HR Gene Variants Identified in Mexican Patients with Alopecia Areata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample
2.2. Molecular and Bioinformatics Analysis
2.3. Statistical Analysis
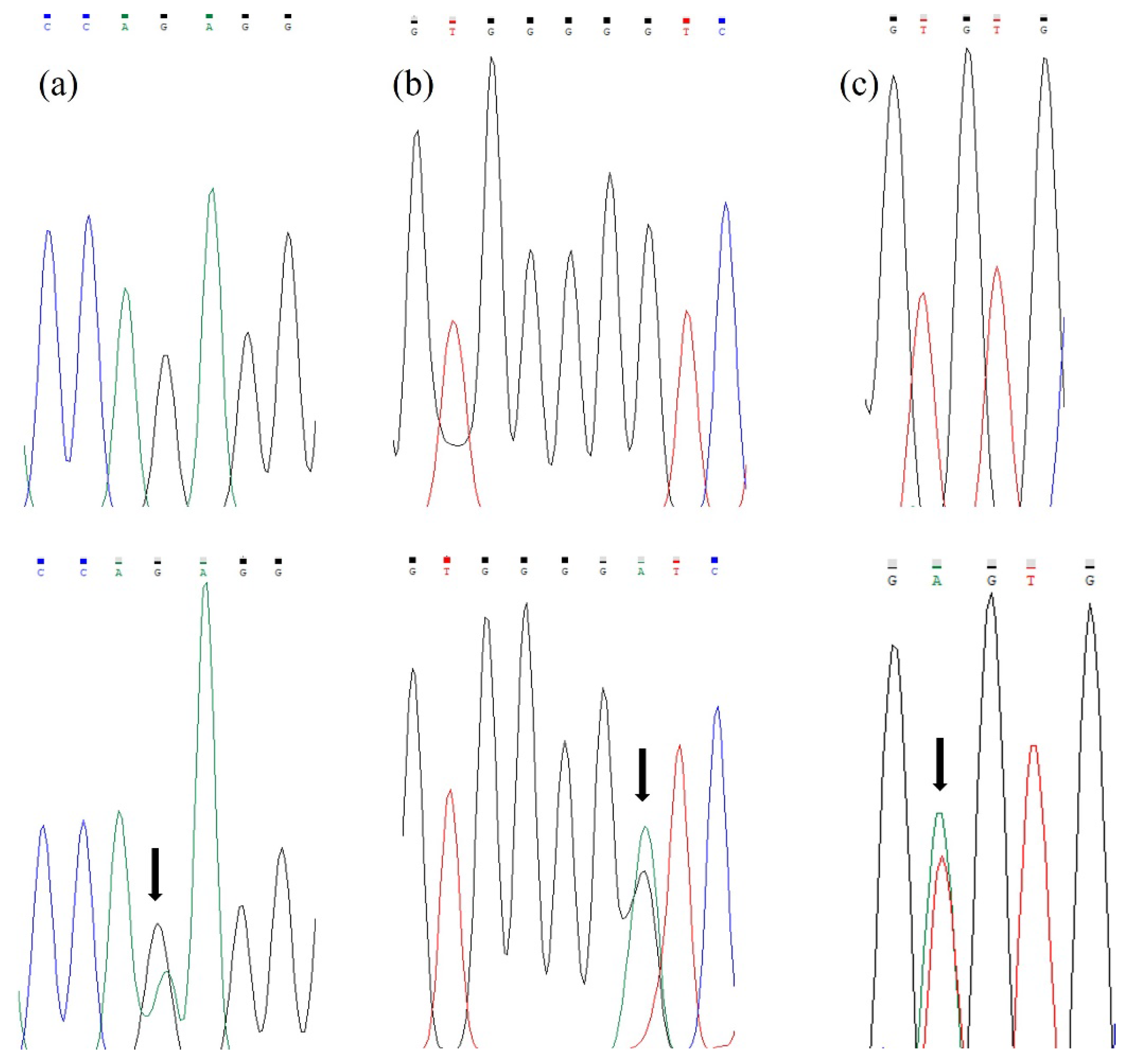
3. Results
3.1. Clinical and Demographic Data
3.2. Molecular and Bioinformatics Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sterkens, A.; Lambert, J.; Bervoets, A. Alopecia areata: A review on diagnosis, immunological etiopathogenesis and treatment options. Clin. Exp. Med. 2021, 21, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Chiang, A.; Ortenzio, F.; Juhasz, M.L.W.; Yu, V.; Mesinkovska, N.A. Balance of tofacitinib efficacy and disease flare in the treatment of alopecia universalis: A case report and review of the literature. JAAD Case Rep. 2018, 4, 733–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajabi, F.; Drake, L.A.; Senna, M.M.; Rezaei, N. Alopecia areata: A review of disease pathogenesis. Br. J. Dermatol. 2018, 179, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Alzolibani, A.A. Epidemiologic and genetic characteristics of alopecia areata (part 1). Acta Dermatovenerol. Alp Pannonica Adriat 2011, 20, 191–198. [Google Scholar] [PubMed]
- Strazzulla, L.C.; Wang, E.H.C.; Avila, L.; Lo Sicco, K.; Brinster, N.; Christiano, A.M.; Shapiro, J. Alopecia areata: Disease characteristics, clinical evaluation, and new perspectives on pathogenesis. J. Am. Acad. Dermatol. 2018, 78, 1–12. [Google Scholar] [CrossRef]
- Olguín, M.G.; Martín del Campo, A.; Rodríguez, M.; Peralta, M.L. Factores psicológicos asociados con la alopecia areata. Dermatol. Rev. Mex. 2013, 57, 171–177. [Google Scholar]
- Simakou, T.; Butcher, J.P.; Reid, S.; Henriquez, F.L. Alopecia areata: A multifactorial autoimmune condition. J. Autoimmun. 2019, 98, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Oka, A.; Takagi, A.; Komiyama, E.; Yoshihara, N.; Mano, S.; Hosomichi, K.; Suzuki, S.; Haida, Y.; Motosugi, N.; Hatanaka, T.; et al. Alopecia areata susceptibility variant in MHC region impacts expressions of genes contributing to hair keratinization and is involved in hair loss. EBioMedicine 2020, 57, 102810. [Google Scholar] [CrossRef]
- Kutner, A.; Friedman, A. Hair loss in the dermatology office: An update on alopecia areata. J. Drugs Dermatol. 2013, 12, 588–593. [Google Scholar]
- Peloquin, L.; Castelo-Soccio, L. Alopecia Areata: An Update on Treatment Options for Children. Paediatr Drugs 2017, 19, 411–422. [Google Scholar] [CrossRef]
