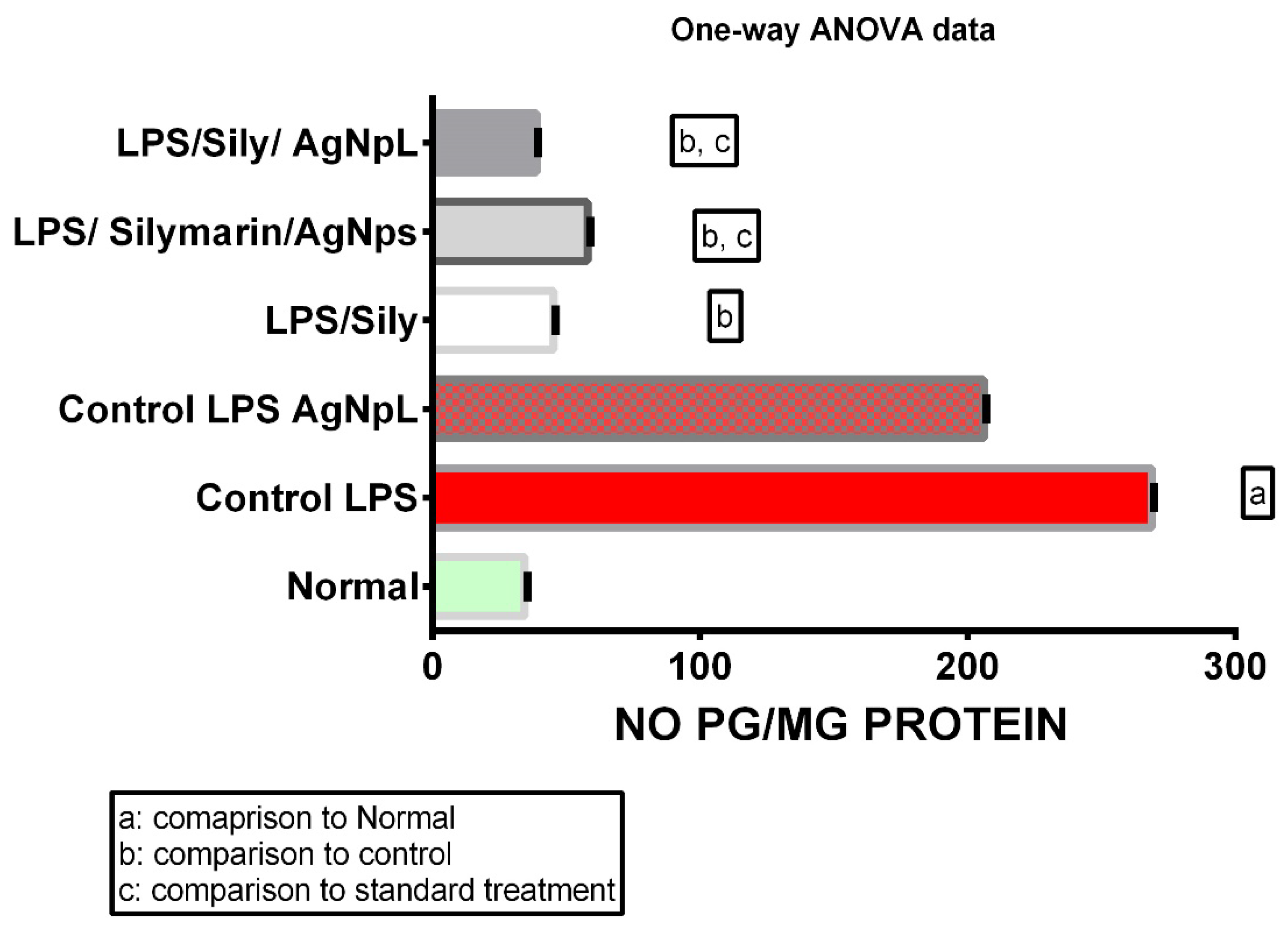
3.3. NO
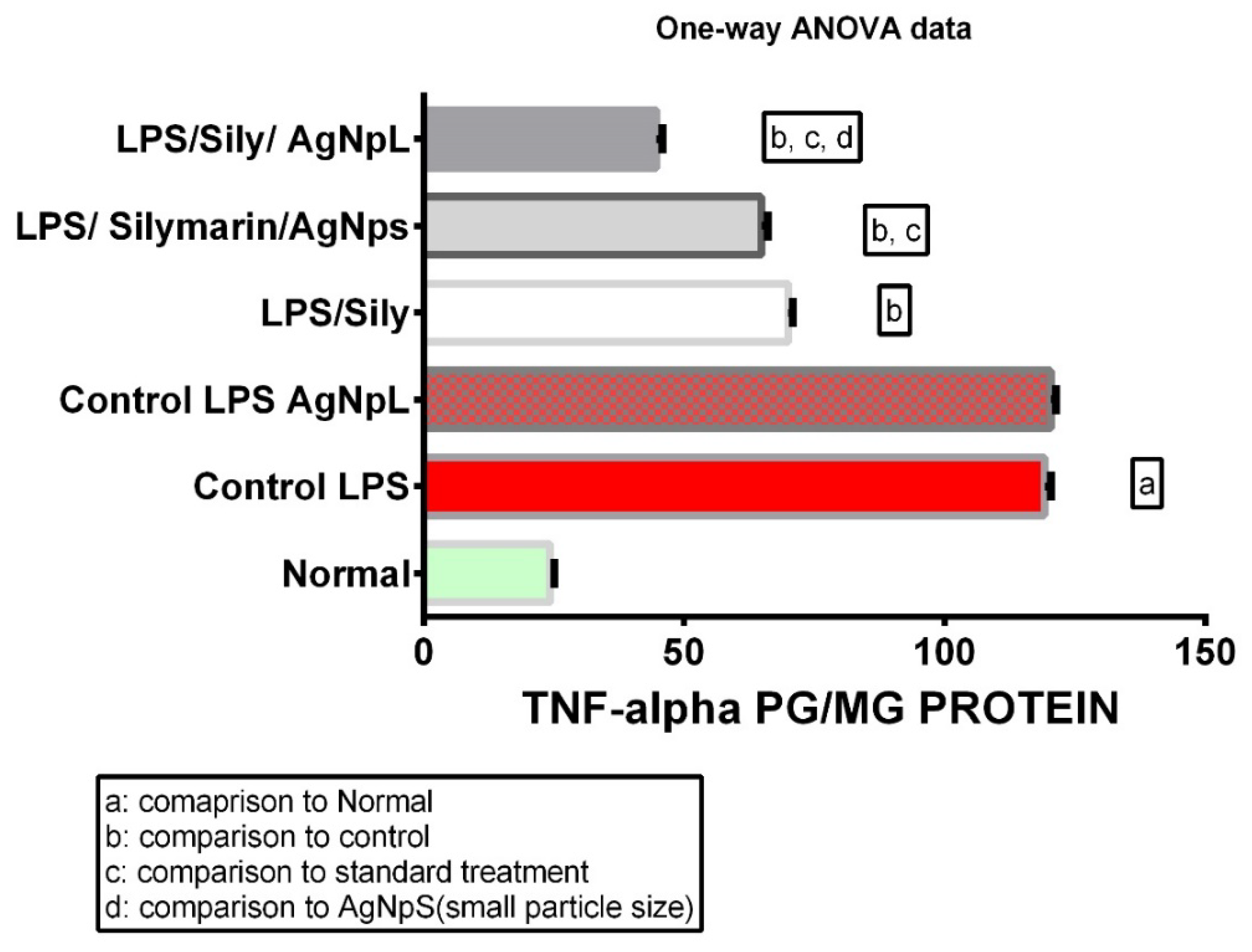
3.4. TNF-α
3.5. COX-2 Gene Expression

4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [Green Version]
- Oves, M.; Rauf, M.A.; Aslam, M.; Qari, H.A.; Sonbol, H.; Ahmad, I.; Zaman, G.S.; Saeed, M. Green synthesis of silver nanoparticles by Conocarpus Lancifolius plant extract and their antimicrobial and anticancer activities. Saudi J. Biol. Sci. 2022, 29, 460–471. [Google Scholar] [CrossRef]
- Surendran, S.P.; Thomas, R.G.; Moon, M.-J.; Jeong, Y.Y. Nanoparticles for the treatment of liver fibrosis. Int. J. Nanomed. 2017, 12, 6997–7006. [Google Scholar] [CrossRef] [Green Version]
- Ahamed, M.S.; AlSalhi, M.K.J.; Siddiqui, M. Silver nanoparticle applications and human health. Clin. Chim. Acta 2010, 411, 1841–1848. [Google Scholar] [CrossRef]
- Jaswal, T.; Gupta, J. A review on the toxicity of silver nanoparticles on human health. Mater. Today Proc. 2021. [Google Scholar] [CrossRef]
- Lee, S.H.; Jun, B.-H. Silver Nanoparticles: Synthesis and Application for Nanomedicine. Int. J. Mol. Sci. 2019, 20, 865. [Google Scholar] [CrossRef] [Green Version]
- Ajdary, M.; Moosavi, M.A.; Rahmati, M.; Falahati, M.; Mahboubi, M.; Mandegary, A.; Jangjoo, S.; Mohammadinejad, R.; Varma, R.S. Health Concerns of Various Nanoparticles: A Review of Their in Vitro and in Vivo Toxicity. Nanomaterials 2018, 8, 634. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Li, M.; Lin, X.; Li, Y.; Song, H.; Chen, H. AgNPs Aggravated Hepatic Steatosis, Inflammation, Oxidative Stress, and Epigenetic Changes in Mice with NAFLD Induced by HFD. Front. Bioeng. Biotechnol. 2022, 10, 912178. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Milić, M.; Leitinger, G.; Pavicic, I.; Avdičević, M.Z.; Dobrović, S.; Goessler, W.; Vrček, I.V. Cellular uptake and toxicity effects of silver nanoparticles in mammalian kidney cells. J. Appl. Toxicol. 2015, 35, 581–592. [Google Scholar] [CrossRef] [Green Version]
- Mauricio, M.D.; Guerra-Ojeda, S.; Marchio, P.; Valles, S.L.; Aldasoro, M.; Escribano-Lopez, I.; Herance, J.R.; Rocha, M.; Vila, J.M.; Victor, V.M. Nanoparticles in Medicine: A Focus on Vascular Oxidative Stress. Oxidative Med. Cell. Longev. 2018, 2018, 6231482. [Google Scholar] [CrossRef] [Green Version]
