Quantitative In Silico Evaluation of Allergenic Proteins from Anacardium occidentale, Carya illinoinensis, Juglans regia and Pistacia vera and Their Epitopes as Precursors of Bioactive Peptides †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Sequences
2.2. BIOPEP-UWM Database of Bioactive Peptides
- a—the number of fragments with given activity in a protein sequence,
- N—the number of amino acid residues of protein chain.
- d—number of hydrolyzed peptide bonds in a protein/peptide chain,
- D—total number of peptide bonds in a protein/peptide chain.
- d—the number of peptides with a given activity (e.g., ACE inhibitors) released by a given enzyme (e.g., trypsin),
- N—the number of amino acid residues in protein.
- (1)
- IPR036312, IPR016140, IPR000617;
- (2)
- IPR006045, IPR014710, IPR011051, IPR006792;
- (3a)
- IPR006045, IPR014710, IPR011051;
- (3b)
- IPR022379, IPR006044, IPR006045, IPR014710, IPR011051.
2.3. Statistical Analysis
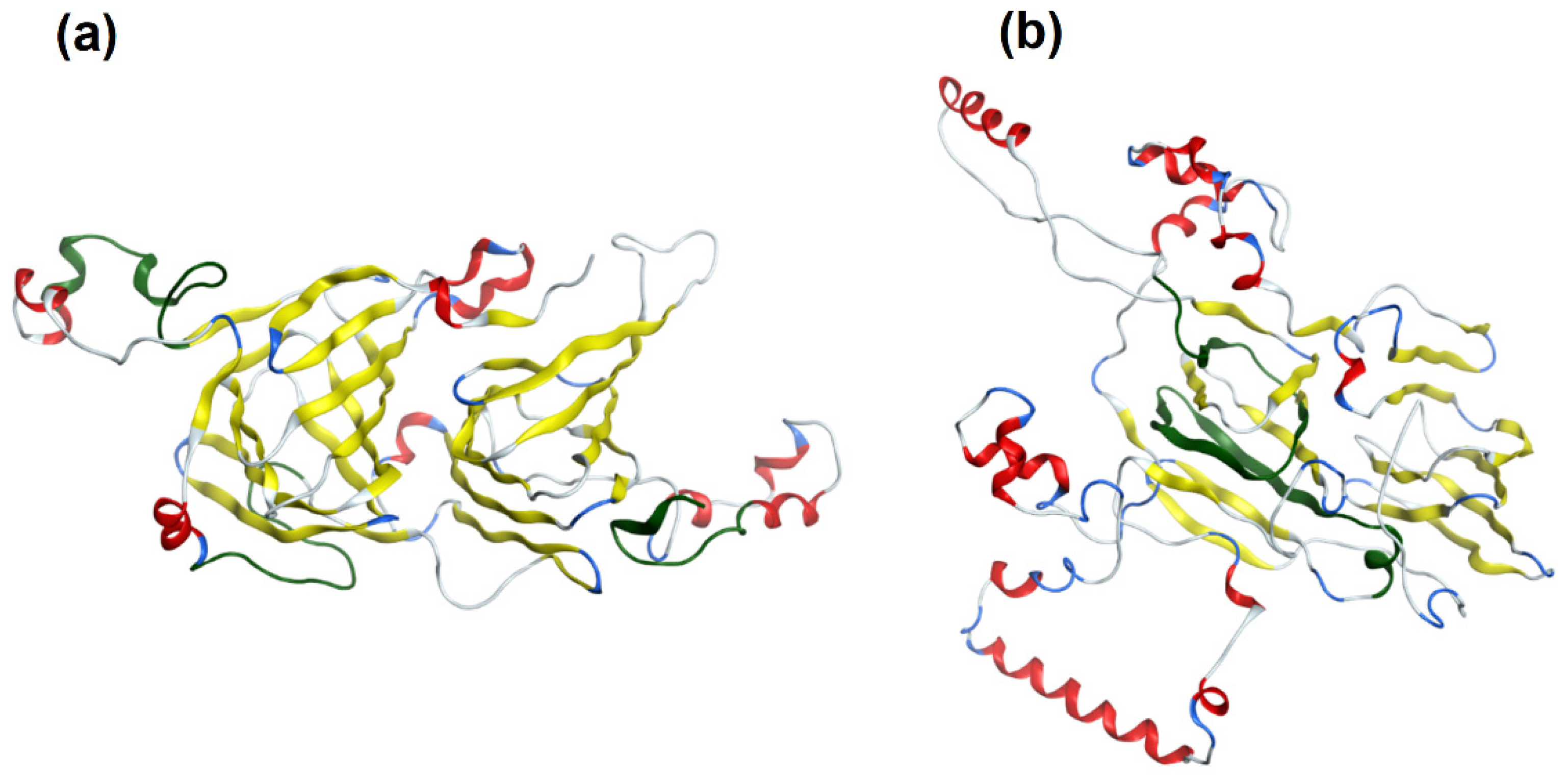
2.4. Protein Modeling
3. Results and Discussion
3.1. Tree Nut Epitopes
3.2. Tree Nut Allergen Protein Domains
3.3. Correlation of Bioactive Fragments between Allergen Protein Domains and Epitopes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iwaniak, A.; Darewicz, M.; Minkiewicz, P. Peptides Derived from Foods as Supportive Diet Components in the Prevention of Metabolic Syndrome. Compr. Rev. Food Sci. Food Saf. 2018, 17, 63–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barati, M.; Javanmardi, F.; Mousavi Jazayeri, S.M.H.; Jabbari, M.; Rahmani, J.; Barati, F.; Nickho, H.; Davoodi, S.H.; Roshanravan, N.; Mousavi Khaneghah, A. Techniques, perspectives, and challenges of bioactive peptide generation: A comprehensive systematic review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1488–1520. [Google Scholar] [CrossRef] [PubMed]
- Manzanares, P.; Gandía, M.; Garrigues, S.; Marcos, J.F. Improving Health-Promoting Effects of Food-Derived Bioactive Peptides through Rational Design and Oral Delivery Strategies. Nutrients 2019, 11, 2545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, A.; Ogita, H. Pathophysiological Implications of Dipeptidyl Peptidases. Curr. Protein Pept. Sci. 2017, 18, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Shimizu, A.; Kurita, S.; Zankov, D.P.; Takeuchi, K.; Yasuda-Yamahara, M.; Kume, S.; Ishida, T.; Ogita, H. Novel Therapeutic Role for Dipeptidyl Peptidase III in the Treatment of Hypertension. Hypertension 2016, 68, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Câmara, J.S.; Albuquerque, B.R.; Aguiar, J.; Corrêa, R.C.G.; Gonçalves, J.L.; Granato, D.; Pereira, J.A.M.; Barros, L.; Ferreira, I. Food Bioactive Compounds and Emerging Techniques for Their Extraction: Polyphenols as a Case Study. Foods 2020, 10, 37. [Google Scholar] [CrossRef]
