BCL11A Expression in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Cell Cultures
2.3. Tissue Microarrays (TMAs)
2.4. Immunohistochemistry (IHC)
2.5. Evaluation of Immunohistochemical Reactions
2.6. RNA Isolation and cDNA Synthesis
2.7. Real-Time PCR
2.8. Immunofluorescence (IF)
2.9. Statistical Analysis
3. Results
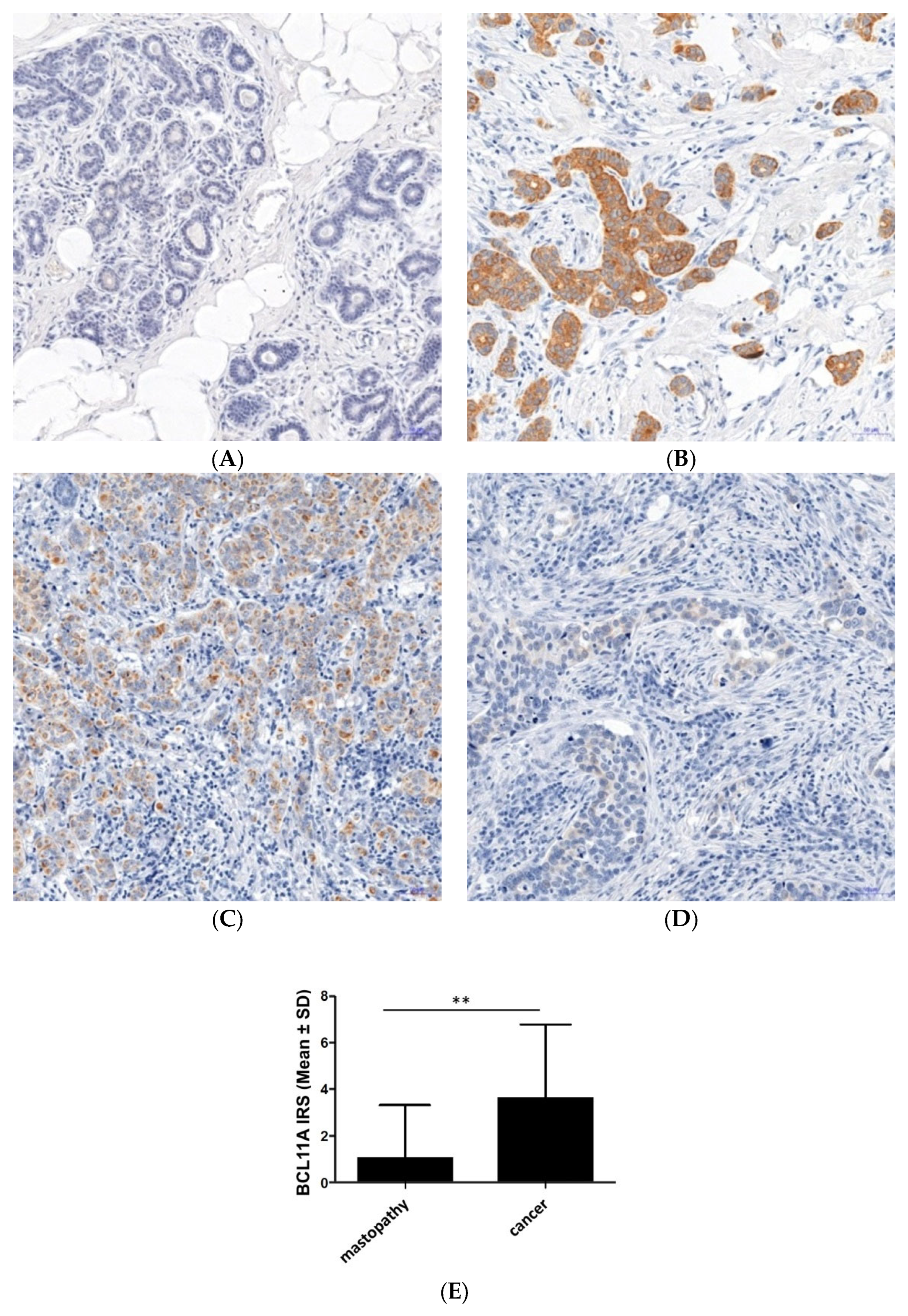
3.1. Expression of BCL11A in Breast Cancer Cells and Mastopathy Samples
3.2. The Relationship between BCL11A Expression and Clinicopathological Factors
3.3. The Relationship between BCL11A Expression and the Presence of Receptors for Steroid Hormones and HER2
3.4. BCL11A mRNA Expression in Breast Cancer and Cell Lines
3.5. The Relationship between BCL11A Expression and Overall Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, U.; Didkowska, J. Zachorowania i Zgony Na Nowotwory Złośliwe w Polsce. Krajowy Rejestr Nowotworów, Centrum Onkologii—Instytut Im. Marii Skłodowskiej—Curie. Available online: http://onkologia.org.pl/raporty (accessed on 21 August 2019).