- Naguib, M.; Mahmoud, U.M.; Mekkawy, I.A.; Sayed, A.E.-D.H. Hepatotoxic effects of silver nanoparticles on Clarias gariepinus; Biochemical, histopathological, and histochemical studies. Toxicol. Rep. 2020, 7, 133–141. [Google Scholar] [CrossRef]
- Yaqoob, A.A.; Umar, K.; Ibrahim, M.N.M. Silver nanoparticles: Various methods of synthesis, size affecting factors and their potential applications—A review. Appl. Nanosci. 2020, 10, 1369–1378. [Google Scholar] [CrossRef]
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.K.M.A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- McShan, D.; Ray, P.C.; Yu, H. Molecular toxicity mechanism of nanosilver. J. Food Drug Anal. 2014, 22, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Ranjan, R.; Kumar, A.; Sinha, M.P.; Srivastava, R.; Subarna, S.; Mandal, S.K. Hepatoprotective activity of Silver Nanoparticles synthesized using aqueous leaf extract of Punica granatum against induced hepatotoxicity in rats. Nova Biol. Reper. 2021, 7, 381–389. [Google Scholar] [CrossRef]
- Arora, S.; Jain, J.M.; Rajwade, J.; Paknikar, K.M. Cellular responses induced by silver nanoparticles: In vitro studies. Toxicol. Lett. 2008, 179, 93–100. [Google Scholar] [CrossRef]
- Mohammed, H.A.; Khan, R.A. Anthocyanins: Traditional Uses, Structural and Functional Variations, Approaches to Increase Yields and Products’ Quality, Hepatoprotection, Liver Longevity, and Commercial Products. Int. J. Mol. Sci. 2022, 23, 2149. [Google Scholar] [CrossRef]
- Abenavoli, L.; Izzo, A.A.; Milić, N.; Cicala, C.; Santini, A.; Capasso, R. Milk thistle (Silybum marianum): A concise overview on its chemistry, pharmacological, and nutraceutical uses in liver diseases. Phytother. Res. 2018, 32, 2202–2213. [Google Scholar] [CrossRef]
- Mukhtar, S.; Xiaoxiong, Z.; Qamer, S.; Saad, M.; Mubarik, M.S.; Mahmoud, A.H.; Mohammed, O.B. Hepatoprotective activity of silymarin encapsulation against hepatic damage in albino rats. Saudi J. Biol. Sci. 2021, 28, 717–723. [Google Scholar] [CrossRef]