- Bolling, B.W.; Chen, C.Y.; McKay, D.L.; Blumberg, J.B. Tree nut phytochemicals: Composition, antioxidant capacity, bioactivity, impact factors. A systematic review of almonds, Brazils, cashews, hazelnuts, macadamias, pecans, pine nuts, pistachios and walnuts. Nutr. Res. Rev. 2011, 24, 244–275. [Google Scholar] [CrossRef] [Green Version]
- Karami, Z.; Akbari-Adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Cavazos, A.; Gonzalez de Mejia, E. Identification of Bioactive Peptides from Cereal Storage Proteins and Their Potential Role in Prevention of Chronic Diseases. Compr. Rev. Food Sci. Food Saf. 2013, 12, 364–380. [Google Scholar] [CrossRef]
- Gu, X.; Hou, Y.-K.; Li, D.; Wang, J.-Z.; Wang, F.-J. Separation, Purification, and Identification of Angiotensin I–Converting Enzyme Inhibitory Peptides from Walnut (Juglans regia L.) Hydrolyzate. Int. J. Food Prop. 2015, 18, 266–276. [Google Scholar] [CrossRef]
- Wang, F.-J.; Yin, X.-Y.; Regenstein, J.M.; Wang, J.-Z. Separation and purification of angiotensin-I-converting enzyme (ACE) inhibitory peptides from walnuts (Juglans regia L.) meal. Eur. Food Res. Technol. 2016, 242, 911–918. [Google Scholar] [CrossRef]
- Liu, D.; Guo, Y.; Ma, H. Production, bioactivities and bioavailability of bioactive peptides derived from walnut origin by-products: A review. Crit. Rev. Food Sci. Nutr. 2022, 1–16. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, L.; Song, W.; Zhang, C.; Hua, Y.; Chen, Y.; Li, X. Separation, identification and molecular binding mechanism of dipeptidyl peptidase IV inhibitory peptides derived from walnut (Juglans regia L.) protein. Food Chem. 2021, 347, 129062. [Google Scholar] [CrossRef]
- Chen, N.; Yang, H.; Sun, Y.; Niu, J.; Liu, S. Purification and identification of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Peptides 2012, 38, 344–349. [Google Scholar] [CrossRef]