- Khaled, W.T.; Choon Lee, S.; Stingl, J.; Chen, X.; Raza Ali, H.; Rueda, O.M.; Hadi, F.; Wang, J.; Yu, Y.; Chin, S.-F.; et al. BCL11A Is a Triple-Negative Breast Cancer Gene with Critical Functions in Stem and Progenitor Cells. Nat. Commun. 2015, 6, 5987. [Google Scholar] [CrossRef]
- Yeo, S.K.; Guan, J. Breast Cancer: Multiple Subtypes within a Tumor? Breast Cancer Stratification and Its Role in Guiding Therapeutic Decisions. Trends Cancer 2018, 3, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Luo, N.; Hu, Y.; Li, X.; Zhang, K. MiR-137 Suppresses Triple-Negative Breast Cancer Stemness and Tumorigenesis by Perturbing BCL11A-DNMT1 Interaction. Cell. Physiol. Biochem. 2018, 47, 2147–2158. [Google Scholar] [CrossRef] [PubMed]
- Nowacka-Zawisza, M.; Krajewska, W.M. Potrójnie Negatywny Rak Piersi: Molekularna Charakterystyka i Potencjalne Strategie Terapeutyczne. Postępy Hig. I Med. Doświadczalnej 2013, 67, 1090–1097. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.; Ahmed, S.; Inzhakova, G.; Shi, J.; Avila, C.; Polikoff, J.; Bernstein, L.; Enger, S.; Press, M. Impact of Breast Cancer Subtypes and Treatment on Survival: An Analysis Spanning Two Decades. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1848–1855. [Google Scholar] [CrossRef]
- Tomao, F.; Papa, A.; Zaccarelli, E.; Rossi, L.; Caruso, D.; Minozzi, M.; Vici, P.; Frati, L.; Tomao, S. Triple-Negative Breast Cancer: New Perspectives for Targeted Therapies. OncoTargets Ther. 2015, 8, 177–193. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J.; Albain, K.S.; André, F.; Bergh, J.; et al. Personalizing the Treatment of Women with Early Breast Cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef]
- Angius, A.; Pira, G.; Cossu-Rocca, P.; Sotgiu, G.; Saderi, L.; Muroni, M.R.; Virdis, P.; Piras, D.; Vincenzo, R.; Carru, C.; et al. Deciphering Clinical Significance of BCL11A Isoforms and Protein Expression Roles in Triple—Negative Breast Cancer Subtype. J. Cancer Res. Clin. Oncol. 2022, 1–13. [Google Scholar] [CrossRef]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.J.; Thürlimann, B.; Senn, H.J.; André, F.; Baselga, J.; et al. Tailoring Therapies-Improving the Management of Early Breast Cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yamazaki, Y.; Saiki, Y.; Moriyama, M.; Largaespada, D.A.; Jenkins, N.A.; Copeland, N.G. Evi9 Encodes a Novel Zinc Finger Protein That Physically Interacts with BCL6, a Known Human B-Cell Proto-Oncogene Product. Mol. Cell. Biol. 2000, 20, 3178–3186. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ippolito, G.C.; Wall, J.K.; Niu, T.; Probst, L.; Lee, B.-S.; Pulford, K.; Banham, A.H.; Stockwin, L.; Shaffer, A.L.; et al. Functional Studies of BCL11A: Characterization of the Conserved BCL11A-XL Splice Variant and Its Interaction with BCL6 in Nuclear Paraspeckles of Germinal Center B Cells. Mol. Cancer 2006, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Keller, J.R.; Ortiz, M.; Tessarollo, L.; Rachel, R.A.; Nakamura, T.; Jenkins, N.A.; Copeland, N.G. Bcl11a Is Essential for Normal Lymphoid Development. Nat. Immunol. 2003, 4, 525–532. [Google Scholar] [CrossRef]