- Pratt, C.H.; King, L.E., Jr.; Messenger, A.G.; Christiano, A.M.; Sundberg, J.P. Alopecia areata. Nat. Rev. Dis. Primers 2017, 3, 17011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maatough, A.; Whitfield, G.K.; Brook, L.; Hsieh, D.; Palade, P.; Hsieh, J.C. Human Hairless Protein Roles in Skin/Hair and Emerging Connections to Brain and Other Cancers. J. Cell Biochem. 2018, 119, 69–80. [Google Scholar] [CrossRef]
- Heidary, H.; Mardi, A.; Mousavi, S.M.; Khazaie, G.; Golab, F. Hairless Gene Nonsense Mutations in Alopecia Universalis: A Case Report. Iran. J. Public Health 2021, 50, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Panteleyev, A.A.; Botchkareva, N.V.; Sundberg, J.P.; Christiano, A.M.; Paus, R. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am. J. Pathol. 1999, 155, 159–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juárez-Rendón, K.J.; Rivera-Sánchez, G.; Reyes-López, M.Á.; García-Ortiz, J.E.; Bocanegra-García, V.; Guardiola-Avila, I.; Altamirano-García, M.L. Alopecia Areata. Current situation and perspectives. Arch. Argent Pediatr. 2017, 115, e404–e411. [Google Scholar] [PubMed]
- Benavides, F.; Oberyszyn, T.M.; VanBuskirk, A.M.; Reeve, V.E.; Kusewitt, D.F. The hairless mouse in skin research. J. Dermatol. Sci. 2009, 53, 10–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.S.; Rauf, S.; Naeem, M.; Khan, M.N.; Mir, A. Identification of novel mutation in the HR gene responsible for atrichia with papular lesions in a Pakistani family. J. Dermatol. 2013, 40, 927–928. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Faiyaz ul Haque, M.; Brancolini, V.; Tsou, H.C.; ul Haque, S.; Lam, H.; Aita, V.M.; Owen, J.; de Blaquiere, M.; Frank, J.; et al. Alopecia universalis associated with a mutation in the human hairless gene. Science 1998, 279, 720–724. [Google Scholar] [CrossRef]
- Nucara, S.; Colao, E.; Mangone, G.; Baudi, F.; Fabiani, F.; Nocera, D.; Passafaro, G.; Longo, T.; Laria, A.E.; Malatesta, P.; et al. Identification of a new mutation in the gene coding for hairless protein responsible for alopecia universalis: The importance of direct gene sequencing. Dermatol. Online J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Potter, G.B.; Beaudoin, G.M.; DeRenzo, C.L.; Zarach, J.M.; Chen, S.H.; Thompson, C.C. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes Dev. 2001, 15, 2687–2701. [Google Scholar] [CrossRef] [Green Version]