- Jahanbani, R.; Ghaffari, S.M.; Salami, M.; Vahdati, K.; Sepehri, H.; Sarvestani, N.N.; Sheibani, N.; Moosavi-Movahedi, A.A. Antioxidant and Anticancer Activities of Walnut (Juglans regia L.) Protein Hydrolysates Using Different Proteases. Plant Foods Hum. Nutr. 2016, 71, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.C.; Yang, S.J.; Hong, D.; Yang, J.P.; Liu, M.; Lin, Y.; Huang, C.H.; Wang, C.J. A simple and convenient method for the preparation of antioxidant peptides from walnut (Juglans regia L.) protein hydrolysates. Chem. Cent. J. 2016, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Yang, S.; Yang, J.; Lee, Y.; Kou, J.; Wang, C. Neuroprotective and memory-enhancing effects of antioxidant peptide from walnut (Juglans regia L.) protein hydrolysates. Nat. Prod. Commun. 2019, 14, 1934578X19865838. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Yang, X.; Chen, J.; Peng, T.; Yin, X.; Liu, W.; Liang, M.; Wan, J.; Yang, X. Antioxidative Effects and Mechanism Study of Bioactive Peptides from Defatted Walnut (Juglans regia L.) Meal Hydrolysate. J. Agric. Food Chem. 2019, 67, 3305–3312. [Google Scholar] [CrossRef]
- Wang, S.; Zheng, L.; Zhao, T.; Zhang, Q.; Liu, Y.; Sun, B.; Su, G.; Zhao, M. Inhibitory Effects of Walnut (Juglans regia) Peptides on Neuroinflammation and Oxidative Stress in Lipopolysaccharide-Induced Cognitive Impairment Mice. J. Agric. Food Chem. 2020, 68, 2381–2392. [Google Scholar] [CrossRef]
- Wang, S.; Su, G.; Zhang, X.; Song, G.; Zhang, L.; Zheng, L.; Zhao, M. Characterization and Exploration of Potential Neuroprotective Peptides in Walnut (Juglans regia) Protein Hydrolysate against Cholinergic System Damage and Oxidative Stress in Scopolamine-Induced Cognitive and Memory Impairment Mice and Zebrafish. J. Agric. Food Chem. 2021, 69, 2773–2783. [Google Scholar] [CrossRef]
- Feng, Y.X.; Wang, Z.C.; Chen, J.X.; Li, H.R.; Wang, Y.B.; Ren, D.F.; Lu, J. Separation, identification, and molecular docking of tyrosinase inhibitory peptides from the hydrolysates of defatted walnut (Juglans regia L.) meal. Food Chem. 2021, 353, 129471. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, L.; Du, Q.; Ren, J.W.; Chen, Q.H.; Li, D.; Mao, R.X.; Liu, X.R.; Li, Y. Small Molecule Oligopeptides Isolated from Walnut (Juglans regia L.) and Their Anti-Fatigue Effects in Mice. Molecules 2018, 24, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Jia, J.; Fang, M.; Zhang, L.; Guo, M.; Xie, J.; Xia, Y.; Zhou, L.; Wei, D. In vitro and in vivo ACE inhibitory of pistachio hydrolysates and in silico mechanism of identified peptide binding with ACE. Process Biochem. 2014, 49, 898–904. [Google Scholar] [CrossRef]