- Satterwhite, E.; Sonoki, T.; Willis, T.G.; Harder, L.; Nowak, R.; Arriola, E.L.; Liu, H.; Price, H.P.; Gesk, S.; Steinemann, D.; et al. The BCL11 Gene Family: Involvement of BCL11A in Lymphoid Malignancies. Blood 2001, 98, 3413–3420. [Google Scholar] [CrossRef]
- Yin, C.C.; Lin, K.I.C.; Ketterling, R.P.; Knudson, R.A.; Medeiros, L.J.; Barron, L.L.; Huh, Y.O.; Luthra, R.; Keating, M.J.; Abruzzo, L.V. Chronic Lymphocytic Leukemia with t(2;14)(P16;Q32) Involves the BCL11A and IgH Genes and Is Associated With Atypical Morphologic Features and Unmutated IgV H Genes. Am. J. Clin. Pathol. 2009, 131, 663–670. [Google Scholar] [CrossRef]
- Kątnik, E.; Gomułkiewicz, A.; Dzięgiel, P. BIAŁKO BCL11A—Rola biologiczna I znaczenie W procesie nowotworowym. Adv. Cell Biol. 2018, 4, 293–318. [Google Scholar]
- Zhang, N.; Jiang, B.-Y.; Zhang, X.-C.; Xie, Z.; Su, J.; Zhang, Q.; Han, J.-F.; Tu, H.-Y.; Wu, Y.-L. The BCL11A-XL Expression Predicts Relapse in Squamous Cell Carcinoma and Large Cell Carcinoma. J. Thorac. Dis. 2015, 7, 1630–1636. [Google Scholar] [CrossRef]
- Kuo, T.Y.; Hsueh, Y.P. Expression of Zinc Finger Transcription Factor Bcl11A/Evi9/CTIP1 in Rat Brain. J. Neurosci. Res. 2007, 85, 1628–1636. [Google Scholar] [CrossRef]
- Li, S.; Teegarden, A.; Bauer, E.M.; Choi, J.; Messaddeq, N.; Hendrix, D.A.; Ganguli-Indra, G.; Leid, M.; Indra, A.K. Transcription Factor CTIP1/BCL11A Regulates Epidermal Differentiation and Lipid Metabolism During Skin Development. Sci. Rep. 2017, 7, 13427. [Google Scholar] [CrossRef]
- Ippolito, G.C.; Dekker, J.D.; Wang, Y.H.; Lee, B.K.; Shaffer, A.L.; Lin, J.; Wall, J.K.; Lee, B.S.; Staudt, L.M.; Liu, Y.J.; et al. Dendritic Cell Fate Is Determined by BCL11A. Proc. Natl. Acad. Sci. USA 2014, 111, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Saiki, Y.; Yamazaki, Y.; Yoshida, M.; Katoh, O.; Nakamura, T. Human EVI9, a Homologue of the Mouse Myeloid Leukemia Gene, Is Expressed in the Hematopoietic Progenitors and down-Regulated during Myeloid Differentiation of HL60 Cells. Genomics 2000, 70, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, J.; Khaled, W.; Burke, S.; Li, P.; Chen, X.; Yang, W.; Jenkins, N.A.; Copeland, N.G.; Zhang, S.; et al. Bcl11a Is Essential for Lymphoid Development and Negatively Regulates P53. J. Exp. Med. 2012, 209, 2467–2483. [Google Scholar] [CrossRef]
- Wu, X.; Satpathy, A.T.; Kc, W.; Liu, P.; Murphy, T.L.; Murphy, K.M. Bcl11a Controls Flt3 Expression in Early Hematopoietic Progenitors and Is Required for PDC Development In Vivo. PLoS ONE 2013, 8, e64800. [Google Scholar] [CrossRef]
