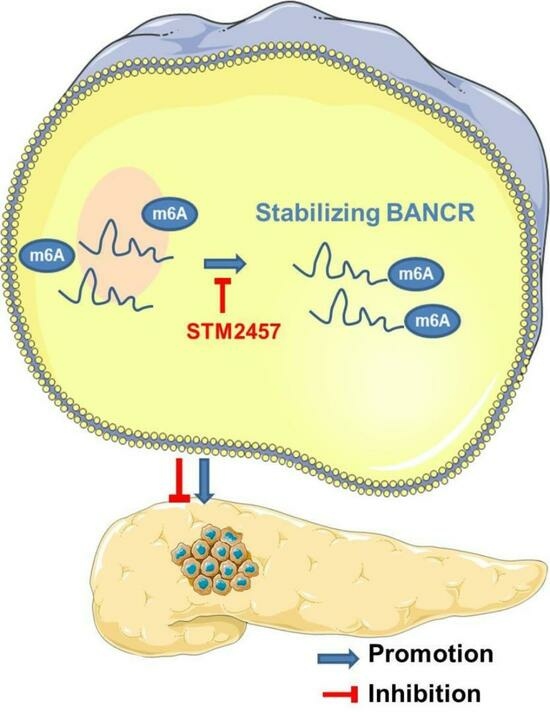
STM2457 Inhibits the Invasion and Metastasis of Pancreatic Cancer by Down-Regulating BRAF-Activated Noncoding RNA N6-Methyladenosine Modification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Chemicals
2.2. Real-Time qRT-PCR
2.3. Western Blotting
2.4. Immunofluorescence In Situ Hybridization
2.5. RNA Isolation and Quantitative Real-Time PCR
2.6. MeRIP-qPCR
2.7. Cell Proliferation Assays
2.8. Colony Formation Assay
2.9. Wound Healing Assay
2.10. Cell Migration and Invasion Assay
2.11. Statistical Analysis
3. Results
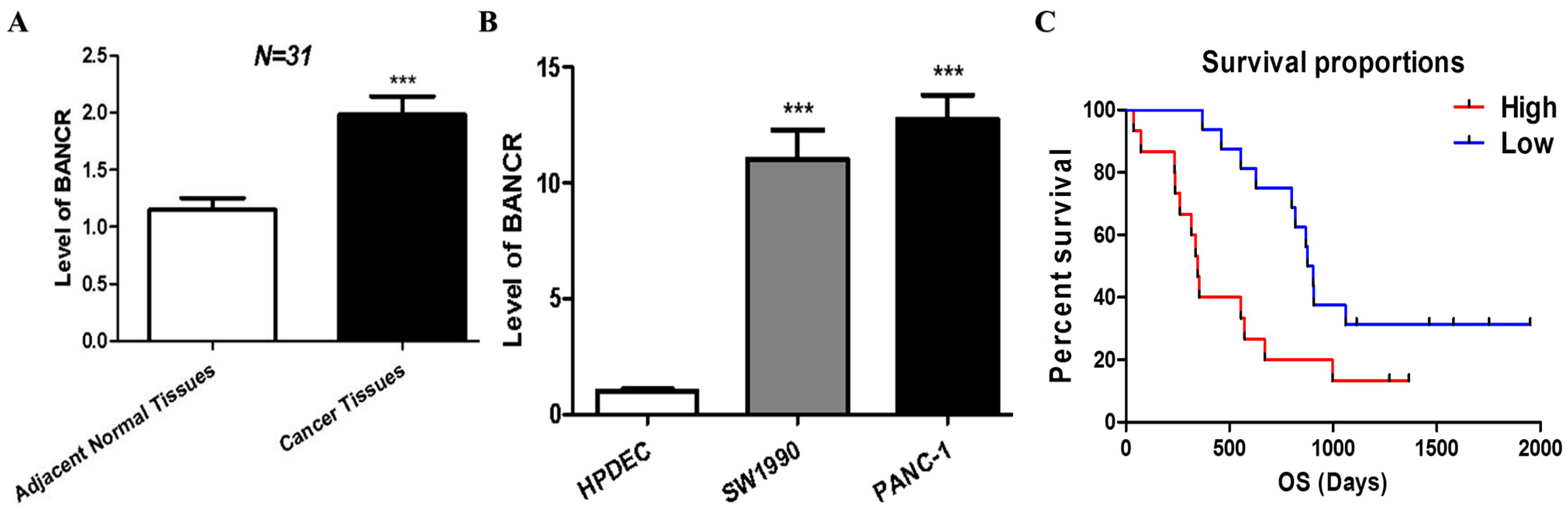
3.1. BANCR Was Overexpressed in Pancreatic Cancer Tissues and Cells, Which Are Associated with Poor Clinical Outcomes
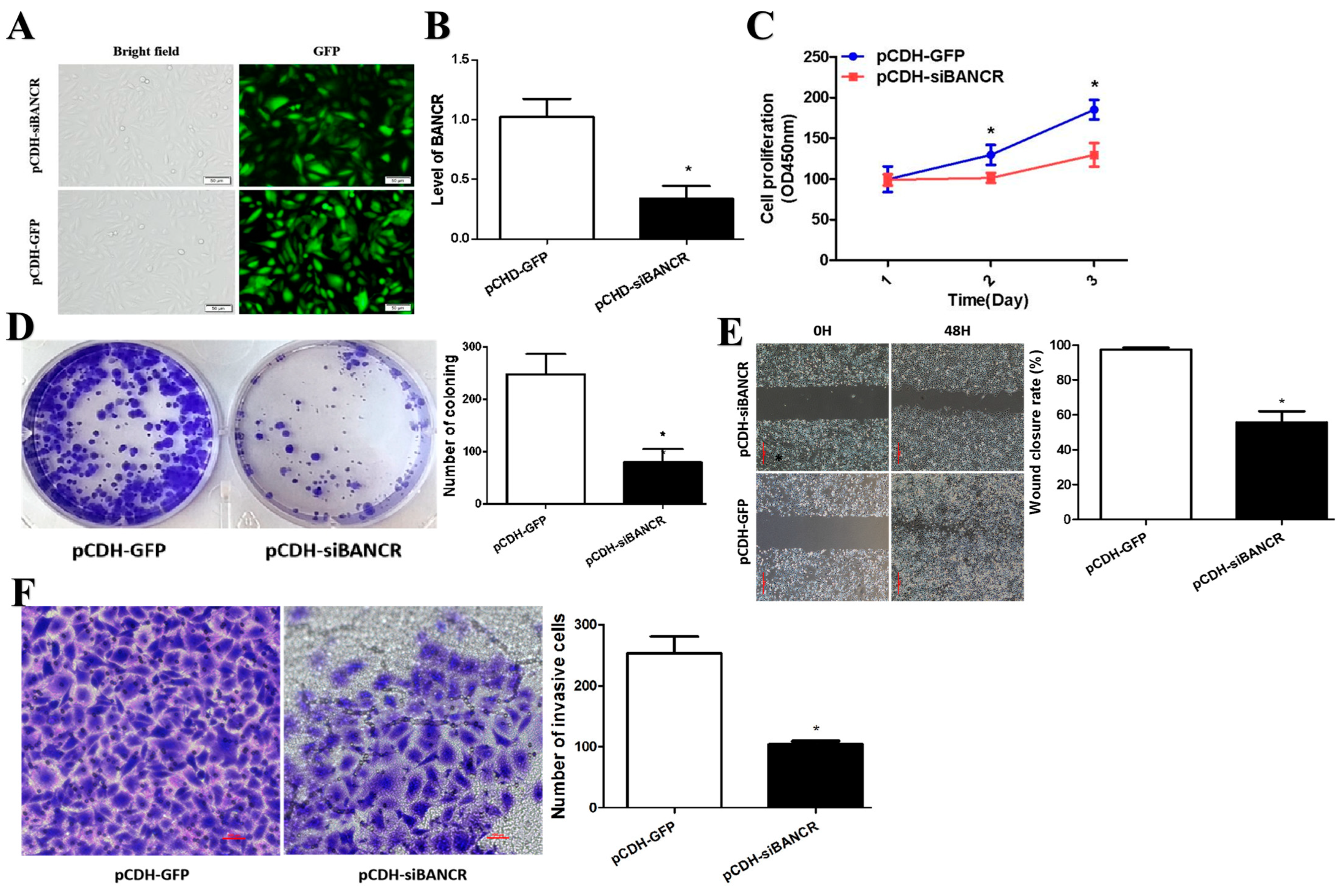
3.2. BANCR Promotes Pancreatic Cancer Cell Proliferation, Migration, and Invasion
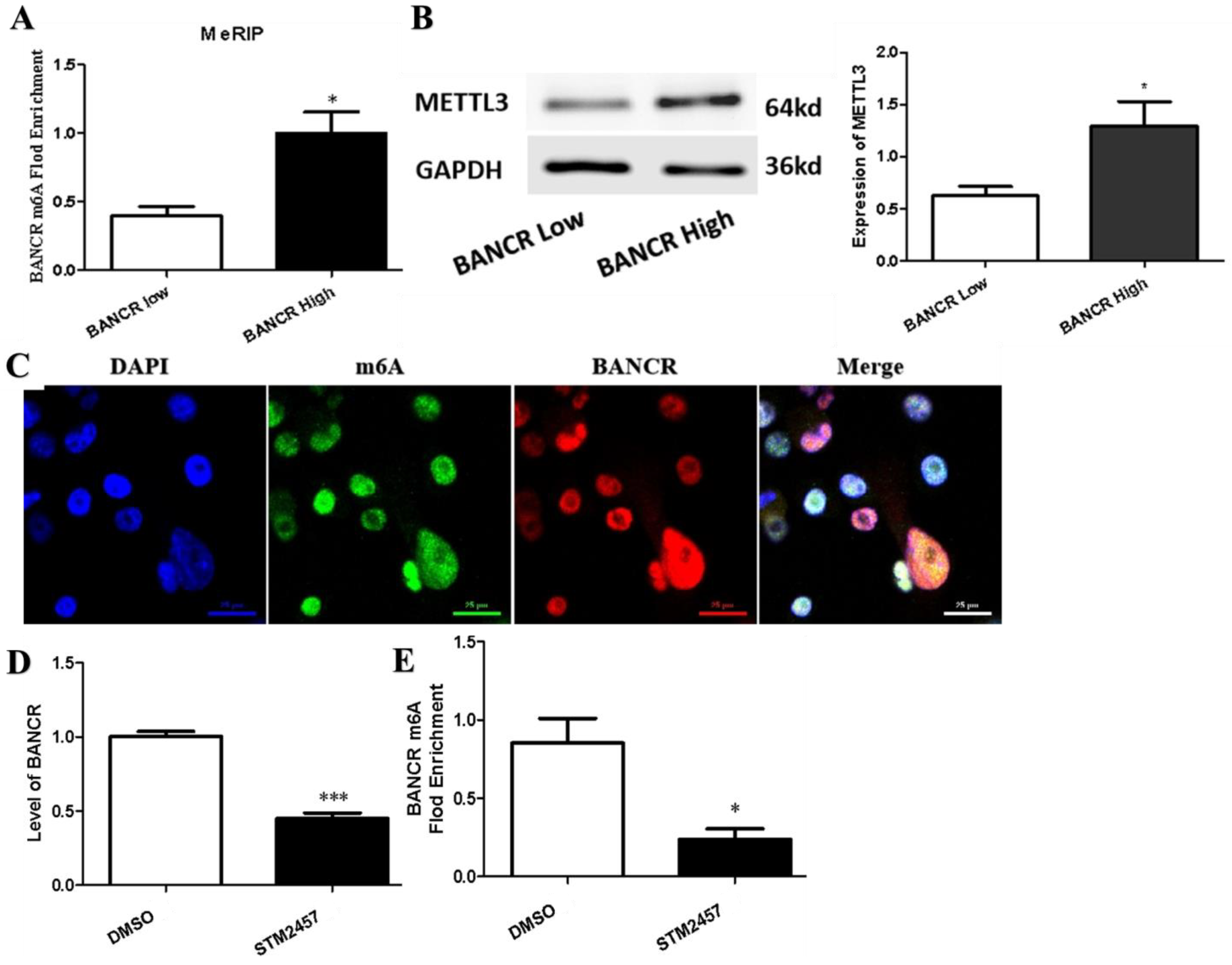
3.3. m6A Modification Was Associated with the Up-Regulation of BANCR in PC Tissues and Cells
3.4. STM2457 Exerts Anticancer Activities in Pancreatic Cancer Cells In Vitro
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Avula, L.R.; Hagerty, B.; Alewine, C. Molecular mediators of peritoneal metastasis in pancreatic cancer. Cancer Metastasis Rev. 2020, 39, 1223–1243. [Google Scholar] [CrossRef] [PubMed]
- Zeni, L.B.; Russi, R.F.; Fialho, A.F.; Fonseca, A.L.P.; Sombrio, L.S.; Rocha, I.C. Morbidity and mortality of pancreatic tumors undergoing surgical treatment. Arq. Bras. Cir. Dig. 2014, 27, 275–279. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long noncoding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Flockhart, R.J.; Webster, D.E.; Qu, K.; Mascarenhas, N.; Kovalski, J.; Kretz, M.; Khavari, P.A. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res. 2012, 22, 1006–1014. [Google Scholar] [CrossRef]