- Yao, G.-l.; Chai, Y.; Chen, J.; Wu, Y.-G. Separation and identification of ACE inhibitory peptides from cashew nut (Anacardium occidentale Linnaeus) protein. Int. J. Food Prop. 2017, 20, S981–S991. [Google Scholar] [CrossRef] [Green Version]
- Mares-Mares, E.; Gutierrez-Vargas, S.; Perez-Moreno, L.; Ordonez-Acevedo, L.G.; Barboza-Corona, J.E.; Leon-Galvan, M.F. Characterization and Identification of Cryptic Biopeptides in Carya illinoinensis (Wangenh K. Koch) Storage Proteins. Biomed. Res. Int. 2017, 2017, 1549156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, J.P.; Sicherer, S. Food allergy: Epidemiology, pathogenesis, diagnosis, prevention, and treatment. Curr. Opin. Immunol. 2020, 66, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.M.; Jiang, J.; Gupta, R.S. Epidemiology and Burden of Food Allergy. Curr. Allergy Asthma Rep. 2020, 20, 6. [Google Scholar] [CrossRef]
- Suther, C.; Moore, M.D.; Beigelman, A.; Zhou, Y. The Gut Microbiome and the Big Eight. Nutrients 2020, 12, 3728. [Google Scholar] [CrossRef]
- Willison, L.N.; Sathe, S.K.; Roux, K.H. Production and analysis of recombinant tree nut allergens. Methods 2014, 66, 34–43. [Google Scholar] [CrossRef]
- Breiteneder, H.; Mills, E.N. Molecular properties of food allergens. J. Allergy Clin. Immunol. 2005, 115, 14–23, quiz 24. [Google Scholar] [CrossRef]
- Geiselhart, S.; Hoffmann-Sommergruber, K.; Bublin, M. Tree nut allergens. Mol. Immunol. 2018, 100, 71–81. [Google Scholar] [CrossRef]
- Palladino, C.; Breiteneder, H. Peanut allergens. Mol. Immunol. 2018, 100, 58–70. [Google Scholar] [CrossRef]
- Smeekens, J.M.; Bagley, K.; Kulis, M. Tree Nut Allergies: Allergen Homology, Cross-reactivity, and Implications for Therapy. Clin. Exp. Allergy 2018, 48, 762–772. [Google Scholar] [CrossRef]
- Dreskin, S.C.; Koppelman, S.J.; Andorf, S.; Nadeau, K.C.; Kalra, A.; Braun, W.; Negi, S.S.; Chen, X.; Schein, C.H. The importance of the 2S albumins for allergenicity and cross-reactivity of peanuts, tree nuts, and sesame seeds. J. Allergy Clin. Immunol. 2021, 147, 1154–1163. [Google Scholar] [CrossRef]
- Fuhrmann, V.; Huang, H.J.; Akarsu, A.; Shilovskiy, I.; Elisyutina, O.; Khaitov, M.; van Hage, M.; Linhart, B.; Focke-Tejkl, M.; Valenta, R.; et al. From Allergen Molecules to Molecular Immunotherapy of Nut Allergy: A Hard Nut to Crack. Front. Immunol. 2021, 12, 742732. [Google Scholar] [CrossRef]
- Shi, Y. Identifying Linear B-cell Epitopes Based on Incorporated Sequence Information. Curr. Proteom. 2018, 15, 190–195. [Google Scholar] [CrossRef]