- Bauer, D.E.; Orkin, S.H. Hemoglobin Switching’s Surprise: The Versatile Transcription Factor BCL11A Is a Master Repressor of Fetal Hemoglobin. Curr. Opin. Genet. Dev. 2015, 33, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Pulford, K.; Gesk, S.; Ehrlich, S.; Banham, A.H.; Lyne, L.; Martin-Subero, J.I.; Siebert, R.; Dyer, M.J.S.; Möller, P.; et al. Gains of the Proto-Oncogene BCL11A and Nuclear Accumulation of BCL11AXL Protein Are Frequent in Primary Mediastinal B-Cell Lymphoma. Leukemia 2006, 20, 1880–1882. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Wang, Y.; Shi, S.; Zhu, H.; Xiao, F.; Yang, J.; Yang, A.; Hao, X. Inhibition of FOXQ1 Induces Apoptosis and Suppresses Proliferation in Prostate Cancer Cells by Controlling BCL11A/MDM2 Expression. Oncol. Rep. 2016, 36, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, H.; Arao, T.; Tanaka, K.; Tamura, D.; Aomatsu, K.; Kudo, K.; Sakai, K.; De Velasco, M.A.; Matsumoto, K.; Fujita, Y.; et al. FOXQ1 Is Overexpressed in Colorectal Cancer and Enhances Tumorigenicity and Tumor Growth. Cancer Res. 2010, 70, 2053–2063. [Google Scholar] [CrossRef]
- Li, T.; Ning, K.; Le, J.; Minjia, T.; Thomas, L.; Yingming, Z.; Richard, B.; Wei, G. Tumor Suppression in the Absence of P53-Mediated Cell Cycle Arrest, Apoptosis, and Senescence. Cell 2013, 149, 1269–1283. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, X.; Su, J.; Meng, W.; Yang, X.; Yang, J.; Zhou, Q.; Chen, Z.; Chen, Z.; Xie, Z.; et al. BCL11A Overexpression Predicts Survival and Relapse in Non-Small Cell Lung Cancer and Is Modulated by MicroRNA-30a and Gene Amplification. Mol. Cancer 2013, 12, 61. [Google Scholar] [CrossRef]
- Szemraj, J.; Rozpończyk, E.; Bartkowiak, J.; Greger, J.; Oszajca, K. Znaczenie Białka MDM2 w Cyklu Komórkowym. Postep. Biochem. 2005, 51, 44–51. [Google Scholar]
- Godlewski, M.; Kobylińska, A. Programowana Śmierć Komórek—Strategia Utrzymania Komórkowej Homeostazy Organizmu. Postępy Hig. I Med. Doświadczalnej 2016, 70, 1229–1244. [Google Scholar]
- Gao, Y.; Wu, H.; He, D.; Hu, X.; Li, Y. Downregulation of BCL11A by SiRNA Induces Apoptosis in B Lymphoma Cell Lines. Biomed. Rep. 2013, 1, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Ke, D.; Yang, R.; Jing, L. Combined Diagnosis of Breast Cancer in the Early Stage by MRI and Detection of Gene Expression. Exp. Ther. Med. 2018, 16, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Santuario-Facio, S.K.; Cardona-Huerta, S.; Perez-Paramo, Y.X.; Trevino, V.; Hernandez-Cabrera, F.; Rojas-Martinez, A.; Uscanga-Perales, G.; Martinez-Rodriguez, J.L.; Martinez-Jacobo, L.; Padilla-Rivas, G.; et al. A New Gene Expression Signature for Triple-Negative Breast Cancer Using Frozen Fresh Tissue before Neoadjuvant Chemotherapy. Mol. Med. 2017, 23, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wei, W.; Huang, X.; Xie, X.; Kong, Y.; Dai, D.; Yang, L.; Wang, J.; Tang, H.; Xie, X. CircEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics 2018, 8, 4003–4015. [Google Scholar] [CrossRef] [PubMed]
- Moody, R.R.; Lo, M.-C.; Meagher, J.L.; Lin, C.-C.; Stevers, N.O.; Tinsley, S.L.; Jung, I.; Matvekas, A.; Stuckey, J.A.; Sun, D. Probing the Interaction between the Histone Methyltransferase/Deacetylase Subunit RBBP4/7 and the Transcription Factor BCL11A in Epigenetic Complexes. J. Biol. Chem. 2018, 293, 2125–2136. [Google Scholar] [CrossRef]