- Palma, E.; Doornebal, E.J.; Chokshi, S. Precision-cut liver slices: A versatile tool to advance liver research. Hepatol. Int. 2019, 13, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Olinga, P.; Schuppan, D. Precision-cut liver slices: A tool to model the liver ex vivo. J. Hepatol. 2013, 58, 1252–1253. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Montemayor, C.; Cordero-Pérez, P.; Salazar-Aranda, R.; Waksman-Minsky, N. Models of hepatoprotective activity assessment. Med. Univ. 2015, 17, 222–228. [Google Scholar] [CrossRef] [Green Version]
- Adães, S.; Mendonça, M.; Santos, T.N.; Castro-Lopes, J.M.; Ferreira-Gomes, J.; Neto, F.L. Intra-Articular Injection of Collagenase in the Knee of Rats as an Alternative Model to Study Nociception Associated with Osteoarthritis. Arthritis Res. Ther. 2014, 16, 1–17. [Google Scholar] [CrossRef]
- Dewyse, L.; Reynaert, H.; van Grunsven, L.A. Best Practices and Progress in Precision-Cut Liver Slice Cultures. Int. J. Mol. Sci. 2021, 22, 7137. [Google Scholar] [CrossRef]
- Gad, S.S.; Abdelrahim, D.S.; Ismail, S.H.; Ibrahim, S.M. Selenium and silver nanoparticles: A new approach for treatment of bacterial and viral hepatic infections via modulating oxidative stress and DNA fragmentation. J. Biochem. Mol. Toxicol. 2022, 36, e22972. [Google Scholar] [CrossRef]
- Westra, I.M.; Oosterhuis, D.; Groothuis, G.M.M.; Olinga, P. Precision-Cut Liver Slices as a Model for the Early Onset of Liver Fibrosis to Test Antifibrotic Drugs. Toxicol. Appl. Pharmacol. 2014, 274, 328–338. [Google Scholar] [CrossRef]
- Gandolfi, A.J.; Wijeweera, J.; Brendel, K. Use of Precision-Cut Liver Slices as an in Vitro Tool for Evaluating Liver Function. Toxicol. Pathol. 1996, 24, 58–61. [Google Scholar] [CrossRef]
- Maldonado, R.F.; Sa-Correia, I.; Valvano, M.A. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016, 40, 480–493. [Google Scholar] [CrossRef]
- Page, M.J.; Kell, D.B.; Pretorius, E. The Role of Lipopolysaccharide-Induced Cell Signalling in Chronic Inflammation. Chronic Stress 2022, 6, 24705470221076390. [Google Scholar] [CrossRef]
- Seemann, S.; Zohles, F.; Lupp, A. Comprehensive comparison of three different animal models for systemic inflammation. J. Biomed. Sci. 2017, 24, 60. [Google Scholar] [CrossRef]
- Marambio-Jones, C.; Hoek, E.M.V. A review of the antibacterial effects of silver nanomaterials and potential implications for human health and the environment. J. Nanopart. Res. 2010, 12, 1531–1551. [Google Scholar] [CrossRef]