- Hasan, M.M.; Khatun, M.S.; Kurata, H. iLBE for Computational Identification of Linear B-cell Epitopes by Integrating Sequence and Evolutionary Features. Genom. Proteom. Bioinform. 2020, 18, 593–600. [Google Scholar] [CrossRef]
- Yao, B.; Zheng, D.; Liang, S.; Zhang, C. SVMTriP: A Method to Predict B-Cell Linear Antigenic Epitopes. Methods Mol. Biol. 2020, 2131, 299–307. [Google Scholar] [CrossRef]
- Ras-Carmona, A.; Pelaez-Prestel, H.F.; Lafuente, E.M.; Reche, P.A. BCEPS: A Web Server to Predict Linear B Cell Epitopes with Enhanced Immunogenicity and Cross-Reactivity. Cells 2021, 10, 2744. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Dziuba, J.; Michalska, J. Bovine meat proteins as potential precursors of biologically active peptides—A computational study based on the BIOPEP database. Food Sci. Technol. Int. 2011, 17, 39–45. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; He, S.; Sun, H.; Wang, Y.; Zhang, Y. Advances in epitope mapping technologies for food protein allergens: A review. Trends Food Sci. Technol. 2021, 107, 226–239. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Dziuba, J.; Iwaniak, A.; Minkiewicz, P. Computer-aided characteristics of proteins as potential precursors of bioactive peptides. Polimery 2003, 48, 50–53. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, P.M.; Petersen, D.; Dambmann, C. Improved Method for Determining Food Protein Degree of Hydrolysis. J. Food Sci. 2001, 66, 642–646. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assuncao, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carriere, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Cabanos, C.; Urabe, H.; Tandang-Silvas, M.R.; Utsumi, S.; Mikami, B.; Maruyama, N. Crystal structure of the major peanut allergen Ara h 1. Mol. Immunol. 2011, 49, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Guo, F.; Chen, Y.W.; Howard, A.; Zhang, Y.Z. Crystal structure of Ara h 3, a major allergen in peanut. Mol. Immunol. 2009, 46, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Elizur, A.; Appel, M.Y.; Nachshon, L.; Levy, M.B.; Epstein-Rigbi, N.; Pontoppidan, B.; Lidholm, J.; Goldberg, M.R. Clinical and Molecular Characterization of Walnut and Pecan Allergy (NUT CRACKER Study). J. Allergy Clin. Immunology. Pract. 2020, 8, 157–165.e152. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.M.; Xiao, H.; Nowak-Wegrzyn, A.; Zhou, P. IgE-binding epitope mapping of tropomyosin allergen (Exo m 1) from Exopalaemon modestus, the freshwater Siberian prawn. Food Chem. 2020, 309, 125603. [Google Scholar] [CrossRef]