- Lakhani, S.; Ellis, I.; Schnitt, S.; Tan, P.; van de Vijver, M. WHO Classification of Tumours of the Breast, 4th ed.; IARC Press: Lyon, France, 2012; ISBN 978-92-832-2433-4. [Google Scholar]
- Dzięgiel, P.; Owczarek, T.; Plazuk, E.; Gomułkiewicz, A.; Majchrzak, M.; Podhorska-Okołów, M.; Driouch, K.; Lidereau, R.; Ugorski, M. Ceramide Galactosyltransferase (UGT8) Is a Molecular Marker of Breast Cancer Malignancy and Lung Metastases. Br. J. Cancer 2010, 103, 524–531. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H. Recommendation for Uniform Definition of an Immunoreactive Score (IRS) for Immunohistochemical Estrogen Receptor Detection (ER-ICA) in Breast Cancer Tissue. Pathologe 1987, 8, 138–140. [Google Scholar]
- Nowak, M.; Madej, J.A.; Pula, B.; Dziegiel, P.; Ciaputa, R. Expression of Matrix Metalloproteinase 2 (MMP-2), E-Cadherin and Ki-67 in Metastatic and Non-Metastatic Canine Mammary Carcinomas. Ir. Vet. J. 2016, 69, 9. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer. World Health Organisation Latest Global Cancer Data. 2018, pp. 13–15. Available online: https://www.iarc.who.int/wp-content/uploads/2018/09/pr263_E.pdf (accessed on 10 February 2023).
- Zhang, C.; Wang, J.; Zhang, L.; Lu, Y.; Ji, T.; Xu, L.; Ling, L. Shikonin Reduces Tamoxifen Resistance through Long Non-Coding RNA Uc.57. Oncotarget 2017, 8, 88658–88669. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Pan, R.; Zhou, D.; Ye, G.; Tan, W. BCL11A Enhances Stemness and Promotes Progression by Activating Wnt/β-Catenin Signaling in Breast Cancer. Cancer Manag. Res. 2019, 11, 2997–3007. [Google Scholar] [CrossRef]
- Lulli, V.; Romania, P.; Morsilli, O.; Cianciulli, P.; Gabbianelli, M.; Testa, U.; Giuliani, A.; Marziali, G. MicroRNA-486-3p Regulates γ-Globin Expression in Human Erythroid Cells by Directly Modulating BCL11A. PLoS ONE 2013, 8, e60436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-J.; Hu, Y.; Luo, N.; Li, X.; Chen, F.-Y.; Yuan, J.-Q.; Guo, L. MiR-574-5p Attenuates Proliferation, Migration and EMT in Triple-negative Breast Cancer Cells by Targeting BCL11A and SOX2 to Inhibit the SKIL/TAZ/CTGF Axis. Int. J. Oncol. 2020, 56, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.-J.; Shao, Y.-C.; Huang, S.-R.; Zeng, Y.-F.; Yuan, X.-N.; Xu, J.-J.; Yin, W.-N.; Wei, L.; Zhang, J.-W. Hypoxia-Associated Prognostic Markers and Competing Endogenous RNA Co-Expression Networks in Breast Cancer. Front. Oncol. 2020, 10, 579868. [Google Scholar] [CrossRef]
- Hayashi, N.; Manyam, G.C.; Gonzalez-Angulo, A.M.; Niikura, N.; Yamauchi, H.; Nakamura, S.; Hortobágyi, G.N.; Baggerly, K.A.; Ueno, N.T. Reverse-Phase Protein Array for Prediction of Patients at Low Risk of Developing Bone Metastasis from Breast Cancer. Oncologist 2014, 19, 909–914. [Google Scholar] [CrossRef]