- Tolaymat, T.M.; El Badawy, A.M.; Genaidy, A.; Scheckel, K.; Luxton, T.P.; Suidan, M. An evidence-based environmental perspective of manufactured silver nanoparticle in syntheses and applications: A systematic review and critical appraisal of peer-reviewed scientific papers. Sci. Total Environ. 2010, 408, 999–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Graaf, I.A.; Olinga, P.; de Jager, M.H.; Merema, M.T.; de Kanter, R.; van de Kerkhof, E.G.; Groothuis, G.M.M. Preparation and incubation of precision-cut liver and intestinal slices for application in drug metabolism and toxicity studies. Nat. Protoc. 2010, 5, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, M.A.; Vicente, A.A.; Pastrana, L.M. Nanotechnology in Food Packaging: Opportunities and Challenges. Nanomater. Food Packag. 2018, 1–11. [Google Scholar] [CrossRef]
- Rosário, F.; Creylman, J.; Verheyen, G.; Van Miert, S.; Santos, C.; Hoet, P.; Oliveira, H. Impact of Particle Size on Toxicity, Tissue Distribution and Excretion Kinetics of Subchronic Intratracheal Instilled Silver Nanoparticles in Mice. Toxics 2022, 10, 260. [Google Scholar] [CrossRef]
- Alajmi, R.A.; Al-Megrin, W.A.; Metwally, D.; Al-Subaie, H.; Altamrah, N.; Barakat, A.M.; Moneim, A.E.A.; Al-Otaibi, T.T.; El-Khadragy, M. Anti-Toxoplasma activity of silver nanoparticles green synthesized with Phoenix dactylifera and Ziziphus spina-christi extracts which inhibits inflammation through liver regulation of cytokines in Balb/c mice. Biosci. Rep. 2019, 39, BSR20190379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parang, Z.; Moghadamnia, D. Effects of silver nanoparticles on the functional tests of liver and its histological changes in adult male rats. Nanomed. Res. J. 2018, 3, 146–153. [Google Scholar] [CrossRef]
- Gliga, A.R.; Skoglund, S.; Wallinder, I.O.; Fadeel, B.; Karlsson, H.L. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: The role of cellular uptake, agglomeration and Ag release. Part. Fibre Toxicol. 2014, 11, 11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Feng, T.; Zhou, X.; Sullivan, P.M.; Hu, F.; Lou, Y.; Yu, J.; Feng, J.; Liu, H.; Chen, Y. Inactivation of TMEM106A promotes lipopolysaccharide-induced inflammation via the MAPK and NF-κB signaling pathways in macrophages. Clin. Exp. Immunol. 2020, 203, 125–136. [Google Scholar] [CrossRef]