- Dessailly, B.H.; Redfern, O.C.; Cuff, A.; Orengo, C.A. Exploiting structural classifications for function prediction: Towards a domain grammar for protein function. Curr. Opin. Struct. Biol. 2009, 19, 349–356. [Google Scholar] [CrossRef] [Green Version]
- Levitt, M. Nature of the protein universe. Proc. Natl. Acad. Sci. USA 2009, 106, 11079–11084. [Google Scholar] [CrossRef] [Green Version]
- Scaiewicz, A.; Levitt, M. The language of the protein universe. Curr. Opin. Genet. Dev. 2015, 35, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Wetlaufer, D.B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl. Acad. Sci. USA 1973, 70, 697–701. [Google Scholar] [CrossRef] [Green Version]
- Nardo, A.E.; Anon, M.C.; Parisi, G. Large-scale mapping of bioactive peptides in structural and sequence space. PLoS ONE 2018, 13, e0191063. [Google Scholar] [CrossRef] [Green Version]
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Sokołowska, J.; Starowicz, P.; Bucholska, J.; Hrynkiewicz, M. Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science. Int. J. Mol. Sci. 2015, 16, 20748–20773. [Google Scholar] [CrossRef] [Green Version]
- Dziuba, M.; Dziuba, J.; Minkiewicz, P. Design of food protein proteolysis with a view to obtaining bioactive peptides. Pol. J. Nat. Sci. 2006, 21, 999–1020. [Google Scholar]
- Dziuba, M.; Darewicz, M. Food Proteins as Precursors of Bioactive Peptides—Classification Into Families. Food Sci. Technol. Int. 2007, 13, 393–404. [Google Scholar] [CrossRef]
- Iwaniak, A.; Dziuba, J. Analysis of Domains in Selected Plant and Animal Food Proteins-Precursors of Biologically Active Peptides—In Silico Approach. Food Sci. Technol. Int. 2009, 15, 179–191. [Google Scholar] [CrossRef]






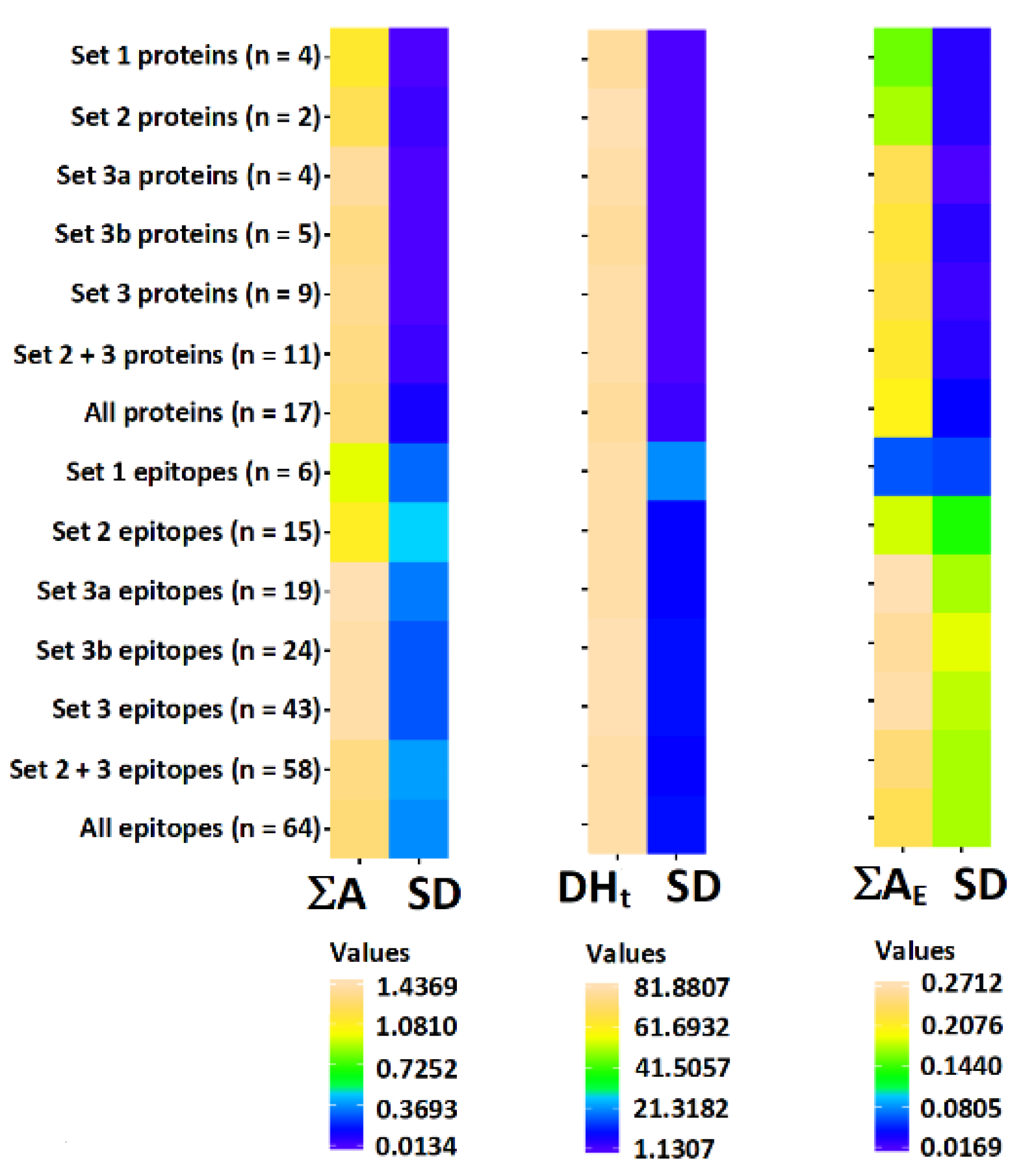
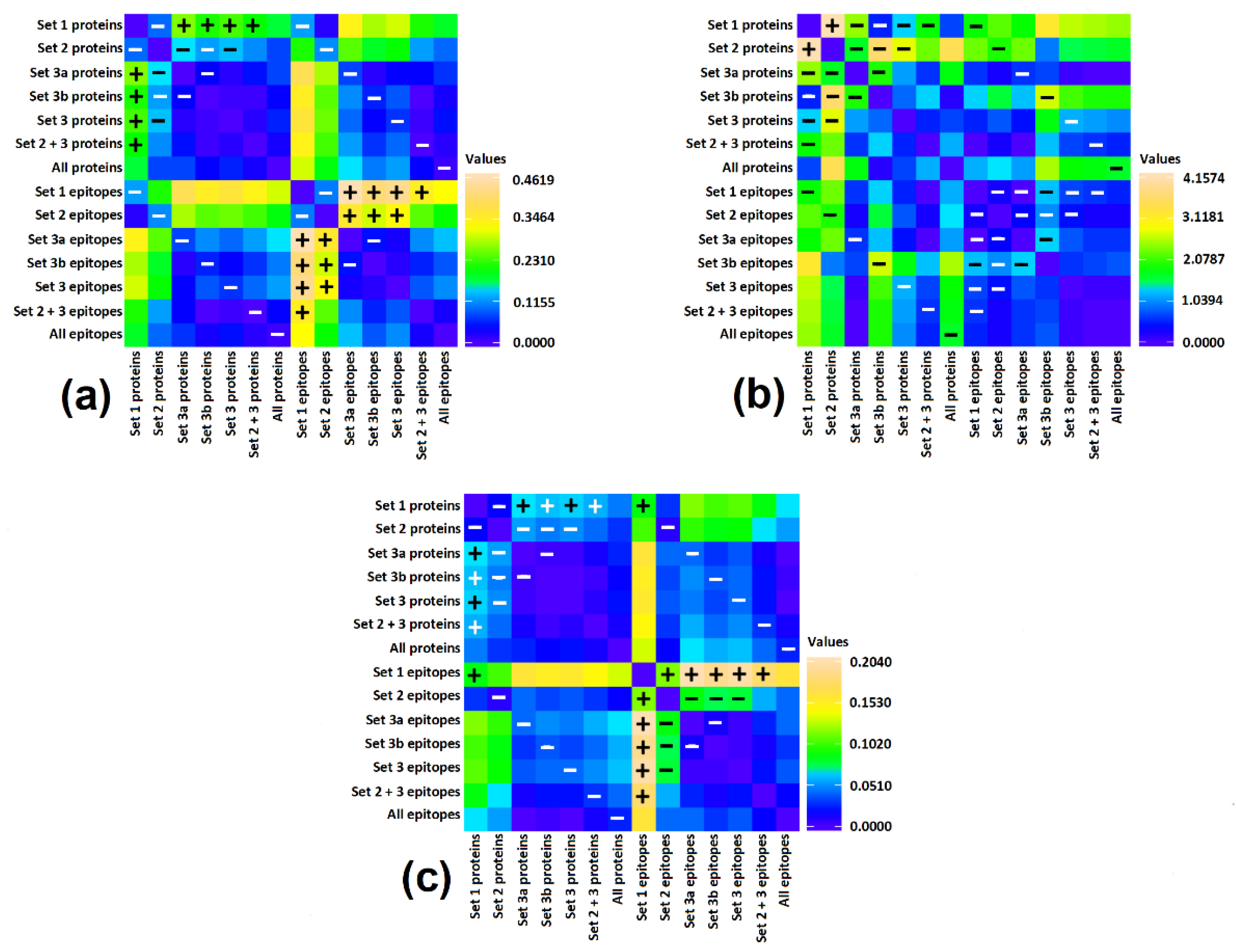
| No | Species | Protein Type | Allergen Name | UniProt Access. No | InterPro Domain IDs 1 | Set of Domains |
|---|---|---|---|---|---|---|
| 1 | Anacardium occidentale | 7S vicilin | Ana o 1.0101 | Q8L5L5 | IPR006045, IPR014710, IPR011051 | 3a |
| 2 | Anacardium occidentale | 7S vicilin | Ana o 1.0102 | Q8L5L6 | IPR006045, IPR014710, IPR011051 | 3a |
| 3 | Anacardium occidentale | 11S legumin | Ana o 2.0101 | Q8GZP6 | IPR022379, IPR006044, IPR006045, IPR014710, IPR011051 | 3b |
| 4 | Anacardium occidentale | 2S albumin | Ana o 3.0101 | Q8H2B8 | IPR036312, IPR016140, IPR000617 | 1 |