- Pulford, K.; Banham, A.H.; Lyne, L.; Jones, M.; Ippolito, G.C.; Liu, H.; Tucker, P.W.; Roncador, G.; Lucas, E.; Ashe, S.; et al. The BCL11AXL Transcription Factor: Its Distribution in Normal and Malignant Tissues and Use as a Marker for Plasmacytoid Dendritic Cells. Leukemia 2006, 20, 1439–1441. [Google Scholar] [CrossRef]







| Parameters | All Cases | |
|---|---|---|
| N = 200 | % | |
| Age | ||
| <58 | 110 | 55.0% |
| ≥58 | 90 | 45.0% |
| Grade of histological malignancy (G) | ||
| G1 | 73 | 36.5% |
| G2 | 73 | 36.5% |
| G3 | 43 | 21.5% |
| no data | 11 | 5.5% |
| Size of the primary tumor (T) | ||
| pT1 | 110 | 55.0% |
| pT2 | 58 | 29.0% |
| pT3 | 4 | 2.0% |
| pT4 | 20 | 10.0% |
| no data | 8 | 4.0% |
| Tumor size (cm) | ||
| <2 cm | 94 | 47.0% |
| ≥2 cm | 98 | 49.0% |
| no data | 8 | 4.0% |
| Lymph node metastases (N) | ||
| pN0 | 68 | 34.0% |
| pN1 | 27 | 13.5% |
| pN2 | 17 | 8.5% |
| pN3 | 6 | 3.0% |
| no data | 82 | 41.0% |
| Cancer stage | ||
| I | 44 | 22.0% |
| II | 40 | 20.0% |
| III | 37 | 18.5% |
| no data | 79 | 39.5% |
| ER status | ||
| positive | 153 | 76.5% |
| negative | 43 | 21.5% |
| no data | 4 | 2.0% |
| PR status | ||
| positive | 134 | 67.0% |
| negative | 57 | 28.5% |
| no data | 9 | 4.5% |
| HER2 status | ||
| positive | 97 | 48.5% |
| negative | 95 | 47.5% |
| no data | 8 | 4.0% |
| Triple-negative (TN) breast cancer | ||
| yes | 16 | 8.0% |
| no | 178 | 89.0% |
| no data | 6 | 3.0% |
| Survival | ||
| survivors | 146 | 73.0% |
| deceased | 52 | 26.0% |
| no data | 2 | 1.0% |
| Parameters | All Cases | |
|---|---|---|
| N = 22 | % | |
| Age | ||
| <61 | 7 | 31.8% |
| >61 | 8 | 36.4% |
| no data | 7 | 31.8% |
| Grade of histological malignancy (G) | ||
| G1 | 5 | 22.7% |
| G2 | 9 | 40.9% |
| G3 | 8 | 36.4% |
| Size of the primary tumor (T) | ||
| pT1 | 11 | 50.0% |
| pT2 | 10 | 45.5% |
| pT3 | 1 | 4.5% |
| Lymph node metastases (N) | ||
| pN0 | 9 | 40.9% |
| pN1 | 11 | 50.0% |
| pN2 | 1 | 4.5% |
| pN3 | 1 | 4.5% |
| Cancer stage | ||
| I | 3 | 13.6% |
| II | 10 | 45.5% |
| III | 3 | 13.6% |
| no data | 6 | 27.3% |
| ER status | ||
| positive | 14 | 63.6% |
| negative | 6 | 27.3% |
| no data | 2 | 9.1% |
| PR status | ||
| positive | 12 | 54.5% |
| negative | 8 | 36.4% |
| no data | 2 | 9.1% |
| HER2 status | ||
| positive | 10 | 45.5% |
| negative | 10 | 45.5% |
| no data available | 2 | 9.1% |
| Triple-negative (TN) breast cancer | ||
| yes | 4 | 18.2% |
| no | 16 | 72.7% |
| no data | 2 | 9.1% |
| Survival | ||
| survivors | 5 | 22.7% |
| deceased | 7 | 31.8% |
| no data | 10 | 45.5% |
| Factor A | Factor B | ||
|---|---|---|---|
| Points | Percentage of Positive Cells | Points | Reaction Intensity |
| 0 | 0% | ||
| 1 | ≤10% | 0 | lack |
| 2 | 11–50% | 1 | low |
| 3 | 51–80% | 2 | medium |
| 4 | >80% | 3 | strong |
| Parameters | All Cases (N = 200) | |
|---|---|---|
| Mean Expression Value ± SD | p | |
| Age | 0.1685 | |
| <58 | 3.836 ± 3.235 | |
| ≥58 | 3.433 ± 2.986 | |
| Grade of histological malignancy (G) | <0.0001 | |
| G1 | 5.432 ± 2.956 | |