- Ni, J.; Zhao, Y.; Su, J.; Liu, Z.; Fang, S.; Li, L.; Deng, J.; Fan, G. Toddalolactone Protects Lipopolysaccharide-Induced Sepsis and Attenuates Lipopolysaccharide-Induced Inflammatory Response by Modulating HMGB1-NF-κB Translocation. Front. Pharmacol. 2020, 11, 109. [Google Scholar] [CrossRef] [Green Version]
- Farhana, A.; Khan, Y.S. Biochemistry, Lipopolysaccharide; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Rimkunas, V.M.; Graham, M.J.; Crooke, R.M.; Liscum, L. TNF-α plays a role in hepatocyte apoptosis in Niemann-Pick type C liver disease. J. Lipid Res. 2009, 50, 327–333. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-B.; Shin, J.-S.; Han, H.-S.; Lee, H.-H.; Park, J.C.; Lee, K.-T. Kaempferol 7-O-β-D-glucoside isolated from the leaves of Cudrania tricuspidata inhibits LPS-induced expression of pro-inflammatory mediators through inactivation of NF-κB, AP-1, and JAK-STAT in RAW 264.7 macrophages. Chem. Biol. Interact. 2018, 284, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Cao, Z.; Song, X.; Zhang, X.; Mai, B.; Wen, T.; Lin, J.; Chen, J.; Chi, Y.; Su, T.; et al. Rhoifolin Alleviates Inflammation of Acute Inflammation Animal Models and LPS-Induced RAW264.7 Cells via IKKβ/NF-κB Signaling Pathway. Inflammation 2020, 43, 2191–2201. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yang, X.; Wu, Z.; Wang, H.; Li, Q.; Mei, H.; You, R.; Zhang, Y. Dendrobium officinale polysaccharide protected CCl4-induced liver fibrosis through intestinal homeostasis and the LPS-TLR4-NF-κB signaling pathway. Front. Pharmacol. 2020, 11, 240. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Su, C.; Zhao, S.; Li, J.; Yu, F. Combination therapy of Ulinastatin with Thrombomodulin alleviates endotoxin (LPS)-induced liver and kidney injury via inhibiting apoptosis, oxidative stress and HMGB1/TLR4/NF-κB pathway. Bioengineered 2022, 13, 2951–2970. [Google Scholar] [CrossRef] [PubMed]
- Gillessen, A.; Schmidt, H.H.-J. Silymarin as Supportive Treatment in Liver Diseases: A Narrative Review. Adv. Ther. 2020, 37, 1279–1301. [Google Scholar] [CrossRef] [Green Version]