| 5 | Carya illinoinensis | 2S albumin | Car i 1.0101 | Q84XA9 | IPR036312, IPR016140, IPR000617 | 1 |
| 6 | Carya illinoinensis | 7S viclin | Car i 2.0101 | B3STU4 | IPR006045, IPR014710, IPR011051, IPR006792 | 2 |
| 7 | Carya illinoinensis | 11S legumin | Car i 4.0101 | B5KVH4 | IPR022379, IPR006044, IPR006045, IPR014710, IPR011051 | 3b |
| 8 | Juglans regia | 2S albumin | Jug r 1.0101 | P93198 | IPR036312, IPR016140, IPR000617 | 1 |
| 9 | Juglans regia | 7S vicilin | Jug r 2.0101 | Q9SEW4 | IPR006045, IPR014710, IPR011051, IPR006792 | 2 |
| 10 | Juglans regia | non-specific lipid transfer protein type 1 (nsLTP1) | Jug r 3 | C5H617 | IPR036312, IPR016140, IPR000528 | - |
| 11 | Juglans regia | 11S legumin | Jug r 4.0101 | Q2TPW5 | IPR022379, IPR006044, IPR006045, IPR014710, IPR011051 | 3b |
| 12 | Pistacia vera | 2S albumin | Pis v 1.0101 | B7P072 | IPR036312, IPR016140, IPR000617 | 1 |
| 13 | Pistacia vera | 11S legumin | Pis v 2.0101 | B7P073 | IPR022379, IPR006044, IPR006045, IPR014710, IPR011051 | 3b |
| 14 | Pistacia vera | 11S legumin | Pis v 2.0201 | B7P074 | IPR022379, IPR006044, IPR006045, IPR014710, IPR011051 | 3b |
| 15 | Pistacia vera | 7S vicilin | Pis v 3.0101 | B4X640 | IPR006045, IPR014710, IPR011051 | 3a |
| 16 | Pistacia vera | manganese superoxide dismutase | Pis v 4.0101 | B2BDZ8 | IPR001189, IPR019833, IPR019832, IPR019831, IPR036324, IPR036314 | - |
| 17 | Pistacia vera | 11S legumin | Pis v 5.0101 | B7SLJ1 | IPR006044, IPR006045, IPR014710, IPR011051 | 3b |
| Epitopes (ID According to the Immune Epitope Database) 1 | Allergen Containing Epitopes, Annotated in Immune Epitope Database | Allergen Containing Epitopes, Found in the UniProt Database Using BLAST |
|---|---|---|
| 157220, 157381, 157603, 157808, 157835 | Car i 4.0101 | Jug r 4.0101 |
| 174135 | Jug r 1.0101 | Car i 1.0101 |
| 157811, 158509, 241161, 241344, 241355, 241557 | Jug r 2.0101 | Car i 2.0101 |
| 114508, 114569, 114610, 114627, 114695 | Jug r 4.0101 | Car i 4.0101 |
| ID | Name | Score for Entire Proteins | Score for Epitopes | ||||
|---|---|---|---|---|---|---|---|
| ∑A | DHt | ∑AE | ∑A | DHt | ∑AE | ||
| IPR000528 | Plant lipid transfer protein/Par allergen | + (s) | − (s) | + (s) | nd | nd | nd |
| IPR000617 | Napin/Bra allergen | − | − | − | − | 0 | − |
| IPR001189 | Manganese/iron superoxide dismutase | + (s) | 0 (s) | 0 (s) | nd | nd | nd |
| IPR006044 | 11-S seed storage protein, plant | 0 | 0 | 0 | 0 | 0 | 0 |
| IPR006045 | Cupin 1 | + | 0 | + | + | 0 | + |