- Miao, X.; Liu, Y.; Fan, Y.; Wang, G.; Zhu, H. LncRNA BANCR Attenuates the Killing Capacity of Cisplatin on Gastric Cancer Cell Through the ERK1/2 Pathway. Cancer Manag. Res. 2021, 13, 287–296. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, L.; Xu, J. LncRNA BANCR promotes proliferation of hepatocellular carcinoma Huh-7 cells by activating Wnt/β-catenin signaling pathway. Minerva Gastroenterol. 2021, 68, 341–343. [Google Scholar] [CrossRef]
- Stojanović, S.; Šelemetjev, S.; Đorić, I.; Rončević, J.; Janković Miljuš, J.; Živaljević, V.; Išić Denčić, T. Elevated BANCR expression levels have different effects on papillary thyroid carci-noma progression depending on the presence of the BRAFV600E mutation. EJSO-Eur. J. Surg. Oncol. 2020, 46, 1835–1842. [Google Scholar] [CrossRef]
- Hao, S.; Han, W.; Ji, Y.; Sun, H.; Shi, H.; Ma, J.; Yip, J.; Ding, Y. BANCR positively regulates the HIF-1α/VEGF-C/VEGFR-3 pathway in a hypoxic microenvironment to promote lymphangiogenesis in pancreatic cancer cells. Oncol. Lett. 2022, 24, 422. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhong, J.; Zeng, Z.; Huang, K.; Ye, Z.; Deng, S.; Chen, H.; Xu, F.; Li, Q.; Zhao, G. Hypoxia-induced feedback of HIF-1α and lncRNA-CF129 contributes to pan-creatic cancer progression through stabilization of p53 protein. Theranostics 2019, 9, 4795–4810. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-X.; Zhang, X.-S.; Sui, N. Advances in the profiling of N6-methyladenosine (m6A) modifications. Biotechnol. Adv. 2020, 45, 107656. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, F.; Liu, Y.; Wang, H.; Ni, B. N6-methyladenosine (m6A) in pancreatic cancer: Regulatory mechanisms and future direction. Int. J. Biol. Sci. 2021, 17, 2323–2335. [Google Scholar] [CrossRef]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef]
- Li, Y.; Huang, H.; Zhu, Y.; Xu, B.; Chen, J.; Liu, Y.; Zheng, X.; Chen, L. Increased expression of METTL3 in pancreatic cancer tissues associates with poor survival of the patients. World J. Surg. Oncol. 2022, 20, 283. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hosein, A.N.; Brekken, R.A.; Maitra, A. Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 487–505. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Noncoding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817. [Google Scholar] [CrossRef]
- Yu, X.; Huang, M.; Yang, G. Long non-coding RNA BANCR promotes proliferation, invasion and migration in esophageal squamous cell carcinoma cells via the Raf/MEK/ERK signaling pathway. Mol. Med. Rep. 2021, 23, 465. [Google Scholar] [CrossRef]
- Lou, K.-X.; Li, Z.-H.; Wang, P.; Liu, Z.; Chen, Y.; Wang, X.-L.; Cui, H.-X. Long non-coding RNA BANCR indicates poor prognosis for breast cancer and promotes cell proliferation and invasion. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Du, L.; Yang, X.; Jiang, X.; Duan, W.; Yan, S.; Xie, Y.; Zhu, Y.; Wang, Q.; Wang, L.; et al. Identification of long noncoding RNAs as potential novel diagnosis and prognosis biomarkers in colorectal cancer. J. Cancer Res. Clin. 2016, 142, 2291–2301. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Yang, D.; Liu, Y.; Wang, Y.; Lin, T.; Li, Y.; Yang, S.; Zhang, W.; Zhang, R. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging 2018, 10, 2062–2078. [Google Scholar] [CrossRef]