- Simončič, B.; Klemenčič, D. Preparation and performance of silver as an antimicrobial agent for textiles: A review. Text. Res. J. 2016, 86, 210–223. [Google Scholar] [CrossRef]
- Corrêa, J.M.; Mori, M.; Sanches, H.L.; Cruz, A.; Poiate, E.; Poiate, I.A.V.P. Silver Nanoparticles in Dental Biomaterials. Int. J. Biomater. 2015, 2015, 485275. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.; Ahmad, M.; Swami, B.L.; Ikram, S. A review on plants extract mediated synthesis of silver nanoparticles for antimicrobial applications: A green expertise. J. Adv. Res. 2015, 7, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Shahidi, M.; Vaziri, F.; Haerian, A.; Farzanegan, A.; Jafari, S.; Sharifi, R.; Shirazi, F.S. Proliferative and Anti-Inflammatory Effects of Resveratrol and Silymarin on Human Gingival Fibroblasts: A View to the Future. J. Dent. Tehran Univ. Med. Sci. 2017, 14, 203–211. [Google Scholar]
- Wang, L.; Huang, Q.-H.; Li, Y.-X.; Huang, Y.-F.; Xie, J.-H.; Xu, L.-Q.; Dou, Y.-X.; Su, Z.-R.; Zeng, H.-F.; Chen, J.-N. Protective effects of silymarin on triptolide-induced acute hepatotoxicity in rats. Mol. Med. Rep. 2018, 17, 789–800. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, H.; Yang, Y.; Gou, Y.; Wang, Z.; Yang, D.; Li, C. Protective Effects of Silymarin Against D-Gal/LPS-Induced Organ Damage and Inflammation in Mice. Drug Des. Dev. Ther. 2021, 15, 1903–1914. [Google Scholar] [CrossRef]
- Bereswill, S.; Muñoz, M.; Fischer, A.; Plickert, R.; Haag, L.-M.; Otto, B.; Kühl, A.A.; Loddenkemper, C.; Göbel, U.B.; Heimesaat, M.M. Anti-Inflammatory Effects of Resveratrol, Curcumin and Simvastatin in Acute Small Intestinal Inflammation. PLoS ONE 2010, 5, e15099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.V.; Laux, P.; Luch, A.; Sudrik, C.; Wiehr, S.; Wild, A.-M.; Santomauro, G.; Bill, J.; Sitti, M. Review of emerging concepts in nanotoxicology: Opportunities and challenges for safer nanomaterial design. Toxicol. Mech. Methods 2019, 29, 378–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferdous, Z.; Nemmar, A. Health Impact of Silver Nanoparticles: A Review of the Biodistribution and Toxicity Following Various Routes of Exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, L.; Nienhaus, K.; Nienhaus, G.U. Engineered nanoparticles interacting with cells: Size matters. J. Nanobiotechnol. 