- Komar, A.A. SNPs, silent but not invisible. Science 2007, 315, 466–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Center for Biotechnology Information. ClinVar; [VCV000362525.10]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000362525.10 (accessed on 6 March 2023).
- Malloy, P.J.; Feldman, D. The role of vitamin D receptor mutations in the development of alopecia. Mol. Cell. Endocrinol. 2011, 347, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, C.C. Hairless is a nuclear receptor corepressor essential for skin function. Nucl. Recept. Signal. 2009, 7, e010. [Google Scholar] [CrossRef] [PubMed]
- Trüeb, R.M.; Dias, M.F.R.G. Alopecia Areata: A Comprehensive Review of Pathogenesis and Management. Clin. Rev. Allergy Immunol. 2018, 54, 68–87. [Google Scholar] [CrossRef]
- Bharathi, G.; Ramana, P.V.; Sridevi, K.; Usha, G.; Kumar, R. Clinico Etiological Study of Alopecia AREATA. J. Dent. Med. Sci. 2015, 14, 29–32. [Google Scholar]
- Dainichi, T.; Kabashima, K. Alopecia areata: What’s new in epidemiology, pathogenesis, diagnosis, and therapeutic options? J. Dermatol. Sci. 2017, 86, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Hordinsky, M.; Junqueira, A.L. Alopecia areata update. Semin Cutan. Med. Surg 2015, 34, 72–75. [Google Scholar] [CrossRef] [Green Version]
| Exon | Fragment Size | Forward | Reverse | Amplification Programs |
|---|---|---|---|---|
| 3 | 835 pb | 5′-AGTCAGCTGA AGCGTAACAC-3′ | 5′-CCTTACCTTT CTGCTCATCA-3′ | 95 °C for 1 min (1 cycle), followed by 29 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min; finally, 1 cycle at 72 °C for 7 min. |
| 15 | 299 pb | 5′-AGTGCCAGGA TTACAGGCGT-3′ | 5′-CTGAGGAGGAAAGAGCGCTC-3′ | 95 °C for 1 min (1 cycle), followed by 32 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 1 min; finally, 1 cycle at 72 °C for 7 min. |
| 17 | 297 pb | 5′-CTGGAAAGTC CATGCCCCAT-3′ | 5′-GTCGCTTCTG CCATCCTGAT-3′ | 95 °C for 1 min (1 cycle), followed by 30 cycles at 95 °C for 30 s, 59 °C for 30 s and 72 °C for 1 min; finally, 1 cycle at 72 °C for 7 min. |
| Exon | Variant | Genotype | Genotype Frequency (AA Patients %) | Genotype Frequency (Healthy Individuals %) | p Value | Allele | Allele Frequency (AA Patients %) | Allele Frequency (Healthy Individuals %) | p Value |
|---|---|---|---|---|---|---|---|---|---|
| 3 | c.750G > A p.(Gln250Gln) | GG | 14 (93) | 13 (87) | 0.543 | G | 29 (97) | 28 (93) | 0.554 |