| G2 | 3.055 ± 2.748 | |
| G3 | 1.593 ± 2.498 | |
| Size of the primary tumor (T) | 0.0479 | |
| pT1 | 4.318 ± 3.187 | |
| pT2 | 2.879 ± 2.908 | |
| pT3 | 1.500 ± 1.291 | |
| pT4 | 2.800 ± 3.172 | |
| Tumor size (cm) | <0.0001 | |
| <2 cm | 4.511 ± 3.224 | |
| ≥2 cm | 2.888 ± 2.864 | |
| Status of lymph node metastases (N) | 0.6905 | |
| pN0 | 3.081 ± 3.100 | |
| pN1 | 4.407 ± 3.522 | |
| pN2 | 2.588 ± 2.210 | |
| pN3 | 2.500 ± 3.209 | |
| Cancer stage | 0.0837 | |
| I | 3.784 ± 3.133 | |
| II | 3.325 ± 3.482 | |
| III | 2.378 ± 2.586 | |
| ER status | <0.0001 | |
| positive | 4.196 ± 3.086 | |
| negative | 1.744 ± 2.564 | |
| PR status | < 0.0001 | |
| positive | 4.377 ± 3.146 | |
| negative | 1.904 ± 2.337 | |
| HER2 status | 0.8222 | |
| positive | 3.500 ± 3.230 | |
| negative | 3.657 ± 3.120 | |
| Triple-negative (TN) breast cancer | <0.0001 | |
| yes | 1.000 ± 1.414 | |
| no | 3.921 ± 3.153 | |
| Parameters | All Cases (N = 22) | |
|---|---|---|
| RQ ± SD | p | |
| Age | 0.4587 | |
| <61 | 41.591 ± 3.459 | |
| >61 | 9.506 ± 7.516 | |
| Grade of histological malignancy (G) | 0.4339 | |
| G1 | 17.258 ± 5.559 | |
| G2 | 50.933 ± 3.855 | |
| G3 | 11.561 ± 8.356 | |
| Size of the primary tumor (T) | 0.9955 | |
| pT1 | 21.232 ± 5.821 | |
| pT2-pT3 | 25.351 ± 6.761 | |
| Lymph node metastases (N) | 0.3899 | |
| pN0 | 34.514 ± 5.117 | |
| pN1-pN3 | 17.660 ± 6.792 | |
| Cancer stage | 0.2427 | |
| I | 32.810 ± 4.581 | |
| II-III | 20.045 ± 6.902 | |
| ER status | 0.2313 | |
| positive | 14.028 ± 6.577 | |
| negative | 54.702 ± 4.375 | |
| PR status | 0.2189 | |
| positive | 11.830 ± 6.339 | |
| negative | 50.350 ± 4.786 | |
| HER2 status | 0.1839 | |
| positive | 10.593 ± 6.808 | |
| negative | 42.073 ± 4.808 | |
| Triple-negative (TN) breast cancer | 0.0890 | |
| yes | 78.886 ± 2.917 | |
| no | 15.205 ± 6.516 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kątnik, E.; Gomułkiewicz, A.; Piotrowska, A.; Grzegrzółka, J.; Rusak, A.; Kmiecik, A.; Ratajczak-Wielgomas, K.; Dzięgiel, P. BCL11A Expression in Breast Cancer. Curr. Issues Mol. Biol. 2023, 45, 2681-2698. https://doi.org/10.3390/cimb45040175
Kątnik E, Gomułkiewicz A, Piotrowska A, Grzegrzółka J, Rusak A, Kmiecik A, Ratajczak-Wielgomas K, Dzięgiel P. BCL11A Expression in Breast Cancer. Current Issues in Molecular Biology. 2023; 45(4):2681-2698. https://doi.org/10.3390/cimb45040175
Chicago/Turabian StyleKątnik, Ewa, Agnieszka Gomułkiewicz, Aleksandra Piotrowska, Jędrzej Grzegrzółka, Agnieszka Rusak, Alicja Kmiecik, Katarzyna Ratajczak-Wielgomas, and Piotr Dzięgiel. 2023. "BCL11A Expression in Breast Cancer" Current Issues in Molecular Biology 45, no. 4: 2681-2698. https://doi.org/10.3390/cimb45040175
APA StyleKątnik, E., Gomułkiewicz, A., Piotrowska, A., Grzegrzółka, J., Rusak, A., Kmiecik, A., Ratajczak-Wielgomas, K., & Dzięgiel, P. (2023). BCL11A Expression in Breast Cancer. Current Issues in Molecular Biology, 45(4), 2681-2698. https://doi.org/10.3390/cimb45040175