2014, 12, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recordati, C.; De Maglie, M.; Bianchessi, S.; Argentiere, S.; Cella, C.; Mattiello, S.; Cubadda, F.; Aureli, F.; D’Amato, M.; Raggi, A.; et al. Tissue distribution and acute toxicity of silver after single intravenous administration in mice: Nano-specific and size-dependent effects. Part. Fibre Toxicol. 2015, 13, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Component | Volume |
|---|---|
| First strand buffer | 5 μL |
| 10 mM dNTPs | 2 μL |
| RNase inhibitor (40 U/μL) | 1 μL |
| MMLV-RT enzyme (50 U/μL) | 1 μL |
| DEPC-treated water | 10 μL |
| Primer Sequence | |
|---|---|
| COX2 | Forward primer 5′-GCAAATCCTTGCTGTTCCAATC-3′ Reverse primer 5′-GGAGAAGGCTTCCCAGCTTTTG-3′ |
| βeta actin | Forward primer 5′-TGTTTGAGACCTTCAACACC-3′ Reverse primer 5′-CGCTCATTGCCGATAGTGAT-3′ |
| PCR Reaction Mix Component | Volume |
|---|---|
| Forward primer | 1 μL |
| Reverse primer | 1 μL |
| SYBR green mix | 12.5 μL |
| cDNA template | 5 μL |
| RNase-free water | 5.5 μL |
| Total volume | 25 μL |
| Thermal Cycling Condition | ||
|---|---|---|
| Stage | Temp. | Time (s.) |
| Hold | 50 °C | 120 |
| One cycle | ||
| Denaturation | 95 °C | 15 |
| Annealing | 60 °C | 60 |
| Extension | 72 °C | 60 |
| 40 cycles | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elfaky, M.A.; Sirwi, A.; Ismail, S.H.; Awad, H.H.; Gad, S.S. Hepatoprotective Effect of Silver Nanoparticles at Two Different Particle Sizes: Comparative Study with and without Silymarin. Curr. Issues Mol. Biol. 2022, 44, 2923-2938. https://doi.org/10.3390/cimb44070202
Elfaky MA, Sirwi A, Ismail SH, Awad HH, Gad SS. Hepatoprotective Effect of Silver Nanoparticles at Two Different Particle Sizes: Comparative Study with and without Silymarin. Current Issues in Molecular Biology. 2022; 44(7):2923-2938. https://doi.org/10.3390/cimb44070202
Chicago/Turabian StyleElfaky, Mahmoud A., Alaa Sirwi, Sameh H. Ismail, Heba H. Awad, and Sameh S. Gad. 2022. "Hepatoprotective Effect of Silver Nanoparticles at Two Different Particle Sizes: Comparative Study with and without Silymarin" Current Issues in Molecular Biology 44, no. 7: 2923-2938. https://doi.org/10.3390/cimb44070202