- Hussen, B.M.; Azimi, T.; Abak, A.; Hidayat, H.J.; Taheri, M.; Ghafouri-Fard, S. Role of lncRNA BANCR in Human Cancers: An Updated Review. Front. Cell Dev. Biol. 2021, 9, 689992. [Google Scholar] [CrossRef]
- Yang, X.; Hu, X.; Liu, J.; Wang, R.; Zhang, C.; Han, F.; Chen, Y.; Ma, D. N6-methyladenine modification in noncoding RNAs and its function in cancer. Biomark. Res. 2020, 8, 61. [Google Scholar] [CrossRef]
- Wu, X.; Ye, W.; Gong, Y. The Role of RNA Methyltransferase METTL3 in Normal and Malignant Hematopoiesis. Front. Oncol. 2022, 12, 873903. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Q.; Zhou, Q. METTL3-m6A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 2023, 559, 216122. [Google Scholar] [CrossRef]
- Sun, Y.; Shen, W.; Hu, S.; Lyu, Q.; Wang, Q.; Wei, T.; Zhu, W.; Zhang, J. METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy. J. Exp. Clin. Cancer Res. 2023, 42, 65. [Google Scholar] [CrossRef] [PubMed]

| Parameter | No. of Patients (n = 31) | Level of BANCR (Mean ± SD) | p Value |
|---|---|---|---|
| Sex | |||
| Male | 21 | 2.135 ± 1.012 | 0.174 |
| Female | 10 | 1.663 ± 0.478 | |
| Age (years) | |||
| >65 | 18 | 2.252 ± 0.903 | 0.046 * |
| ≤65 | 13 | 1.609 ± 0.766 | |
| Tumor Size (cm) | |||
| T1 | 4 | 1.033 ± 0.240 | 0.057 |
| T2 | 11 | 1.867 ± 0.770 | |
| T3 | 8 | 2.460 ± 1.180 | |
| T4 | 8 | 2.138 ± 0.587 | |
| Lymph Node Metastasis | |||
| N0 | 11 | 1.445 ± 0.594 | <0.001 *** |
| N1 | 16 | 2.010 ± 0.653 | |
| N2 | 4 | 3.351 ± 1.072 | |
| Distant Metastasis | |||