| GA | 1 (7) | 2 (13) | A | 1 (3) | 2 (7) | ||||
| AA | 0 (0) | 0 (0) | |||||||
| Total | 15 (100) | 15 (100) | 30 (100) | 30 (100) | |||||
| c.1010G > A p.(Gly337Asp) rs12675375 | GG | 7 (47) | 14 (93) | 0.005 * | G | 22 (73) | 29 (97) | 0.011 * | |
| GA | 8 (53) | 1 (7) | A | 8 (27) | 1 (3) | ||||
| AA | 0 (0) | 0 (0) | |||||||
| Total | 15 (100) | 15 (100) | 30 (100) | 30 (100) | |||||
| 17 | c.3215T > A p.(Val1072Glu) | TT | 8 (53) | 15 (100) | 0.003 * | T | 23 (77) | 30 (0) | 0.005 * |
| TA | 7 (47) | 0 (0) | A | 7 (23) | 0 (0) | ||||
| AA | 0 (0) | 0 (0) | |||||||
| Total | 15 (100) | 15 (100) | 30 (100) | 30 (100) |
| Variant | Bioinformatic Server | |||||
|---|---|---|---|---|---|---|
| Polyphen2 | Mutation Taster | SIFT | ||||
| Effect | Score | Effect | Probability | Effect | Score | |
| c.1010G > A p.(Gly337Asp) | Benign | 0.005 | Polymorphism (Probably harmless). | 0.99 | Tolerated | 0.48 |
| c.3215T > A p.(Val1072Glu) | Probably damaging | 0.998 | Disease causing | 0.99 | Damaging | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz-Ramírez, A.; Hernández-Jiménez, M.C.; Guardiola-Avila, I.B.; De Luna-Santillana, E.d.J.; Oliva-Hernández, A.A.; Altamirano-García, M.L.; Juárez-Rendón, K.J. HR Gene Variants Identified in Mexican Patients with Alopecia Areata. Curr. Issues Mol. Biol. 2023, 45, 2965-2971. https://doi.org/10.3390/cimb45040194
Ortiz-Ramírez A, Hernández-Jiménez MC, Guardiola-Avila IB, De Luna-Santillana EdJ, Oliva-Hernández AA, Altamirano-García ML, Juárez-Rendón KJ. HR Gene Variants Identified in Mexican Patients with Alopecia Areata. Current Issues in Molecular Biology. 2023; 45(4):2965-2971. https://doi.org/10.3390/cimb45040194
Chicago/Turabian StyleOrtiz-Ramírez, Andrés, María Cristina Hernández-Jiménez, Iliana Berenice Guardiola-Avila, Erick de Jesús De Luna-Santillana, Amanda Alejandra Oliva-Hernández, María Lourdes Altamirano-García, and Karina Janett Juárez-Rendón. 2023. "HR Gene Variants Identified in Mexican Patients with Alopecia Areata" Current Issues in Molecular Biology 45, no. 4: 2965-2971. https://doi.org/10.3390/cimb45040194
APA StyleOrtiz-Ramírez, A., Hernández-Jiménez, M. C., Guardiola-Avila, I. B., De Luna-Santillana, E. d. J., Oliva-Hernández, A. A., Altamirano-García, M. L., & Juárez-Rendón, K. J. (2023). HR Gene Variants Identified in Mexican Patients with Alopecia Areata. Current Issues in Molecular Biology, 45(4), 2965-2971. https://doi.org/10.3390/cimb45040194