| IPR006792 | Vicilin, N-terminal | − | + | − | − | 0 | 0 |
| IPR011051 | RmlC-like cupin domain superfamily | + | 0 | + | + | 0 | + |
| IPR014710 | RmlC-like jelly roll fold | + | 0 | + | + | 0 | + |
| IPR016140 | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain | 0 | 0 | 0 | − | 0 | − |
| IPR019831 | Manganese/iron superoxide dismutase, N-terminal | + (s) | 0 (s) | 0 (s) | nd | nd | nd |
| IPR019832 | Manganese/iron superoxide dismutase, C-terminal | + (s) | 0 (s) | 0 (s) | nd | nd | nd |
| IPR019833 | Manganese/iron superoxide dismutase, binding site | + (s) | 0 (s) | 0 (s) | nd | nd | nd |
| IPR022379 | 11-S seed storage protein, conserved site | 0 | 0 | 0 | 0 | 0 | 0 |
| IPR036312 | Bifunctional inhibitor/plant lipid transfer protein/seed storage helical domain superfamily | 0 | 0 | 0 | − | 0 | − |
| IPR036314 | Manganese/iron superoxide dismutase, C-terminal domain superfamily | + (s) | 0 (s) | 0 (s) | nd | nd | nd |
| IPR036324 | Manganese/iron superoxide dismutase, N-terminal domain superfamily | + (s) | 0 (s) | 0 (s) | nd | nd | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minkiewicz, P.; Mattison, C.P.; Darewicz, M. Quantitative In Silico Evaluation of Allergenic Proteins from Anacardium occidentale, Carya illinoinensis, Juglans regia and Pistacia vera and Their Epitopes as Precursors of Bioactive Peptides. Curr. Issues Mol. Biol. 2022, 44, 3100-3117. https://doi.org/10.3390/cimb44070214
Minkiewicz P, Mattison CP, Darewicz M. Quantitative In Silico Evaluation of Allergenic Proteins from Anacardium occidentale, Carya illinoinensis, Juglans regia and Pistacia vera and Their Epitopes as Precursors of Bioactive Peptides. Current Issues in Molecular Biology. 2022; 44(7):3100-3117. https://doi.org/10.3390/cimb44070214
Chicago/Turabian StyleMinkiewicz, Piotr, Christopher P. Mattison, and Małgorzata Darewicz. 2022. "Quantitative In Silico Evaluation of Allergenic Proteins from Anacardium occidentale, Carya illinoinensis, Juglans regia and Pistacia vera and Their Epitopes as Precursors of Bioactive Peptides" Current Issues in Molecular Biology 44, no. 7: 3100-3117. https://doi.org/10.3390/cimb44070214
APA StyleMinkiewicz, P., Mattison, C. P., & Darewicz, M. (2022). Quantitative In Silico Evaluation of Allergenic Proteins from Anacardium occidentale, Carya illinoinensis, Juglans regia and Pistacia vera and Their Epitopes as Precursors of Bioactive Peptides. Current Issues in Molecular Biology, 44(7), 3100-3117. https://doi.org/10.3390/cimb44070214