| Yeas | 3 | 2.578 ± 1.330 | 0.231 |
| No | 28 | 1.919 ± 0.846 | |
| Histological Grade | |||
| Well | 9 | 1.656 ± 0.923 | 0.036 * |
| Moderate | 15 | 1.757 ± 0.763 | |
| Poor | 7 | 2.613 ± 0.842 | |
| Clinical Stage | |||
| I | 9 | 1.339 ± 0.503 | 0.024 * |
| II | 9 | 1.915 ± 0.759 | |
| III | 10 | 2.444 ± 0.876 | |
| IV | 3 | 2.578 ± 1.330 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, S.; Sun, H.; Sun, H.; Zhang, B.; Ji, K.; Liu, P.; Nie, F.; Han, W. STM2457 Inhibits the Invasion and Metastasis of Pancreatic Cancer by Down-Regulating BRAF-Activated Noncoding RNA N6-Methyladenosine Modification. Curr. Issues Mol. Biol. 2023, 45, 8852-8863. https://doi.org/10.3390/cimb45110555
Hao S, Sun H, Sun H, Zhang B, Ji K, Liu P, Nie F, Han W. STM2457 Inhibits the Invasion and Metastasis of Pancreatic Cancer by Down-Regulating BRAF-Activated Noncoding RNA N6-Methyladenosine Modification. Current Issues in Molecular Biology. 2023; 45(11):8852-8863. https://doi.org/10.3390/cimb45110555
Chicago/Turabian StyleHao, Shaolong, Haitao Sun, Hao Sun, Bo Zhang, Kailun Ji, Peng Liu, Fang Nie, and Wei Han. 2023. "STM2457 Inhibits the Invasion and Metastasis of Pancreatic Cancer by Down-Regulating BRAF-Activated Noncoding RNA N6-Methyladenosine Modification" Current Issues in Molecular Biology 45, no. 11: 8852-8863. https://doi.org/10.3390/cimb45110555
APA StyleHao, S., Sun, H., Sun, H., Zhang, B., Ji, K., Liu, P., Nie, F., & Han, W. (2023). STM2457 Inhibits the Invasion and Metastasis of Pancreatic Cancer by Down-Regulating BRAF-Activated Noncoding RNA N6-Methyladenosine Modification. Current Issues in Molecular Biology, 45(11), 8852-8863. https://doi.org/10.3390/cimb45110555